Abstract
Xenorhabdus nematophila HB310 secreted the insecticidal protein toxin complex (Tc). The chi60 and chi70 chitinase genes are located on the gene cluster encoding Tc toxins. To clarify the insecticidal activity of chitinases and their relationship with Tc toxins, the insecticidal activity of the chitinases was assessed on Helicoverpa armigera. Then, the chi60 and chi70 genes of X. nematophila HB310 were knocked out by the pJQ200SK suicide plasmid knockout system. The insecticidal activity of Tc toxin from the wild-type strain (WT) and mutant strains was carried out. The results demonstrate that Chi60 and Chi70 had an obvious growth inhibition effect against the second instar larvae of H. armigera with growth-inhibiting rates of 81.99% and 90.51%, respectively. Chi70 had a synergistic effect with the insecticidal toxicity of Tc toxins, but Chi60 had no synergistic effect with Tc toxins. After feeding Chi60 and Chi70, the peritrophic membrane of H. armigera became inelastic, was easily broken and leaked blue dextran. The Δchi60, Δchi70 and Δchi60-chi70 mutant strains were successfully screened. The toxicity of Tc toxins from the WT, Δchi60, Δchi70 and Δchi60-chi70 was 196.11 μg/mL, 757.25 μg/mL, 885.74 μg/mL and 20,049.83 μg/mL, respectively. The insecticidal activity of Tc toxins from Δchi60 and Δchi70 was 3.861 and 4.517 times lower than that of Tc toxins from the WT, respectively, while the insecticidal activity of Tc toxins from the Δchi60-chi70 mutant strain almost disappeared. These results indicate that the presence of chi60 and chi70 is indispensable for the toxicity of Tc toxins.
Keywords:
Xenorhabdus nematophila HB310; chitinases; gene knock out; toxin complex; insecticidal activity Key Contribution:
Chitinases from X. nematophila HB310 could inhibit the growth of H. armigera and destroy the peritrophic membrane of H. armigera. The chitinase gene could be knocked out by homologous recombination. After the two chitinase genes (chi60 and chi70) were simultaneously knocked out, the insecticidal activity of Tc toxins almost disappeared.
1. Introduction
Chitin composed of linear β-1,4-N-acetylglucosamine (GlcNAc) residues is a major component of the intestinal peritrophic membrane (annelids and some arthropods) and exoskeleton (arthropods) [1,2,3,4]. Chitinases (EC 3.2.1.14) are a kind of chitin-degrading glycosidase that play an important role in the hydrolysis of glycosidic bonds in chitin to form soluble chitooligosaccharides [5,6,7]. Chitinases are produced by a variety of microorganisms, with diverse structures and functions. The chitinases bind to the chitin in the exoskeleton or peritrophic membrane, which can lead to structural changes and increase the accessibility of the substrate for the pathogens into the haemocoel of susceptible insects [8,9]. In addition, chitinases could also promote the process of toxin binding to specific receptors and be used to improve the insecticidal activity of toxins [8,10]. Therefore, chitinases have been used in agriculture as an effective virulence factor against pests.
The emergence of Bacillus thuringiensis (Bt)-resistant insects has made it important to identify other novel biopesticides [11,12]. Toxin complex proteins (Tc) comprise a candidate class of molecules [8,13]. Tc toxins were first identified in Photorhabdus luminescens W14 [14,15], which belongs to the Enterobacteriaceae family and lives in a mutualistic symbiosis with entomopathogenic nematodes (EPNs) from the genus Heterorhabditis [16,17]. Tc toxins have high molecular weights and multi-subunit protein complexes, which have high insecticidal activity against various pests [18,19]. Tc toxins consist of three separate components: TcA, TcB and TcC [20,21,22,23,24,25,26]. TcA proteins harbor the cytotoxic effects of Tc toxins, while TcB and TcC proteins modulate and enhance the toxicity of TcA proteins [21,27,28].
Tc toxins are found in P. luminescens and Photorhabdus asymbiotica as well as in other entomopathogenic bacteria, such as Xenorhabdus nematophila [18,29], Serratia entomophila [30,31] and Yersinia entomophaga [15]. In Y. entomophaga, two putative chitinases (Chi1 and Chi2) are contained in the 3D structure of Tc toxins [14,15]. Chi1 and Chi2 proteins are vital for this complex formation [14]. Two chitinase genes (chitinase 60 (chi60) and chitinase 70 (chi70)) were also found in the locus of Tc toxins from X. nematophila [14,32]. The relationship between Tc toxins and chitinases is currently unclear.
The X. nematophila HB310 is symbiotically associated with a strain of the entomopathogenic nematode Steinernema carpocapsae isolated from the soil in Hebei Province, China [33]. In a previous study, the peptides of chitinases from the intracellular proteins of X. nematophila HB310 were identified by matrix-assisted laser desorption-time-of-flight mass spectrometry (MALDI-TOFMS). We also found that the recombinant chitinase 60 (Chi60) and chitinase 70 (Chi70) could enhance the insecticidal activity of the Bt HD73 strain and the Bt Cry1Ac toxin. To clarify the relationship between chitinases and Tc toxins, we determined the insecticidal activity of Chi60 and Chi70 and the pathologic effects on the peritrophic membranes of Helicoverpa armigera (Lepidoptera: Noctuidae). Then, the chi60 and chi70 genes of X. nematophila HB310 were knocked out by the pJQ200SK suicide plasmid knockout system. Moreover, the insecticidal activity of Tc toxins from the wild-type strain (WT), chi60 gene knockout mutants (Δchi60), chi70 gene knockout mutants (Δchi70), and double gene (chi60 and chi70) knockout mutants (Δchi60-chi70) in X. nematophila HB310 were determined against the second instar larvae of H. armigera. This research will help reveal the insecticidal mechanism of Tc toxins and lay a foundation for the development and utilization of insecticidal formulations for entomopathogenic nematode symbiotic bacteria. In addition, insecticidal genes can also be cloned from symbiotic bacteria for the development of transgenic insect-resistant crops, thereby delaying the resistance of pests to Bt transgenic insect-resistant crops.
2. Results
2.1. Sequence Analysis of Chitinases
Chi60 had 535 amino acids with a predicted molecular weight of 59.3 kDa and a PI of 4.51, Chi70 had 648 amino acids with a molecular weight of 72.4 kDa and PI of 4.89. The two amino acid sequences showed low similarity, with a similarity of 24.29% (Figure 1).
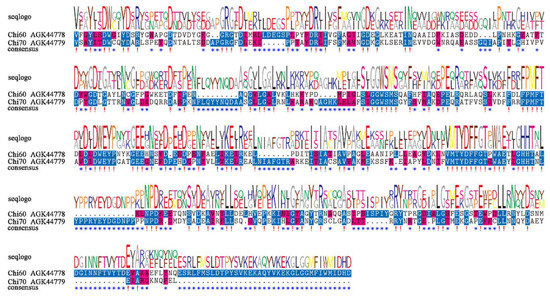
Figure 1.
Sequence alignment of the conserved domains of Chi60 and Chi70 from X. nematophila HB310. !, conserved sites. *, differential sites.
The chitinases from X. nematophila HB310 and other bacteria were used to construct the phylogenetic tree to assess the evolutionary relationships among the chitinases. Then, a schematic representing the structure of all complete chitinase sequences was constructed from the MEME motif analysis results. As shown in Figure 2, the chitinases were divided into two subclasses. Chitinases in the same subclass usually showed a highly similar motif composition. All chitinases contain motif 1–3, motif 5, motif 8 and motif 9. However, motif 4 was unique to the subclass of Chi70. Compared to Chi60, motif 4 and motif 6 were unique to Chi70. Compared to Chi70, motif 7 and motif 10 were unique to Chi60. In addition, the sequence similarity of the two subgroups was very low, which could perform different biological functions as two different subclasses.
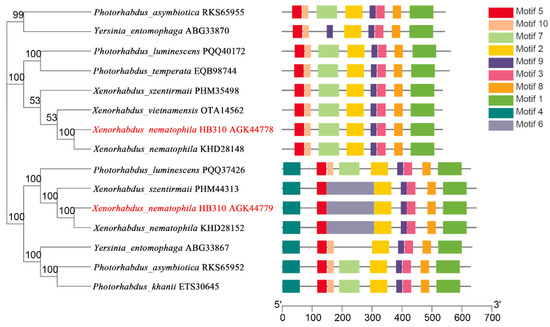
Figure 2.
Phylogenetic relationships and composition of the conserved motif patterns. The phylogenetic tree was constructed based on the full-length sequences of chitinases using the MEGA 5.0 software. The sequence information for each motif was provided in Table S1. The conserved motifs were displayed in different colored boxes, and the length of protein can be estimated using the scale at the bottom.
2.2. Insecticidal Activity of Chitinases and Synergistic Effect with Tc Toxin
The inhibitory effect of chitinases on the growth of second-instar larvae of H. armigera was determined by feeding methods. Chitinases could significantly inhibit the growth of H. armigera (Table 1). At the same concentration, the growth inhibition rates of Chi60 and Chi70 were 81.99% and 90.51%, respectively. The growth inhibition rate of Chi70 against H. armigera was higher than Chi60. Both Chi60 and Chi70 had a lower lethal effect on H. armigera, with a corrected mortality of 13.89% and 4.17%.

Table 1.
The corrected mortality and growth inhibition rate of Chi60 and Chi70 against H. armigera.
In the search for possible synergistic interactions between chitinase and Tc toxins, different combinations were tested against the second-instar larvae of H. armigera—the results of which are shown in Table 2. The LC50 value of Tc toxins against the second instar larvae of H. armigera is 196.11 μg/mL, however, the LC50 value of Tc toxins mixed with Chi70 was 146.47 μg/mL. Chi70 had a synergistic effect on Tc toxins, while Chi60 did not exhibit synergistic toxicity to Tc toxins against H. armigera with an LC50 value of 185.85 μg/mL.

Table 2.
The synergistic effect of Chi60 and Chi70 with Tc toxins against H. armigera.
2.3. Pathological Effect of Chitinases on the Peritrophic Membrane of H. armigera
In order to clarify the pathological effect of chitinases on the peritrophic membrane of the fifth-instar larvae from H. armigera, the effect of chitinases on the damage and permeability of the peritrophic membrane was determined by feeding method. The peritrophic membrane of the fifth-instar larvae of H. armigera after phosphate-buffered solution (PBS, pH 7.2) treatment was intact, translucent, elastic and swinging in water without breaking (Figure 3a). After feeding with Chi60 or Chi70, the peritrophic membrane broke into inelastic fragments when it swung in water, and there was obvious tissue fragmentation (Figure 3b,c).
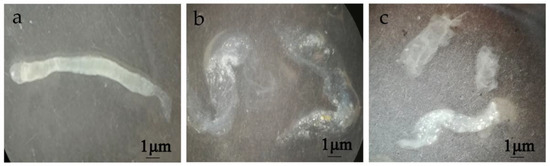
Figure 3.
Changes of the peritrophic membrane of H. armigera after different treatments. (a) Treatment with PBS (pH 7.2) (negative control). (b) Treatment with Chi60—100 μg of Chi60 was fed per insect for 48 h, the peritrophic membrane broke into inelastic fragments. (c) Treatment with Chi70—100 μg of Chi70 was fed per insect for 48 h, the peritrophic membrane obviously broke into inelastic fragments.
The effect of chitinases on the permeability of the peritrophic membrane from H. armigera was determined by feeding methods. The peritrophic membrane of fifth-instar larvae H. armigera was intact after PBS (pH 7.2) treatment, and there was no exudation of blue dextran (Figure 4a). The peritrophic membrane of H. armigera displayed obvious exudation after feeding Chi60 or Chi70 (Figure 4b,c).
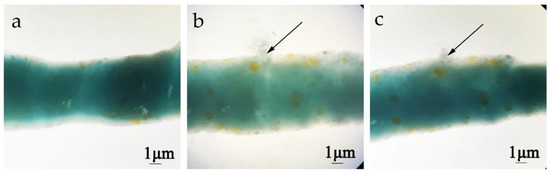
Figure 4.
Changes in the peritrophic membrane permeability of H. armigera after different treatments. (a) Treatment with PBS (pH 7.2) and blue dextran 2000 (negative control). (b) Treatment with Chi60—100 μg of Chi60 and 10 μg blue dextran 2000 were fed per insect for 48 h, the peritrophic membrane displayed obvious exudation. (c) Treatment with Chi70—100 μg of Chi70 and 10 μg blue dextran 2000 were fed per insect for 48 h, the peritrophic membrane displayed obvious exudation.
2.4. Homologous Recombination Vector Construction
Six DNA fragments were successfully amplified by PCR and cloned using the methods described (Figure S1). The results indicate that the upstream and downstream fragments of the chi60 gene and chi70 gene were successfully amplified from the genomic DNA of X. nematophila HB310, respectively. The DNA sequencing identified that the size of upstream and downstream fragments from the chi60 gene were 1069 bp and 1223 bp, and the size of upstream and downstream fragments from the chi70 gene were 1147 bp and 1081 bp, respectively. The Kmr (1300 bp) and tetA (1300 bp) were successfully amplified from the plasmids of pYBA-1132 and pTKLP-tet, respectively.
The whole fragments were successfully amplified by fusion PCR (Figure S2). The results indicate that chi60-Kmr was successfully fused with a size of 3900 bp, and amplified by Kmr and the upstream and downstream fragments of the chi60 gene. The chi70-tetA was successfully fused with a size of 4175 bp, and amplified by the tetA and the upstream and downstream fragments of the chi70 gene.
Verification of the correctness of the constructed homologous recombination vector by double digestion. The plasmid DNA of pJQ200SK-chi60-Kmr and pJQ200SK-chi70-tetA were digested with Xba I and Xho I, generating two fragments (Figure S3). These results indicate that the 3900 bp chi60-Kmr and 4175 bp chi70-tetA were successfully amplified by fusion PCR, respectively.
2.5. Identification of Single Gene Knockout Mutants
Δchi60 and Δchi70 were screened by homologous recombination between the recombinant E. coli S17-1 λ pir containing the target genes and the WT. As shown in Figure 5, the fragments of Δchi60 or Δchi70 were smaller than that of the WT, which were amplified by the primer of 60-up-F/60-down-R or 70-up-F/70-down-R, respectively. The results indicate that the chi60 or chi70 gene from the WT had been successfully replaced by the resistance gene from the homologous recombination strain, respectively.
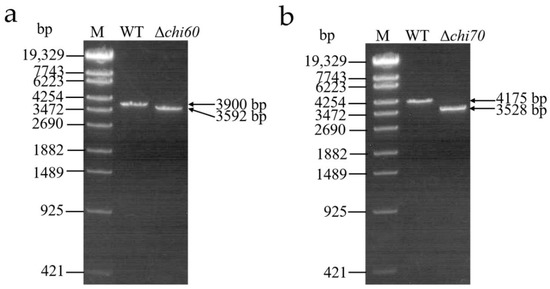
Figure 5.
PCR identification of single gene knockout mutants. (a) PCR identification of chi60 gene knockout mutants. (b) PCR identification of chi70 gene knockout mutants. Lane M, λ-EcoT14 I digest DNA marker. Lane WT, the wild-type strain. Lane Δchi60, the mutant strain with chi60 gene knocked out. Lane Δchi70, the mutant strain with chi70 gene knocked out.
2.6. Identification of Double Gene Knockout Mutants
Δchi60-chi70 were screened by homologous recombination between the recombinant E. coli S17-1 λ pir containing chi60-Kmr and Δchi70. Two pairs of primers (60-up-F/60-down-R and 70-up-F/70-down-R) were simultaneously used to detect the mutants. Δchi60-chi70 was successfully screened (Figure 6).
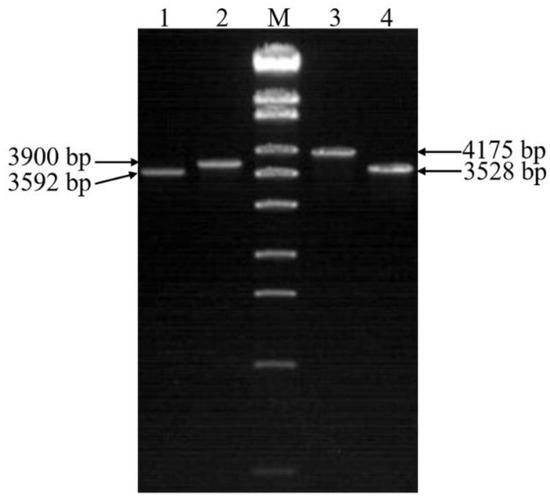
Figure 6.
PCR identification of double gene knockout mutants. Lane M, λ-EcoT14 I digest (19,329, 7743, 6223, 4254, 3472, 2690, 1882, 1489, 925, 421, and 74 bp). Lane 1, the mutant strain with chi60 gene knocked out. Lane 2 and lane 3, the wild-type strain. Lane 4, the mutant strain with chi70 gene knocked out.
2.7. Western Blot Analysis
To verify the correctness of gene knockout, the Western blot analysis of the Tc toxins from the WT and knockout mutants was performed (Figure 7). Two bands of 78 kDa (Chi70) and 65 kDa (Chi60) were found in Tc toxins from the WT. The band of 65 kDa disappeared when the chi60 gene was knocked out, and the band of 78 kDa disappeared when the chi70 gene was knocked out. It was found that both bands of 78 kDa and 65 kDa disappeared when the chi60 and chi70 were simultaneously knocked out. Western blot analysis also showed that the chi60 gene and chi70 gene were successfully knocked out.
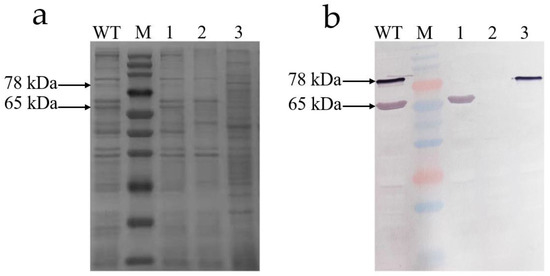
Figure 7.
SDS-PAGE (12%) and Western blotting analysis of Tc toxins from WT, Δchi60, Δchi70 and Δchi60-chi70. (a) SDS-PAGE stained with Coomassie brilliant blue analysis. (b) Western blotting analysis. Lane M, multicolor prestained protein marker (250, 150, 100, 70, 50, 40, 35, 25, 20, 15, and 10 kDa). Lane WT, the wild-type strain. Lane 1, the mutant strain with chi70 gene knocked out. Lane 2, the mutant strain with chi60 and chi70 gene knocked out simultaneously. Lane 3, the mutant strain with the chi60 gene knocked out.
2.8. Insecticidal Activity of Tc Toxins
To clarify the relationship of insecticidal activity between chitinases and Tc toxins, the insecticidal activity of Tc toxins from the WT and gene knockout mutants to the second instar larvae of H. armigera was tested by feeding method. The LC50 of Tc toxins to the second-instar larvae of H. armigera was shown in Table 3. The LC50 value of Tc toxins from the WT against the second-instar larvae of H. armigera was 196.11 μg/mL. The LC50 values of Tc toxins from Δchi60 and Δchi70 were 757.25 μg/mL and 885.74 μg/mL, respectively. The virulence of Tc toxins from Δchi60 and Δchi70 were significantly lower than that of the WT, and the toxicity of Tc toxins from Δchi60-chi70 almost disappeared with the LC50 of 20,049.83 μg/mL.

Table 3.
Insecticidal activity of Tc toxins from WT and mutant strains against H. armigera.
3. Discussion
The chitinolytic mechanism of bacteria primarily consists of chitinase [17], which specifically degrades chitin and prevents chitin biosynthesis. In this study, chitinases could significantly inhibit the growth of the second-instar larvae of H. armigera and damage the peritrophic membrane of H. armigera. In previous studies, the chitinase from Bacillus subtilis could effectively inhibit the growth of Spodoptera litura (Lepidoptera: Noctuidae) [34]. Chitinase purified from Pseudomonas fluorescens MP-13 demonstrated 100% mortality against Helopeltis theivora (Heteroptera: Miridae) [35]. Among the seven chitinases isolated from Bacillus firmus, Bacillus licheniformis, Thermomyces lanuginosus and Streptomyces sp., most of the chitinases can delay the pupation of Sesamia calamistis (Lepidoptera: Noctuidae) and Chilo partellus (Lepidoptera: Pyralidae) [36]. This indicates that the insecticidal activities of chitinases from different microorganisms have some differences. However, in this study, the inhibition of Chi60 was significantly lower than that of Chi70, which could be related to the inclusion of Chi60 [9]. It is speculated that Chi60 protein becomes a soluble protein after denaturation and renaturation, but the natural conformation could not be completely restored. Therefore, the inhibition of Chi60 in the second-instar larvae of H. armigera was lower than that of Chi70.
Chitinases can accelerate the binding process of toxins to the receptors and cause perforation in the gut peritrophic membrane, which increases accessibility to the substrate and makes it easier for pathogens to enter the haemocoel of susceptible insects [8]. In this study, Chi70 had a synergistic effect on the insecticidal activity of Tc toxins from X. nematophila. Previously, studies reported that chitinases produced by VLBt27, VLBt38, VLBt109 and VLBt135 strains isolated from more than 80 B. thuringiensis strains could enhance the insecticidal activity of insecticides to H. armigera and Brevicoryne brassicae (Hemiptera: Aphididae) [37]. In addition, chitinases from B. thuringiensis could also enhance the insecticidal activity of its crystal protein against Plutella xylostella (Lepidoptera: Plutellidae) [38], Lymantria dispar (Lepidoptera: Lymantriidae) [39], Spodoptera exigua (Lepidoptera: Noctuidae) and H. armigera [40]. However, in this study, the synergistic effect of Chi70 on the insecticidal activity of Tc toxins was lower than its synergistic effect on Bt Cry1Ac toxins [8]. This could be due to the action mode of X. nematophila, which was carried into the host insect hemocoel depending on the nematode and did not need themselves to destroy the cuticle or peritrophic membrane of the insect midgut. It is speculated that some functions of X. nematophila could degenerate during the evolutionary process, while the toxicity of toxins or secondary metabolites could decrease.
The chi60 and chi70 genes from X. nematopbila are vital to the insecticidal activity of Tc toxins. In a previous study, the insecticidal activity of the Tc toxins disappeared after the knockout of the chi1 and chi2 genes in the toxin complex locus of Y. entomophaga MH96 [14,41]. In this study, Δchi60, Δchi70, and Δchi60-chi70 were constructed by homologous recombination using pJQ200SK plasmids containing the sacB gene. The insecticidal activities of Tc toxins from the mutant strains were lower than that of the WT when chi60 or chi70 were knocked out. However, the insecticidal activity of Tc toxins from Δchi60-chi70 almost disappeared after chi60 and chi70 were simultaneously knocked out. It is speculated that the chitinases (Chi60 and Chi70) from X. nematophila and the chitinases (Chi1 and Chi2) from Y. entomophaga have similar functions.
The chitinases from X. nematophila, P. luminescens, P. asymbiotica and Y. entomophaga MH96 had the same location on the loci of Tc toxins [14]. The chitinases sizes vary widely within 20–90 kDa, and bacterial chitinases had a size range of 20–60 kDa [42]. Chitinases from X. nematophila and Y. entomophaga all had molecular weights higher than 60 kDa. Based on the data of GH-18 domains in the phylogenetic tree, Chi60 from X. nematophila and Chi1 from Y. entomophaga were clustered into the same branch, while Chi70 from X. nematophila and Chi2 from Y. entomophaga were clustered into the same branch [8]. In Y. entomophaga, the genetic knockout of chi1 and chi2 genes forms no complex even though the remaining genes are still expressed [41]. In this study, the chi60 and chi70 genes of X. nematophila were successfully knocked out alone and simultaneously, though whether the knockout of the chi60 and chi70 genes influenced the formation of the toxin complex must be further verified. The main research at present is the effect of the chitinase gene knockout on the insecticidal activity of Tc toxins. The next step is to further study the effect of chitinase gene knockout on the structure of Tc toxins. In addition, it is also necessary to pay attention to the development of biological pesticides, and effectively develop the Tc toxins into a new biological pesticide and produce it on a large scale. At the same time, further exploration of transgenic insect-resistant plants with Tc toxin genes is needed.
4. Conclusions
Chi60 and Chi70 had an obvious growth inhibition effect against the second-instar larvae of H. armigera. Chi60 and Chi70 could destroy the peritrophic membrane of the fifth-instar larvae of H. armigera. Chi70 had a synergistic effect with the insecticidal toxicity of Tc toxins, but Chi60 had no synergistic effect with Tc toxins.
The Δchi60, Δchi70, and Δchi60-chi70 were successfully screened using homologous recombination. The insecticidal activity of Tc toxins from WT, Δchi60, Δchi70, and Δchi60-chi70 were 196.11 μg/mL, 757.25 μg/mL, 885.74 μg/mL and 20,049.83 μg/mL, respectively. The insecticidal activity of Tc toxins from Δchi60-chi70 almost disappeared.
These results will help reveal the insecticidal mechanism of Tc toxins and lay a foundation for the development and utilization of insecticidal formulations for entomopathogenic nematode symbiotic bacteria.
5. Materials and Methods
5.1. Insects, Microorganisms and Proteins
H. armigera larvae were obtained from the Pest Biocontrol Laboratory, Hebei Agricultural University, China. The larvae were fed with an artificial diet (13% maize meal, 6.5% soybean powder, 5.8% dry yeast, 0.2% sorbic acid, 0.2% methyl-para-hydroxybenzoate, 4.7% vitamin C, 0.2% compound vitamin B, 3.2% sucrose, 1.3% agar and 64.9% sterilized distilled water) and reared at 28 °C and 70% relative humidity (RH) under a 16 h (h) light (L): 8 h dark (D) photoperiod.
X. nematophila HB310 was isolated from Steinernema carpocapsae HB310, which was screened from the soil in Hebei province of China and stored in the Pest Biocontrol Laboratory, Hebei Agricultural University, China [16,33]. The bacteria were incubated in Luria–Bertani (LB) broth for 48 h at 28 °C on a rotary shaker at 200 revolutions per minute (rpm).
The pYBA-1132 plasmid, pTKLP-tet plasmid, pJQ200SK plasmid and Escherichia coli S17-1 λ pir competent cell were contributed by the researcher Guangyue Li of the Chinese Academy of Agricultural Sciences.
The chitinases (Chi60 and Chi70) and Tc toxins were obtained from the Pest Biocontrol Laboratory, Hebei Agricultural University, China. The concentration of chitinases was diluted to 1000 μg/mL. The concentration of Tc toxins was diluted to 5000 μg/mL.
5.2. Sequence Analysis of Chitinase
The ExPASy tools (https://web.expasy.org/compute_pi/, accessed on 1 August 2022) were used to predict the isoelectric points (pI) and molecular weights (MWs) of chitinases. The conserved domains of chitinases were predicted by the hmmsearch tool [43]. The MEME online program for protein sequence (http://meme.nbcr.net/meme/intro.html, accessed on 2 August 2022) was used to identify the conserved motifs of chitinases, which the optimized parameters being any number of repetitions, a maximum number of 10 motifs and an optimum of 6–200 residues. The full-length amino acid sequences of chitinases from different bacteria were aligned using ClustalW with default parameters [44]. After sequence alignments, the phylogenetic tree was constructed by MEGA5.0 software using the neighbor-joining method with the following parameters: Poisson model, pairwise deletion and 1000 bootstrap replications [45]. The protein names and sequences of chitinases that were used in this analysis were listed in Table S2. All sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 2 August 2022).
5.3. Assay for Insecticidal Activity of Chitinases and Pathological Effect
5.3.1. Assay for Insecticidal Activity of Chitinase
Chitinase was diluted to 1000 μg/mL and mixed with the artificial diet at a dose of 100 μL protein per gram of artificial diet. The same volume of 10 mM PBS (pH 7.2) (Coolaber, Beijing, China) was used as a control. Approximately 0.1 g artificial diet and one second-instar larva of H. armigera were transferred to each well of a 24-well tissue culture plate and then incubated at 28 °C. The corrected mortality and growth inhibition rate were calculated according to the following formulas at 120 h after treatment:
Growth inhibition rate (%) = ((Weight of control − Weight of treatment)/(Weight of control − Weight of initial)) × 100
Corrected mortality (%) = ((Mortality rate of treatment − Mortality rate of control)/(1 − Mortality rate of control)) × 100
The synergistic effect of Chi60 and Chi70 with Tc toxins against the second-instar larvae of H. armigera was tested. Tc toxins were diluted to 2000, 1000, 500, 250, 125, 62.5 and 31.25 μg/mL. Chi60 and Chi70 were added to every treatment in the same quantity (1000 μg). The method of treatment and the culture condition were consistent with the above methods. Each treatment was replicated three times (n = 72 larvae per concentration). The LC50 of Tc toxin and its mixture with Chi60 and Chi70 was calculated at 72 h after treatment.
5.3.2. Pathological Effect of Chitinase on the Peritrophic Membrane
The chitinase (1000 μg) was added to a 10 g artificial diet. The same quantity of 10 mM PBS (pH 7.2) was used as a control. Approximately 1 g artificial diet and one fifth-instar larva of H. armigera were transferred to each well of a six-well tissue culture plate and then incubated at 28 °C. The peritrophic membrane was extracted and the artificial diet inside was washed at 48 h after feeding. Then, the washed peritrophic membrane was placed on a concave slide, and the morphology was observed under a dissecting microscope.
The chitinase (1000 μg) and blue dextran 2000 (100 μg) (Solarbio, Beijing, China) were added to a 10 g artificial diet. The method of treatment and the culture conditions were consistent with the above methods. The peritrophic membrane was extracted at 48 h after feeding. Then, the peritrophic membrane was placed on a concave slide, and the morphology was observed under a dissecting microscope.
5.4. Knockout of the Chitinase Gene from X. nematophila HB310
5.4.1. Genomic DNA Extraction
The genomic DNA, used as a template for PCR, was extracted from X. nematophila HB310 using the Bacterial Genomic DNA Extraction Kit (TIANGEN, Beijing, China). The quality of genomic DNA was detected using a Touch Screen MD2000C Nano-Spectrophotometer (Biofuture, Shanghai, China) prior to 1% agarose gel electrophoresis.
5.4.2. Homologous Recombination Vector Construction
Six pairs of specific primers based on the gene sequence of chi60 (GenBank access no.: KC701470), chi70 (GenBank access no.: KC701471), kanamycin resistance gene (Kmr) (GenBank access no.: KU221181), and tetracycline resistance gene (tetA) (GenBank access no.: KR071151) are listed in Table S3. Oligo primers 60-up-F/60-up-R and 60-down-F/60-down-R were used to amplify the upstream and downstream fragments of the chi60 gene. Oligo primers 70-up-F/70-up-R and 70-down-F/70-down-R were used to amplify the upstream and downstream fragments of the chi70 gene. Oligo primers Kmr-F/Kmr-R and tet-F/tet-R were used to amplify the kanamycin resistance gene (Kmr) and tetracycline resistance gene (tetA), respectively.
The purified upstream fragment of the chi60 gene, downstream fragment of the chi60 gene and the Kmr gene were used as a template for the fusion amplification of chi60-Kmr. The purified upstream fragment of the chi70 gene, downstream fragment of the chi70 gene, and the tetA gene were used as a template for the fusion amplification of chi70-tetA.
The resulting PCR products were ligated into the pJQ200SK suicide vector. The constructed plasmids were named pJQ200SK-chi60-Kmr and pJQ200SK-chi70-tetA. The plasmids were transformed into E. coli S17-1 λ pir using the heat shock method. Recombinant E. coli was grown in LB medium with ampicillin (100 μg/mL) (Solarbio, Beijing, China) and kanamycin (50 μg/mL) (Solarbio, Beijing, China)/tetracycline (10 μg/mL) (Solarbio, Beijing, China) at 37 °C for 16 h, respectively. The positive clones were subjected to Beijing Genomics Institute for sequencing.
5.4.3. Single Gene Knockout Mutants Screening
Recombinant E. coli S17-1 λ pir containing the suicide plasmid were grown in liquid LB medium at 37 °C. At the same time, X. nematophila HB310 was grown in LB medium at 28 °C. When liquid cultures were grown to an OD600 nm of 0.7, 1 mL cultures were harvested and washed three times using fresh LB medium, respectively. The cells were resuspended in 100 μL LB. Then, the E. coli S17-1 λ pir and X. nematophila were mixed in 1:3 ratio (20 μL E. coli S17-1 λ pir: 60 μL X. nematophila) and spotted onto LB agar plates. Plates were incubated for 3 h at 37 °C and then 28 °C overnight. The bacterial colonies were suspended in the liquid LB medium. The mixture was spread on the LB agar plates containing ampicillin and corresponding resistance. The single colony was transferred into liquid LB medium containing ampicillin and the corresponding resistance for culture. The culture was spread on the LB agar plates containing ampicillin and the corresponding resistance. The single colony was transferred into an LB medium (NaCl free) containing 6% sucrose (Solarbio, Beijing, China). When the transformants were obtained, the genomic DNA was isolated. The genotype of the mutant was confirmed by PCR and the products of PCR were subjected to the Beijing Genomics Institute for sequencing.
5.4.4. Double Gene Knockout Mutant Screening
The Δchi70 was grown in LB medium (NaCl free) containing 6% sucrose with ampicillin and tetracycline at 28 °C, and the recombinant E. coli S17-1 λ pir containing chi60-Kmr gene was grown in liquid LB medium with kanamycin at 37 °C. The method of treatment and the culture conditions were consistent with the above method, except for the antibiotics in the medium from ampicillin and tetracycline to ampicillin, kanamycin and tetracycline.
5.5. Western Blot
The cells of X. nematophila HB310 were centrifuged (4 °C, 10,000 g, 10 min) from culture broth, washed three times with 10 mM PBS (pH 7.2), and suspended in PBS (adding 5 mL PBS per 200 mL bacterial solution). The bacterial cells were lysed by sonication (2 s on, 3 s off, 30 cycles) and centrifuged at 4 °C and 10,000 g for 30 min. The supernatant was collected and filtered with 0.22 μm membrane. The Tc toxins were isolated by precipitation with 85% saturated ammonium sulfate and concentrated using a Centriprep 100 ultrafiltration device with a molecular mass cutoff of 100 kDa (Millipore Corporation, Shanghai, China). The Tc toxins were separated by 6% native polyacrylamide gel electrophoresis (PAGE) using the LIUYI model DYCZ-24F dual vertical electrophoresis apparatus (LIUYI, Beijing, China). The fractions were monitored by a UV detector and collected using a fraction collector, then concentrated by a Centriprep 100 device.
To verify the correctness of the gene knockout, the primary antibody (Chi60 and Chi70 chitinase antiserum) and commercial antibodies (Goat anti-mouse IgG, HRP conjugated) (CWBIO, Beijing, China) were used in the Western blot analysis, which was performed as previously described [46,47].
5.6. Assay for Insecticidal Activity of Tc Toxins
The insecticidal activity of Tc toxins from the WT, Δchi60, Δchi70 and Δchi60-chi70 against the second instar larvae of H. armigera was determined. The Tc toxins of the WT, Δchi60 and Δchi70 were diluted to 2000, 1000, 500, 250, 125 and 62.5 μg/mL, while Tc toxins from Δchi60-chi70 were diluted to 32,000, 16,000, 8000, 4000, 2000, 1000 and 500 μg/mL. The method of treatment and the culture conditions were consistent with the above methods. The LC50 was calculated at 120 h after treatment.
5.7. Data Analysis
The significance of differences in the growth inhibition rate and corrected mortality of chitinase to H. armigera larvae were analyzed by independent samples t-test (SPSS v26.0 software). Mortality data were analyzed by Probit regression (SPSS v26.0 software) to calculate the LC50 for each protein and mixture, with corresponding confidence limits and slopes of regression lines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/toxins14090646/s1, Figure S1: Amplification of the chi60, chi70, Kmr and tetA genes; Figure S2: Amplification of fusion PCR; Figure S3: Double digestion of pJQ200SK-chi60-Kmr and pJQ200SK-chi70-tetA plasmids; Table S1: The sequence information of each motif; Table S2: Amino acid sequences of chitinases used in phylogenetic analyses; Table S3: Primers used in this study.
Author Contributions
J.L., Z.L. and Q.W. conceived the study. J.L., H.B., P.S., Z.N. and Z.D. performed the experiments and analyzed the data. J.L. wrote and edited the original manuscript. Z.L. and Q.W. revised the manuscript and contributed to its improvement. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “China Agriculture Research System, grant number CARS-06-14.5-A25”, and “Hebei Academy of Agriculture and Forestry Sciences (HAAFS) Agriculture Science and Technology Innovation Project, grant number 2022KJCXZX-GZS-4”, and “The Talents Construction Project of Science and Technology Innovation, HAAFS, grant number C21R0401”, and “HAAFS Basic Science and Technology Contract Project, grant number HBNKY-BGZ-02”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank all the personnel who assisted with the experiments. Then, we want to thank to the researcher Guangyue Li from Institute of Plant Protection, Chinese Academy of Agricultural Sciences, who provided the plasmids, competent cell and vector. Finally, we want to thank to the reviewers for their valuable comments and suggestions for improving the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, C.; Shen, N.; Wu, J.; Jiang, M.; Shi, S.; Wang, J.; Wei, Y.; Yang, L. Cloning, expression and characterization of a chitinase from Paenibacillus chitinolyticus strain UMBR 0002. PeerJ 2020, 8, e8964. [Google Scholar] [CrossRef] [PubMed]
- Paek, A.; Kim, M.J.; Park, H.Y.; Yoo, J.G.; Jeong, S.E. Functional expression of recombinant hybrid enzymes composed of bacterial and insect’s chitinase domains in E. coli. Enzyme Microb. Technol. 2020, 136, 109492. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, J.; Liang, Y.; Yan, R.; Xu, X.; Lin, J. Expression and characterization of a chitinase from Serratia marcescens. Protein Expr. Purif. 2020, 171, 105613. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Kim, J.; Son, K.H.; Chung, C.W.; Shin, D.H.; Ku, B.H.; Kim, D.Y.; Park, H.Y. Novel Bi-Modular GH19 chitinase with broad pH stability from a fibrolytic intestinal symbiont of Eisenia fetida, Cellulosimicrobium funkei HY-13. Biomolecules 2021, 11, 1735. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.; Kumar, M.; Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Novel insecticidal chitinase from the insect pathogen Xenorhabdus nematophila. Int. J. Biol. Macromol. 2020, 159, 394–401. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, W.; Zhang, X.; Huang, J.; Wang, W.; Miao, M.; Hu, L.; Wan, C.; Yuan, Y.; Wu, B.; et al. Genome-wide identification and expression analysis of chitinase-like genes in Petunia axillaris. Plants 2022, 11, 1269. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, F.; Xu, G.; Liu, X.; Zhang, Y.; Sun, J.; Yao, B.; Huang, H.; Wu, N.; Tian, J. A novel thermophilic chitinase directly mined from the marine metagenome using the deep learning tool Preoptem. Bioresour. Bioprocess 2022, 9, 54. [Google Scholar] [CrossRef]
- Arora, N.; Ahmad, T.; Rajagopal, R.; Bhatnagar, R.K. A constitutively expressed 36 kDa exochitinase from Bacillus thuringiensis HD-1. Biochem. Biophys. Res. Commun. 2003, 307, 620–625. [Google Scholar] [CrossRef]
- Liu, J.; Nangong, Z.; Zhang, J.; Song, P.; Tang, Y.; Gao, Y.; Wang, Q. Expression and characterization of two chitinases with synergistic effect and antifungal activity from Xenorhabdus nematophila. World J. Microbiol. Biotechnol. 2019, 35, 106. [Google Scholar] [CrossRef]
- Liu, M.; Cai, Q.X.; Liu, H.Z.; Zhang, B.H.; Yan, J.P.; Yuan, Z.M. Chitinolytic activities in Bacillus thuringiensis and their synergistic effects on larvicidal activity. J. Appl. Microbiol. 2002, 93, 374–379. [Google Scholar]
- Bravo, A.; Soberon, M. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 2008, 26, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Leidreiter, F.; Roderer, D.; Meusch, D.; Gatsogiannis, C.; Benz, R.; Raunser, S. Common architecture of Tc toxins from human and insect pathogenic bacteria. Sci. Adv. 2019, 5, x6497. [Google Scholar] [CrossRef]
- Fuchs, T.M.; Bresolin, G.; Marcinowski, L.; Schachtner, J.; Scherer, S. Insecticidal genes of Yersinia spp.: Taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 2008, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Landsberg, M.J.; Jones, S.A.; Rothnagel, R.; Busby, J.N.; Marshall, S.D.; Simpson, R.M.; Lott, J.S.; Hankamer, B.; Hurst, M.R. 3D structure of the Yersinia entomophaga toxin complex and implications for insecticidal activity. Proc. Natl. Acad. Sci. USA 2011, 108, 20544–20549. [Google Scholar] [CrossRef] [PubMed]
- Busby, J.N.; Landsberg, M.J.; Simpson, R.M.; Jones, S.A.; Hankamer, B.; Hurst, M.R.; Lott, J.S. Structural analysis of Chi1 chitinase from Yen-Tc: The multisubunit insecticidal ABC toxin complex of Yersinia entomophaga. J. Mol. Biol. 2012, 415, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, J.; Li, T.; Liu, S.; Song, P.; Nangong, Z.; Wang, Q. PirAB protein from Xenorhabdus nematophila HB310 exhibits a binary toxin with insecticidal activity and cytotoxicity in Galleria mellonella. J. Invertebr. Pathol. 2017, 148, 43–50. [Google Scholar] [CrossRef]
- Liu, J.; Song, P.; Zhang, J.; Nangong, Z.; Liu, X.; Gao, Y.; Wang, Q. Characteristics and function of the chitin binding protein from Xenorhabdus nematophila. Protein Peptide Lett. 2019, 26, 414–422. [Google Scholar] [CrossRef]
- Sergeant, M.; Jarrett, P.; Ousley, M.; Morgan, J.A. Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 2003, 69, 3344–3349. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.H.; Dowling, A.; Waterfield, N.R. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon 2007, 49, 436–451. [Google Scholar] [CrossRef]
- Waterfield, N.; Hares, M.; Hinchliffe, S.; Wren, B.; Ffrench-Constant, R. The insect toxin complex of Yersinia. Adv. Exp. Med. Biol. 2007, 603, 247–257. [Google Scholar]
- Lang, A.E.; Konukiewitz, J.; Aktories, K.; Benz, R. TcdA1 of Photorhabdus luminescens: Electrophysiological analysis of pore formation and effector binding. Biophys. J. 2013, 105, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Gatsogiannis, C.; Lang, A.E.; Meusch, D.; Pfaumann, V.; Hofnagel, O.; Benz, R.; Aktories, K.; Raunser, S. A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature 2013, 495, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Gatsogiannis, C.; Merino, F.; Prumbaum, D.; Roderer, D.; Leidreiter, F.; Meusch, D.; Raunser, S. Membrane insertion of a Tc toxin in near-atomic detail. Nat. Struct. Mol. Biol. 2016, 23, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Gatsogiannis, C.; Merino, F.; Roderer, D.; Balchin, D.; Schubert, E.; Kuhlee, A.; Hayer-Hartl, M.; Raunser, S. Tc toxin activation requires unfolding and refolding of a beta-propeller. Nature 2018, 563, 209–213. [Google Scholar] [CrossRef]
- Meusch, D.; Gatsogiannis, C.; Efremov, R.G.; Lang, A.E.; Hofnagel, O.; Vetter, I.R.; Aktories, K.; Raunser, S. Mechanism of Tc toxin action revealed in molecular detail. Nature 2014, 508, 61–65. [Google Scholar] [CrossRef]
- Roderer, D.; Schubert, E.; Sitsel, O.; Raunser, S. Towards the application of Tc toxins as a universal protein translocation system. Nat. Commun. 2019, 10, 5263. [Google Scholar] [CrossRef]
- Sheets, J.J.; Hey, T.D.; Fencil, K.J.; Burton, S.L.; Ni, W.; Lang, A.E.; Benz, R.; Aktories, K. Insecticidal toxin complex proteins from Xenorhabdus nematophilus: Structure and pore formation. J. Biol. Chem. 2011, 286, 22742–22749. [Google Scholar] [CrossRef]
- Roderer, D.; Raunser, S. Tc toxin complexes: Assembly, membrane permeation, and protein translocation. Annu. Rev. Microbiol. 2019, 73, 247–265. [Google Scholar] [CrossRef]
- Morgan, J.A.; Sergeant, M.; Ellis, D.; Ousley, M.; Jarrett, P. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 2001, 67, 2062–2069. [Google Scholar] [CrossRef]
- Hurst, M.R.; Glare, T.R.; Jackson, T.A.; Ronson, C.W. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 2000, 182, 5127–5138. [Google Scholar] [CrossRef]
- Hurst, M.R.; Jones, S.M.; Tan, B.; Jackson, T.A. Induced expression of the Serratia entomophila Sep proteins shows activity towards the larvae of the New Zealand grass grub Costelytra zealandica. Fems. Microbiol. Lett. 2007, 275, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Stoilova-Mcphie, S.; Baxter, L.; Fulop, V.; Henderson, J.; Rodger, A.; Roper, D.I.; Scott, D.J.; Smith, C.J.; Morgan, J.A. Structural characterisation of the insecticidal toxin XptA1, reveals a 1.15 MDa tetramer with a cage-like structure. J. Mol. Biol. 2007, 366, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Nangong, Z.Y.; Yang, J.; Song, P.; Wang, Y.; Cui, L.; Cui, L. Toxic activity of a protein complex purified from Xenorhabdus nematophila HB310 to Plutella xylostella larvae. Insect Sci. 2012, 19, 329–336. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Revathi, K.; Thanigaivel, A.; Kirubakaran, S.A.; Senthil-Nathan, S. Bacillus subtilis chitinase identified by matrix-assisted laser desorption/ionization time-of flight/time of flight mass spectrometry has insecticidal activity against Spodoptera litura Fab. Pestic. Biochem. Physiol. 2014, 116, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Suganthi, M.; Senthilkumar, P.; Arvinth, S.; Chandrashekara, K.N. Chitinase from Pseudomonas fluorescens and its insecticidal activity against Helopeltis theivora. J. Gen. Appl. Microbiol. 2017, 63, 222–227. [Google Scholar] [CrossRef]
- Okongo, R.N.; Puri, A.K.; Wang, Z.; Singh, S.; Permaul, K. Comparative biocontrol ability of chitinases from bacteria and recombinant chitinases from the thermophilic fungus Thermomyces lanuginosus. J. Biosci. Bioeng. 2019, 127, 663–671. [Google Scholar] [CrossRef]
- Subbanna, A.; Chandrashekara, C.; Stanley, J.; Mishra, K.K.; Mishra, P.K.; Pattanayak, A. Bio-efficacy of chitinolytic Bacillus thuringiensis isolates native to northwestern Indian Himalayas and their synergistic toxicity with selected insecticides. Pestic. Biochem. Physiol. 2019, 158, 166–174. [Google Scholar] [CrossRef]
- Wiwat, C.; Thaithanun, S.; Pantuwatana, S.; Bhumiratana, A. Toxicity of chitinase-producing Bacillus thuringiensis ssp. kurstaki HD-1 (G) toward Plutella xylostella. J. Invertebr. Pathol. 2000, 76, 270–277. [Google Scholar]
- Lertcanawanichakul, M.; Wiwat, C.; Bhumiratana, A.; Dean, D.H. Expression of chitinase-encoding genes in Bacillus thuringiensis and toxicity of engineered B. thuringiensis subsp. aizawai toward Lymantria dispari larvae. Curr. Microbiol. 2004, 48, 175–181. [Google Scholar] [CrossRef]
- Liu, D.; Cai, J.; Xie, C. Purification and partial characterization of a 36-kDa chitinase from Bacillus thuringiensis subsp. colmeri, and its biocontrol potential. Enzyme. Microb. Tech. 2010, 46, 252–256. [Google Scholar] [CrossRef]
- Hurst, M.R.; Jones, S.A.; Binglin, T.; Harper, L.A.; Jackson, T.A.; Glare, T.R. The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J. Bacteriol. 2011, 193, 1966–1980. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Nagpure, A.; Gupta, R.K. Bacterial chitinases: Properties and potential. Crit. Rev. Biotechnol. 2007, 27, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cao, Y.; Zhang, W.; Liu, Z.; Li, Y.; Chen, Y.; Zhang, H.; Yu, F.; Liu, X. The wheat TaIQD3D-6 gene encodes a microtubule-associated protein and regulates cell morphogenesis in Arabidopsis. Plant. Sci. 2022, 324, 111420. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, Y.; Wang, Q.; Shi, H.; Yin, J.; Li, C. Identification and characterization of an antennae-specific glutathione S-transferase from the Indian Meal Moth. Front. Physiol. 2021, 12, 727619. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, K.; Jing, C.; Wu, R.; Wu, G.; Li, M.; Qing, L. Molecular characterization of a novel Conyza canadensis-infecting begomovirus in China. Phytopathol. Res. 2022, 4, 13. [Google Scholar] [CrossRef]
- Ajit, N.S.; Verma, R.; Shanmugam, V. Extracellular chitinases of fluorescent pseudomonads antifungal to Fusarium oxysporum f. Sp. dianthi causing carnation wilt. Curr. Microbiol. 2006, 52, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Kirubakaran, S.I.; Sakthivel, N. Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Expr. Purif. 2007, 52, 159–166. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).