Genotoxicity of 12 Mycotoxins by the SOS/umu Test: Comparison of Liver and Kidney S9 Fraction
Abstract
:1. Introduction
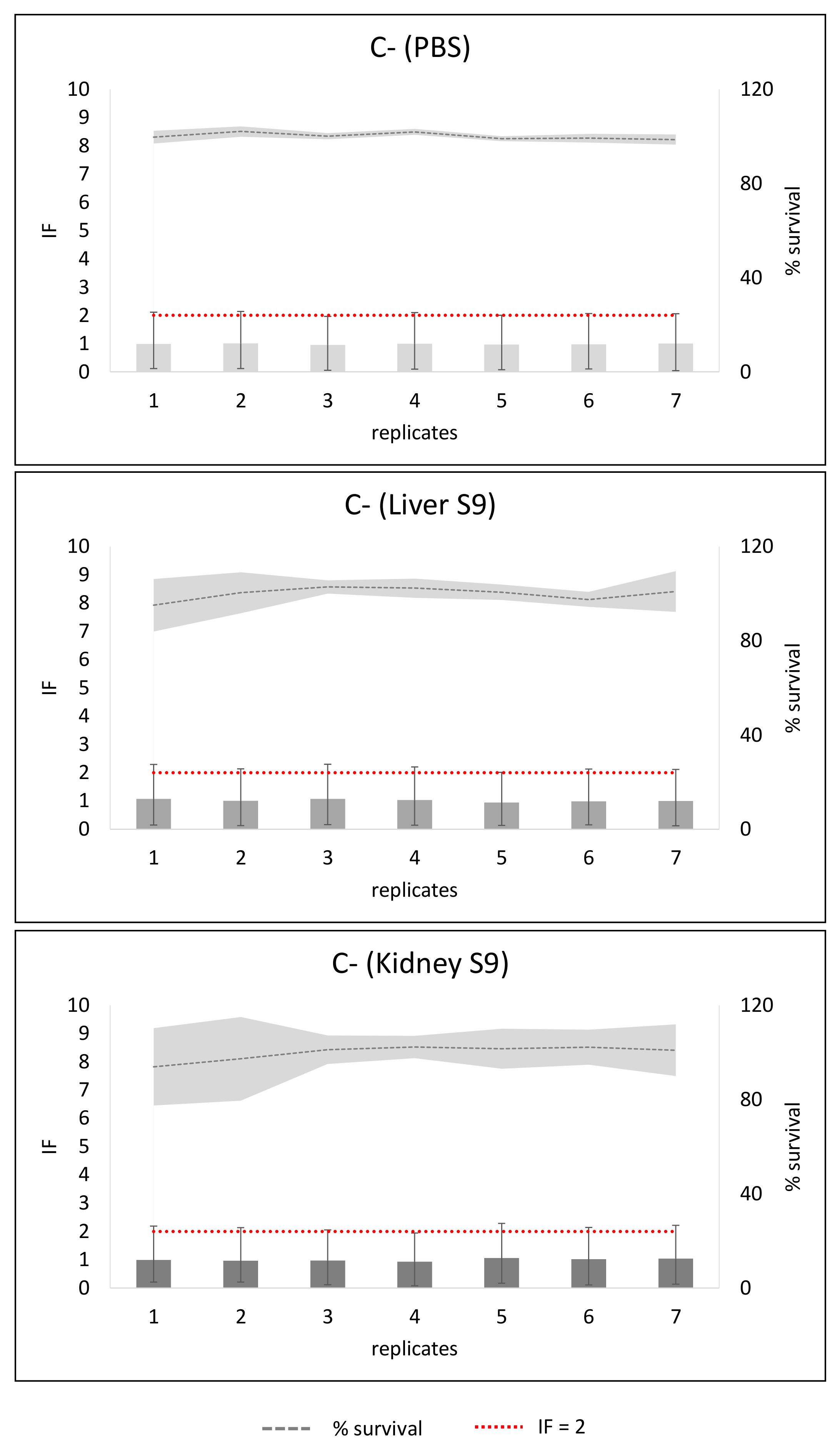
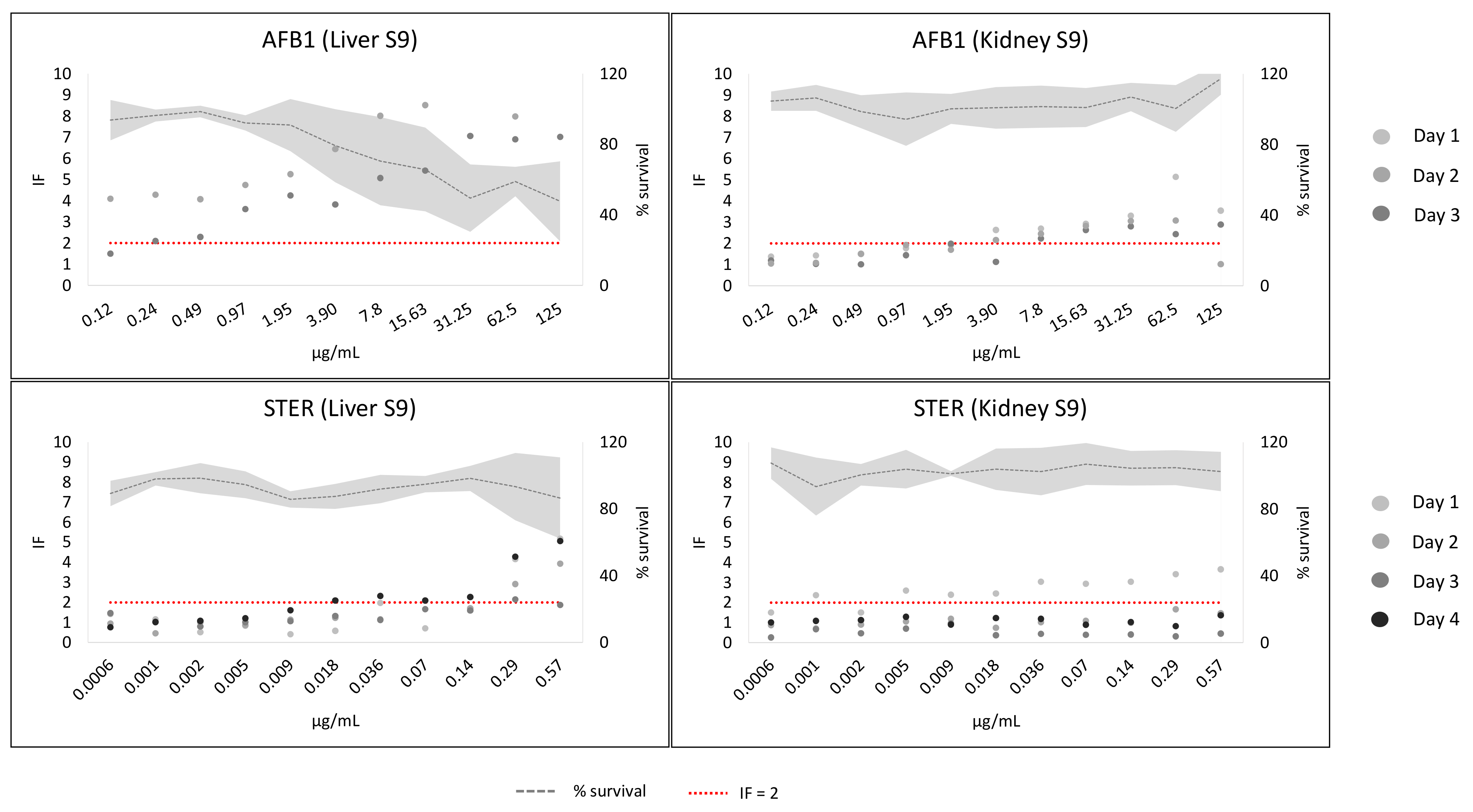
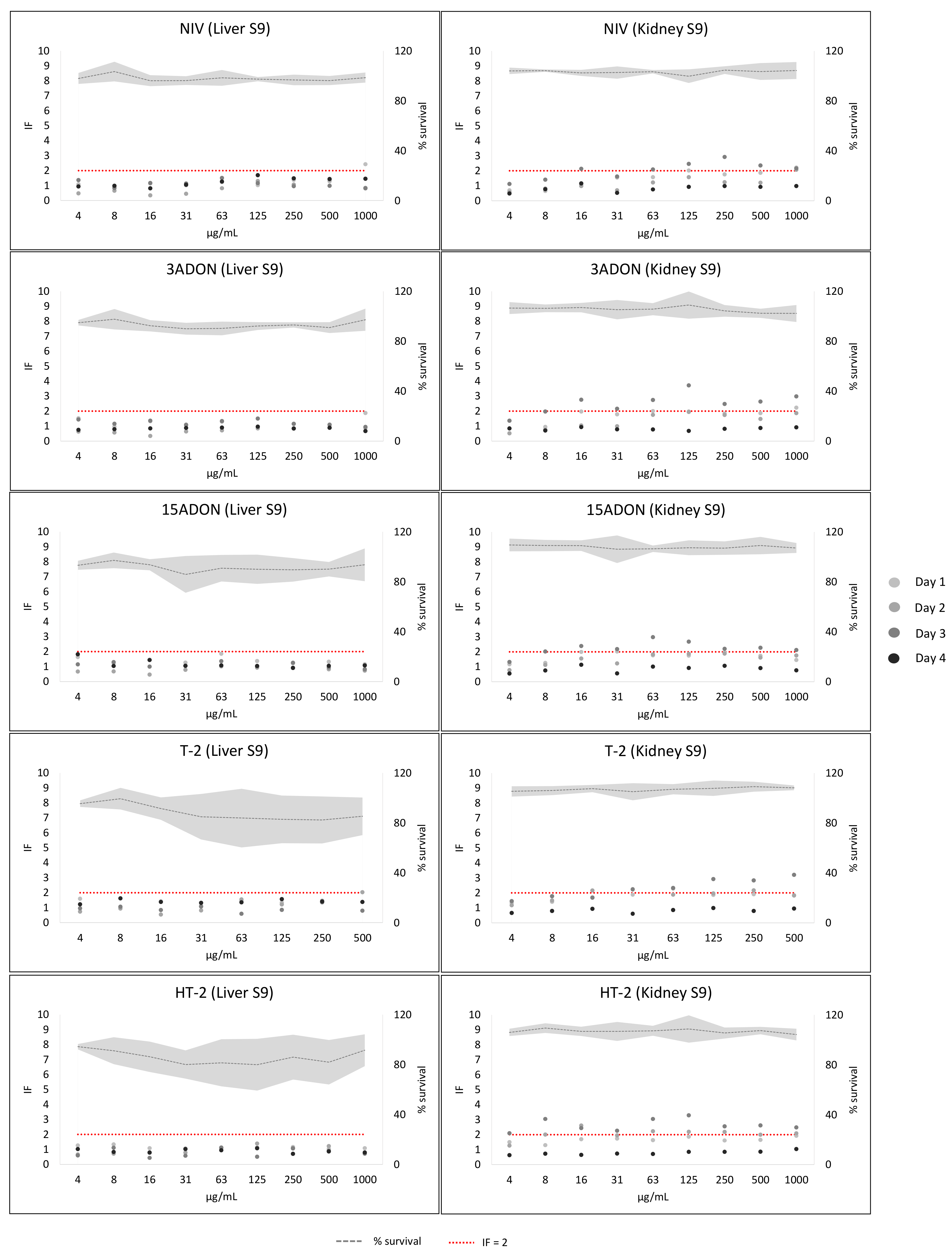
2. Results
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reagents
5.2. Experimental System
5.3. Mycotoxins
5.4. Rat Tissue Fractions and S9 Mix Preparation
5.5. Replicates and Number of Independent Experiments
5.6. SOS/umu Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vettorazzi, A.; López de Cerain, A. Mycotoxins as Food Carcinogens. In Environmental Mycology in Public Health; Elsevier: Amsterdam, The Netherlands, 2016; pp. 261–298. ISBN 9780124114715. [Google Scholar]
- Alonso-Jauregui, M.; Font, M.; González-Peñas, E.; López de Cerain, A.; Vettorazzi, A. Prioritization of Mycotoxins Based on Their Genotoxic Potential with an In Silico-In Vitro Strategy. Toxins 2021, 13, 734. [Google Scholar] [CrossRef] [PubMed]
- OECD. Test. In OECD Guidelines for the Testing of Chemicals, Section 4; OECD: Paris, France, 2020; ISBN 9789264071247. [Google Scholar]
- ICH. ICH Guideline S2 (R1) on Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use; ICH: Geneva, Switzerland, 2008; Volume 2, pp. 1–28. [Google Scholar]
- Yoshikawa, K.; Nohmi, T.; Miyata, R.; Ishidate, M.; Ozawa, N.; Isobe, M.; Watabe, T.; Kada, T.; Kawachi, T. Differences in Liver Homogenates from Donryu, Fischer, Sprague-Dawley and Wistar Strains of Rat in the Drug-Metabolizing Enzyme Assay and the Salmonella/Hepatic S9 Activation Test. Mutat. Res. Mol. Mech. Mutagen. 1982, 96, 167–186. [Google Scholar] [CrossRef]
- Brendt, J.; Crawford, S.E.; Velki, M.; Xiao, H.; Thalmann, B.; Hollert, H.; Schiwy, A. Is a Liver Comparable to a Liver? A Comparison of Different Rat-Derived S9-Fractions with a Biotechnological Animal-Free Alternative in the Ames Fluctuation Assay. Sci. Total Environ. 2021, 759, 143522. [Google Scholar] [CrossRef] [PubMed]
- Ku, W.W.; Bigger, A.; Brambilla, G.; Glatt, H.; Gocke, E.; Guzzie, P.J.; Hakura, A.; Honma, M.; Martus, H.-J.; Obach, R.S.; et al. Strategy for Genotoxicity Testing—Metabolic Considerations. Mutat. Res. Toxicol. Environ. Mutagen. 2007, 627, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Shahin, M.M.; Chopy, C.; Mayet, M.J.; Lequesne, N. Mutagenicity of Structurally Related Aromatic Amines in the Salmonella/Mammalian Microsome Test with Various S-9 Fractions. Food Chem. Toxicol. 1983, 21, 615–619. [Google Scholar] [CrossRef]
- Zvereva, I.; Semenistaya, E.; Krotov, G.; Rodchenkov, G. Comparison of Various In Vitro Model Systems of the Metabolism of Synthetic Doping Peptides: Proteolytic Enzymes, Human Blood Serum, Liver and Kidney Microsomes and Liver S9 Fraction. J. Proteom. 2016, 149, 85–97. [Google Scholar] [CrossRef]
- Barnett, L.M.A.; Cummings, B.S. Nephrotoxicity and Renal Pathophysiology: A Contemporary Perspective. Toxicol. Sci. 2018, 164, 379–390. [Google Scholar] [CrossRef] [Green Version]
- Berndt, W.O. The Role of Transport in Chemical Nephrotoxicity. Toxicol. Pathol. 1998, 26, 52–57. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Schrenk, D.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.R.; Leblanc, J.; Nebbia, C.S.; et al. Risk Assessment of Ochratoxin A in Food. EFSA J. 2020, 18, e06113. [Google Scholar] [CrossRef]
- Obrecht-Pflumio, S.; Chassat, T.; Dirheimer, G.; Marzin, D. Genotoxicity of Ochratoxin A by Salmonella Mutagenicity Test after Bioactivation by Mouse Kidney Microsomes. Mutat. Res. Toxicol. Environ. Mutagen. 1999, 446, 95–102. [Google Scholar] [CrossRef]
- Yasunaga, K.; Kiyonari, A.; Oikawa, T.; Abe, N.; Yoshikawa, K. Evaluation of the Salmonella umu Test with 83 NTP Chemicals. Environ. Mol. Mutagen. 2004, 44, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Reifferscheid, G.; Heil, J. Validation of the SOS/Umu Test Using Test Results of 486 Chemicals and Comparison with the Ames Test and Carcinogenicity Data. Mutat. Res. Toxicol. 1996, 369, 129–145. [Google Scholar] [CrossRef]
- McDaniels, A.E.; Reyes, A.L.; Wymer, L.J.; Rankin, C.C.; Stelma, G.N. Comparison of the Salmonella (Ames) Test, Umu Tests, and the Sos Chromotests for Detecting Genotoxins. Environ. Mol. Mutagen. 1990, 16, 204–215. [Google Scholar] [CrossRef]
- Parmentier, Y.; Bossant, M.-J.; Bertrand, M.; Walther, B. In Vitro Studies of Drug Metabolism. In Comprehensive Medicinal Chemistry II; Elsevier: Amsterdam, The Netherlands, 2007; pp. 231–257. [Google Scholar]
- Maron, D.M.; Ames, B.N. Revised Methods for the Salmonella Mutagenicity Test. Mutat. Res. Mutagen. Relat. Subj. 1983, 113, 173–215. [Google Scholar] [CrossRef]
- Oda, Y.; Nakamura, S.; Oki, I.; Kato, T.; Shinagawa, H. Evaluation of the New System (Umu-Test) for the Detection of Environmental Mutagens and Carcinogens. Mutat. Res. Mutagen. Relat. Subj. 1985, 147, 219–229. [Google Scholar] [CrossRef]
- Oda, Y.; Yamazaki, H.; Watanabe, M.; Nohmi, T.; Shimada, T. Development of High Sensitive Umu Test System: Rapid Detection of Genotoxicity of Promutagenic Aromatic Amines by Salmonella typhimurium Strain NM2009 Possessing High O-Acetyltransferase Activity. Mutat. Res. Mutagen. Relat. Subj. 1995, 334, 145–156. [Google Scholar] [CrossRef]
- Oda, Y.; Funasaka, K.; Kitano, M.; Nakama, A.; Yoshikura, T. Use of a High-Throughputumu-Microplate Test System for Rapid Detection of Genotoxicity Produced by Mutagenic Carcinogens and Airborne Particulate Matter. Environ. Mol. Mutagen. 2004, 43, 10–19. [Google Scholar] [CrossRef]
- Hamer, B.; Bihari, N.; Reifferscheid, G.; Zahn, R.K.; Müller, W.E.; Batel, R. Evaluation of the SOS/Umu-Test Post-Treatment Assay for the Detection of Genotoxic Activities of Pure Compounds and Complex Environmental Mixtures. Mutat. Res. Toxicol. Environ. Mutagen. 2000, 466, 161–171. [Google Scholar] [CrossRef]
- Reifferscheid, G.; Heil, J.; Oda, Y.; Zahn, R.K. A Microplate Version of the SOS/Umu-Test for Rapid Detection of Genotoxins and Genotoxic Potentials of Environmental Samples. Mutat. Res. Mutagen. Relat. Subj. 1991, 253, 215–222. [Google Scholar] [CrossRef]
- Whong, W.-Z.; Wen, Y.-F.; Stewart, J.; Ong, T. Validation of the SOS/Umu Test with Mutagenic Complex Mixtures. Mutat. Res. Lett. 1986, 175, 139–144. [Google Scholar] [CrossRef]
- Prival, M.J.; Mitchell, V.D. Analysis of a Method for Testing Azo Dyes for Mutagenic Activity in Salmonella typhimurium in the Presence of Flavin Mononucleotide and Hamster Liver S9. Mutat. Res. Mutagen. Relat. Subj. 1982, 97, 103–116. [Google Scholar] [CrossRef]
- Prival, M.J.; Bell, S.J.; Mitchell, V.D.; Peiperl, M.D.; Vaughan, V.L. Mutagenicity of Benzidine and Benzidine-Congener Dyes and Selected Monoazo Dyes in a Modified Salmonella Assay. Mutat. Res. Toxicol. 1984, 136, 33–47. [Google Scholar] [CrossRef]
- Prival, M.J.; Davis, V.M.; Peiperl, M.D.; Bell, S.J. Evaluation of Azo Food Dyes for Mutagenicity and Inhibition of Mutagenicity by Methods Using Salmonella typhimurium. Mutat. Res. Toxicol. 1988, 206, 247–259. [Google Scholar] [CrossRef]
- Poapolathep, A.; Sugita-Konishi, Y.; Doi, K.; Kumagai, S. The Fates of Trichothecene Mycotoxins, Nivalenol and Fusarenon-X, in Mice. Toxicon 2003, 41, 1047–1054. [Google Scholar] [CrossRef]
- Schuhmacher-Wolz, U.; Heine, K.; Schneider, K. Report on Toxicity Data on Trichothecene Mycotoxins HT-2 and T-2 Toxins. EFSA Support. Publ. 2010, 7, 65E. [Google Scholar] [CrossRef]
- SCF. Opinion of the Scientific Panel on Food on Fusarium Toxins. Part 5: T-2 Toxin and HT-2 Toxin; SCF: Brussels, Belgium, 2001; Volume 5. [Google Scholar]
- Nakamura, S.; Oda, Y.; Shimada, T.; Oki, I.; Sugimoto, K. SOS-Inducing Activity of Chemical Carcinogens and Mutagens in Salmonella typhimurium TA1535/PSK1002: Examination with 151 Chemicals. Mutat. Res. Lett. 1987, 192, 239–246. [Google Scholar] [CrossRef]
- Matsushima, T.; Kobuna, I.; Fukuoka, F.; Sugimura, T. Metabolism of 4-Nitroquinoline 1-Oxide. IV. Quantitative Determination of In Vivo Conversion of 4-Nitroquinoline 1-Oxide in Subcutaneous Tissue of Rat. Gan 1968, 59, 247–250. [Google Scholar]
- Shirasu, Y. Comparative Carcinogenicity of 4-Nitroquinoline 1-Oxide and 4-Hydroxyaminoquinoline 1-Oxide in 3 Strains of Mice. Exp. Biol. Med. 1965, 118, 812–814. [Google Scholar] [CrossRef]
- Ayrton, A.D.; Neville, S.; Ioannides, C. Cytosolic Activation of 2-Aminoanthracene: Implications in Its Use as Diagnostic Mutagen in the Ames Test. Mutat. Res. Mol. Mech. Mutagen. 1992, 265, 1–8. [Google Scholar] [CrossRef]
- Phillipson, C.E.; Ioannides, C. Activation of Aromatic Amines to Mutagens by Various Animal Species Including Man. Mutat. Res. Toxicol. 1983, 124, 325–336. [Google Scholar] [CrossRef]
- Guobaitis, R.J.; Ellingham, T.J.; Maddock, M.B. The Effects of Pretreatment with Cytochrome P-450 Inducers and Preincubation with a Cytochrome P-450 Effector on the Mutagenicity of Genotoxic Carcinogens Mediated by Hepatic and Renal S9 from Two Species of Marine Fish. Mutat. Res. Mutagen. Relat. Subj. 1986, 164, 59–70. [Google Scholar] [CrossRef]
- IARC. Some Traditional Herbal Medicines, Some Mycotoxins, Naphtalene and Styrene. Monogr. Eval. Carcinog. Risks Hum. 2002, 82, 169–522. [Google Scholar]
- Auffray, Y.; Boutibonnes, P. Induction of SOS Function InEscherichia Coli by Some Mycotoxins. Toxic. Assess. 1988, 3, 371–378. [Google Scholar] [CrossRef]
- Krivobok, S.; Olivier, P.; Marzin, D.R.; Seigle-Murandi, F.; Steiman, R. Study of the Genotoxic Potential of 17 Mycotoxins with the SOS Chromotest. Mutagenesis 1987, 2, 433–439. [Google Scholar] [CrossRef]
- Theumer, M.G.; Henneb, Y.; Khoury, L.; Snini, S.P.; Tadrist, S.; Canlet, C.; Puel, O.; Oswald, I.P.; Audebert, M. Genotoxicity of Aflatoxins and Their Precursors in Human Cells. Toxicol. Lett. 2018, 287, 100–107. [Google Scholar] [CrossRef]
- Baertschi, S.W.; Raney, K.D.; Shimada, T.; Harris, T.M.; Guengerich, F.P. Comparison of Rates of Enzymic Oxidation of Aflatoxin B1, Aflatoxin G1, and Sterigmatocystin and Activities of the Epoxides in Forming Guanyl-N7 Adducts and Inducing Different Genetic Responses. Chem. Res. Toxicol. 1989, 2, 114–122. [Google Scholar] [CrossRef]
- Sakai, M.; Abe, K.-I.; Okumura, H.; Kawamura, O.; Sugiura, Y.; Horie, Y.; Ueno, Y. Genotoxicity of Fungi Evaluated by SOS Microplate Assay. Nat. Toxins 1992, 1, 27–34. [Google Scholar] [CrossRef]
- Wehner, F.C.; Marasas, W.F.O.; Thiel, P.G. Lack of Mutagenicity to Salmonella Typhimurium of Some Fusarium Mycotoxins. Appl. Environ. Microbiol. 1978, 35, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Knasmüller, S.; Bresgen, N.; Kassie, F.; Mersch-Sundermann, V.; Gelderblom, W.; Zöhrer, E.; Eckl, P.M. Genotoxic Effects of Three Fusarium Mycotoxins, Fumonisin B1, Moniliformin and Vomitoxin in Bacteria and in Primary Cultures of Rat Hepatocytes. Mutat. Res. Toxicol. Environ. Mutagen. 1997, 391, 39–48. [Google Scholar] [CrossRef]
- Takakura, N.; Nesslany, F.; Fessard, V.; Le Hegarat, L. Absence of In Vitro Genotoxicity Potential of the Mycotoxin Deoxynivalenol in Bacteria and in Human TK6 and HepaRG Cell Lines. Food Chem. Toxicol. 2014, 66, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ueno, Y. Mode of Action of Trichothecenes. Pure Appl. Chem. 1977, 49, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Kuczuk, M.H.; Benson, P.M.; Heath, H.; Wallace Hayes, A. Evaluation of the Mutagenic Potential of Mycotoxins Using Salmonella Typhimurium and Saccharomyces Cerevisiae. Mutat. Res. Mutagen. Relat. Subj. 1978, 53, 11–20. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Scientific Opinion on Risks for Animal and Public Health Related to the Presence of Nivalenol in Food and Feed. EFSA J. 2013, 11, 3262. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). Scientific Opinion on the Risks for Animal and Public Health Related to the Presence of T-2 and HT-2 Toxin in Food and Feed. EFSA J. 2011, 9, 2481. [Google Scholar] [CrossRef]
- Knutsen, H.K.; Alexander, J.; Barregård, L.; Bignami, M.; Brüschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Grasl-Kraupp, B.; Hogstrand, C.; et al. Risks to Human and Animal Health Related to the Presence of Deoxynivalenol and Its Acetylated and Modified Forms in Food and Feed. EFSA J. 2017, 15, 04718. [Google Scholar] [CrossRef]
- Bach, P.H.; Gregg, N.J.; Delacruz, L. Relevance of a Rat Model of Papillary Necrosis and Upper Urothelial Carcinoma in Understanding the Role of Ochratoxin A in Balkan Endemic Nephropathy and Its Associated Carcinoma. Food Chem. Toxicol. 1992, 30, 205–211. [Google Scholar] [CrossRef]
- Rašić, D.; Mladinić, M.; Želježić, D.; Pizent, A.; Stefanović, S.; Milićević, D.; Konjevoda, P.; Peraica, M. Effects of Combined Treatment with Ochratoxin A and Citrinin on Oxidative Damage in Kidneys and Liver of Rats. Toxicon 2018, 146, 99–105. [Google Scholar] [CrossRef]
- Malaveille, C.; Brun, G.; Bartsch, H. Structure-Activity Studies in E. Coli Strains on Ochratoxin A (OTA) and Its Analogues Implicate a Genotoxic Free Radical and a Cytotoxic Thiol Derivative as Reactive Metabolites. Mutat. Res. Mol. Mech. Mutagen. 1994, 307, 141–147. [Google Scholar] [CrossRef]
- Reiss, J. Detection of Genotoxic Properties of Mycotoxins with the SOS Chromotest. Naturwissenschaften 1986, 73, 677–678. [Google Scholar] [CrossRef]
- Bartholomew, R.M.; Ryan, D.S. Lack of Mutagenicity of Some Phytoestrogens in the Salmonella/Mammalian Microsome Assay. Mutat. Res. Toxicol. 1980, 78, 317–321. [Google Scholar] [CrossRef]
- Auffray, Y.; Boutibonnes, P. Evaluation of the Genotoxic Activity of Some Mycotoxins Using Escherichia Coli in the SOS Spot Test. Mutat. Res. Toxicol. 1986, 171, 79–82. [Google Scholar] [CrossRef]
- Ghédira-Chékir, L.; Maaroufi, K.; Zakhama, A.; Ellouz, F.; Dhouib, S.; Creppy, E.; Bacha, H. Induction of a SOS Repair System in Lysogenic Bacteria by Zearalenone and Its Prevention by Vitamin E. Chem. Biol. Interact. 1998, 113, 15–25. [Google Scholar] [CrossRef]
- Müller, S.; Dekant, W.; Mally, A. Fumonisin B1 and the Kidney: Modes of Action for Renal Tumor Formation by Fumonisin B1 in Rodents. Food Chem. Toxicol. 2012, 50, 3833–3846. [Google Scholar] [CrossRef] [PubMed]
- Park, D.L.; Rua, S.M.; Mirocha, C.J.; Abd-Alla, E.-S.A.M.; Weng, C.Y. Mutagenic Potentials of Fumonisin Contaminated Corn Following Ammonia Decontamination Procedure. Mycopathologia 1992, 117, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M. Assessment of In Vitro Mutagenicity in Salmonella and In Vivo Genotoxicity in Mice of the Mycotoxin Fumonisin B1. Mutagenesis 2000, 15, 469–471. [Google Scholar] [CrossRef] [Green Version]
- Escobar, P.A.; Kemper, R.A.; Tarca, J.; Nicolette, J.; Kenyon, M.; Glowienke, S.; Sawant, S.G.; Christensen, J.; Johnson, T.E.; McKnight, C.; et al. Bacterial Mutagenicity Screening in the Pharmaceutical Industry. Mutat. Res. Mutat. Res. 2013, 752, 99–118. [Google Scholar] [CrossRef]
- Diehl, M.S.; Willaby, S.L.; Snyder, R.D. Comparison of the Results of a Modified Miniscreen and the Standard Bacterial Reverse Mutation Assays. Environ. Mol. Mutagen. 2000, 36, 72–77. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alonso-Jauregui, M.; González-Peñas, E.; López de Cerain, A.; Vettorazzi, A. Genotoxicity of 12 Mycotoxins by the SOS/umu Test: Comparison of Liver and Kidney S9 Fraction. Toxins 2022, 14, 400. https://doi.org/10.3390/toxins14060400
Alonso-Jauregui M, González-Peñas E, López de Cerain A, Vettorazzi A. Genotoxicity of 12 Mycotoxins by the SOS/umu Test: Comparison of Liver and Kidney S9 Fraction. Toxins. 2022; 14(6):400. https://doi.org/10.3390/toxins14060400
Chicago/Turabian StyleAlonso-Jauregui, Maria, Elena González-Peñas, Adela López de Cerain, and Ariane Vettorazzi. 2022. "Genotoxicity of 12 Mycotoxins by the SOS/umu Test: Comparison of Liver and Kidney S9 Fraction" Toxins 14, no. 6: 400. https://doi.org/10.3390/toxins14060400
APA StyleAlonso-Jauregui, M., González-Peñas, E., López de Cerain, A., & Vettorazzi, A. (2022). Genotoxicity of 12 Mycotoxins by the SOS/umu Test: Comparison of Liver and Kidney S9 Fraction. Toxins, 14(6), 400. https://doi.org/10.3390/toxins14060400




