Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis
Abstract
:1. Introduction
2. Results
2.1. Verification of Species Identity
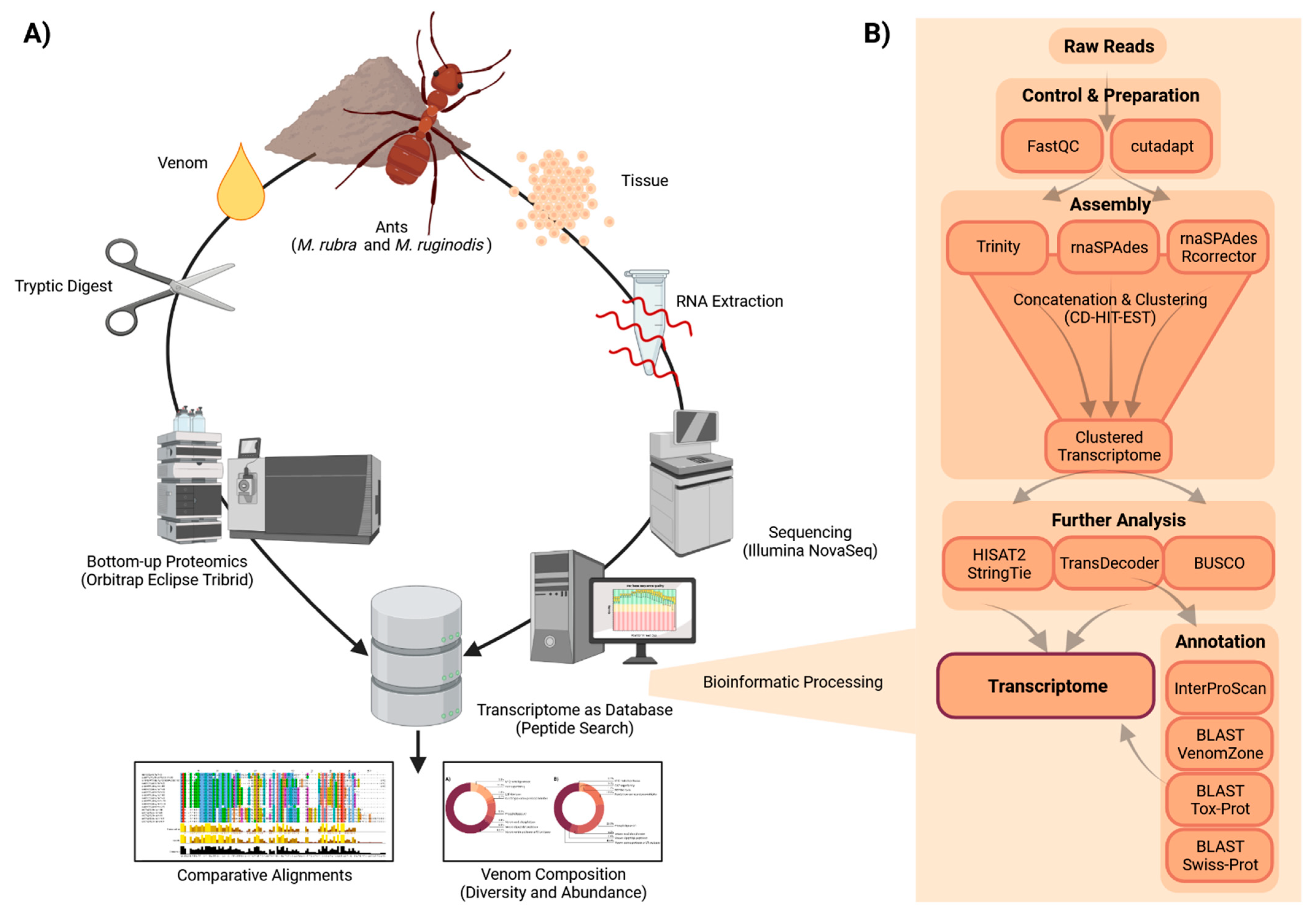
2.2. Proteomic and Transcriptomic Landscapes
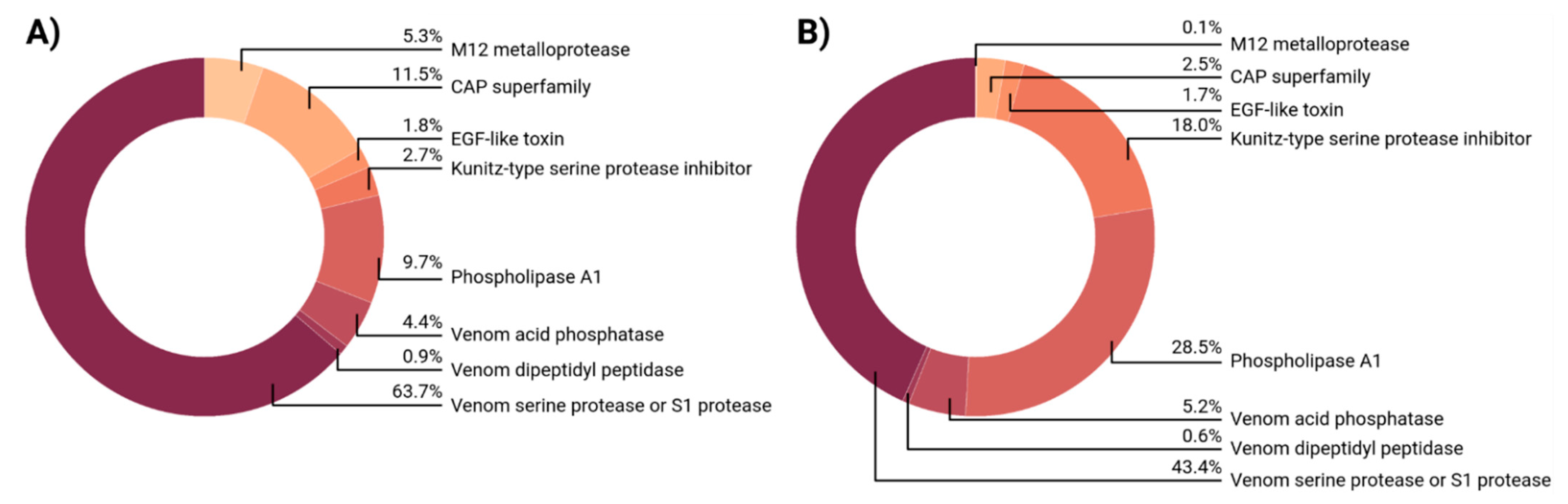
2.3. Venom Composition of Myrmica rubra
2.4. Venom Composition of Myrmica ruginodis
2.5. Diversity and Characteristics of EGF-Like Toxins in Myrmicine Ants
3. Discussion
3.1. Serine Proteases and Kunitz-Type Serine Protease Inhibitors
3.2. Venom Allergens
3.3. EGF-Like Toxins
3.4. Evolution and Function of Venom Proteins in Myrmicine Ants
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Collection of Venom and Venom Glands
5.3. RNA Extraction and Sequencing
5.4. Transcriptome Analysis
5.5. Species Identification by CO1 Sequence Analysis
5.6. Proteomics
5.7. Analysis of EGF-Like Toxins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- California Academy of Science AntWeb. Version 8.69.2. Available online: https://www.antweb.org/ (accessed on 2 March 2022).
- Ward, P.S. The Phylogeny and Evolution of Ants. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 23–43. [Google Scholar] [CrossRef] [Green Version]
- Hölldobler, B.; Wilson, E.O. The Ants; Harvard University Press: Cambridge, MA, USA, 1990; ISBN 978-0-674-04075-5. [Google Scholar]
- Schmidt, J.O. Chemistry, Pharmacology, and Chemical Ecology of Ant Venoms. In Venoms of the Hymenoptera: Biochemical, Pharmacological and Behavioural Aspects; Piek, T., Ed.; Academic Press: London, UK, 1986; pp. 425–508. ISBN 978-0-12-554770-3. [Google Scholar]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.A.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The Toxicogenomic Multiverse: Convergent Recruitment of Proteins Into Animal Venoms. Annu. Rev. Genom. Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schendel, V.; Rash, L.D.; Jenner, R.A.; Undheim, E.A.B. The Diversity of Venom: The Importance of Behavior and Venom System Morphology in Understanding Its Ecology and Evolution. Toxins 2019, 11, 666. [Google Scholar] [CrossRef] [Green Version]
- Robertson, P.L. A Morphological and Functional Study of the Venom Apparatus in Representatives of Some Major Groups of Hymenoptera. Aust. J. Zool. 1968, 16, 133–166. [Google Scholar] [CrossRef]
- Fox, E.G.P.; Adams, R.M.M. On the Biological Diversity of Ant Alkaloids. Annu. Rev. Entomol. 2022, 67, 367–385. [Google Scholar] [CrossRef]
- Touchard, A.; Aili, S.; Fox, E.; Escoubas, P.; Orivel, J.; Nicholson, G.; Dejean, A. The Biochemical Toxin Arsenal from Ant Venoms. Toxins 2016, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Aili, S.R.; Touchard, A.; Escoubas, P.; Padula, M.P.; Orivel, J.; Dejean, A.; Nicholson, G.M. Diversity of Peptide Toxins from Stinging Ant Venoms. Toxicon 2014, 92, 166–178. [Google Scholar] [CrossRef]
- Barassé, V.; Touchard, A.; Téné, N.; Tindo, M.; Kenne, M.; Klopp, C.; Dejean, A.; Bonnafé, E.; Treilhou, M. The Peptide Venom Composition of the Fierce Stinging Ant Tetraponera aethiops (Formicidae: Pseudomyrmecinae). Toxins 2019, 11, 732. [Google Scholar] [CrossRef] [Green Version]
- Touchard, A.; Téné, N.; Song, P.C.T.; Lefranc, B.; Leprince, J.; Treilhou, M.; Bonnafé, E. Deciphering the Molecular Diversity of an Ant Venom Peptidome through a Venomics Approach. J. Proteome Res. 2018, 17, 3503–3516. [Google Scholar] [CrossRef]
- Touchard, A.; Aili, S.R.; Téné, N.; Barassé, V.; Klopp, C.; Dejean, A.; Kini, R.M.; Mrinalini, M.; Coquet, L.; Jouenne, T.; et al. Venom Peptide Repertoire of the European Myrmicine Ant Manica rubida: Identification of Insecticidal Toxins. J. Proteome Res. 2020, 19, 1800–1811. [Google Scholar] [CrossRef] [PubMed]
- Aili, S.R.; Touchard, A.; Hayward, R.; Robinson, S.D.; Pineda, S.S.; Lalagüe, H.; Vetter, I.; Undheim, E.A.B.; Kini, R.M.; Escoubas, P.; et al. An Integrated Proteomic and Transcriptomic Analysis Reveals the Venom Complexity of the Bullet Ant Paraponera clavata. Toxins 2020, 12, 324. [Google Scholar] [CrossRef] [PubMed]
- Touchard, A.; Labrière, N.; Roux, O.; Petitclerc, F.; Orivel, J.; Escoubas, P.; Koh, J.M.S.; Nicholson, G.M.; Dejean, A. Venom Toxicity and Composition in Three Pseudomyrmex Ant Species Having Different Nesting Modes. Toxicon 2014, 88, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Aili, S.R.; Touchard, A.; Petitclerc, F.; Dejean, A.; Orivel, J.; Padula, M.P.; Escoubas, P.; Nicholson, G.M. Combined Peptidomic and Proteomic Analysis of Electrically Stimulated and Manually Dissected Venom from the South American Bullet Ant Paraponera clavata. J. Proteome Res. 2017, 16, 1339–1351. [Google Scholar] [CrossRef]
- Touchard, A.; Koh, J.M.S.; Aili, S.R.; Dejean, A.; Nicholson, G.M.; Orivel, J.; Escoubas, P. The Complexity and Structural Diversity of Ant Venom Peptidomes Is Revealed by Mass Spectrometry Profiling. Rapid. Commun. Mass Spectrom. 2015, 29, 385–396. [Google Scholar] [CrossRef]
- Herzig, V.; King, G.F.; Undheim, E.A.B. Can We Resolve the Taxonomic Bias in Spider Venom Research? Toxicon X 2019, 1, 100005. [Google Scholar] [CrossRef]
- Lüddecke, T.; Vilcinskas, A.; Lemke, S. Phylogeny-Guided Selection of Priority Groups for Venom Bioprospecting: Harvesting Toxin Sequences in Tarantulas as a Case Study. Toxins 2019, 11, 488. [Google Scholar] [CrossRef] [Green Version]
- Drukewitz, S.H.; von Reumont, B.M. The Significance of Comparative Genomics in Modern Evolutionary Venomics. Front. Ecol. Evol. 2019, 7, 163. [Google Scholar] [CrossRef] [Green Version]
- Robinson, S.D.; Mueller, A.; Clayton, D.; Starobova, H.; Hamilton, B.R.; Payne, R.J.; Vetter, I.; King, G.F.; Undheim, E.A.B. A Comprehensive Portrait of the Venom of the Giant Red Bull Ant, Myrmecia gulosa, Reveals a Hyperdiverse Hymenopteran Toxin Gene Family. Sci. Adv. 2018, 4, eaau4640. [Google Scholar] [CrossRef] [Green Version]
- Ceolin Mariano, D.O.; de Oliveira, Ú.C.; Zaharenko, A.J.; Pimenta, D.C.; Rádis-Baptista, G.; Prieto-da-Silva, Á.R.d.B. Bottom-Up Proteomic Analysis of Polypeptide Venom Components of the Giant Ant Dinoponera quadriceps. Toxins 2019, 11, 448. [Google Scholar] [CrossRef] [Green Version]
- Santos, P.P.; Games, P.D.; Azevedo, D.O.; Barros, E.; de Oliveira, L.L.; de Oliveira Ramos, H.J.; Baracat-Pereira, M.C.; Serrão, J.E. Proteomic Analysis of the Venom of the Predatory Ant Pachycondyla striata (Hymenoptera: Formicidae). Arch. Insect Biochem. Physiol. 2017, 96, e21424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heep, J.; Klaus, A.; Kessel, T.; Seip, M.; Vilcinskas, A.; Skaljac, M. Proteomic Analysis of the Venom from the Ruby Ant Myrmica rubra and the Isolation of a Novel Insecticidal Decapeptide. Insects 2019, 10, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanandy, T.; Wilson, R.; Gell, D.; Rose, H.E.; Gueven, N.; Davies, N.W.; Brown, S.G.A.; Wiese, M.D. Towards Complete Identification of Allergens in Jack Jumper (Myrmecia pilosula) Ant Venom and Their Clinical Relevance: An Immunoproteomic Approach. Clin. Exp. Allergy 2018, 48, 1222–1234. [Google Scholar] [CrossRef] [PubMed]
- Pessoa, W.F.B.; Silva, L.C.C.; De Oliveira Dias, L.; Delabie, J.H.C.; Costa, H.; Romano, C.C. Analysis of Protein Composition and Bioactivity of Neoponera villosa Venom (Hymenoptera: Formicidae). Int. J. Mol. Sci. 2016, 17, 513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holding, M.L.; Margres, M.J.; Mason, A.J.; Parkinson, C.L.; Rokyta, D.R. Evaluating the Performance of De Novo Assembly Methods for Venom-Gland Transcriptomics. Toxins 2018, 10, 249. [Google Scholar] [CrossRef] [Green Version]
- von Reumont, B.M. Studying Smaller and Neglected Organisms in Modern Evolutionary Venomics Implementing RNASeq (Transcriptomics)—A Critical Guide. Toxins 2018, 10, 292. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.J.; Undheim, E.A.B. True Lies: Using Proteomics to Assess the Accuracy of Transcriptome-Based Venomics in Centipedes Uncovers False Positives and Reveals Startling Intraspecific Variation in Scolopendra subspinipes. Toxins 2018, 10, 96. [Google Scholar] [CrossRef] [Green Version]
- Eagles, D.A.; Saez, N.J.; Krishnarjuna, B.; Bradford, J.J.; Chin, Y.K.-Y.; Starobova, H.; Mueller, A.; Reichelt, M.E.; Undheim, E.A.B.; Norton, R.S.; et al. A Peptide Toxin in Ant Venom Mimics Vertebrate EGF-like Hormones to Cause Long-Lasting Hypersensitivity in Mammals. Proc. Natl. Acad. Sci. USA 2022, 119, e2112630119. [Google Scholar] [CrossRef]
- Di Cera, E. Serine Proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, B.Y.; Sohn, M.R.; Roh, J.Y.; Je, Y.H.; Kim, N.J.; Kim, I.; Woo, S.D.; et al. Dual Function of a Bee Venom Serine Protease: Prophenoloxidase-Activating Factor in Arthropods and Fibrin(Ogen)Olytic Enzyme in Mammals. PLoS ONE 2010, 5, e10393. [Google Scholar] [CrossRef]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Qiu, Y.; Wan, H.; Sohn, M.R.; Sohn, H.D.; Jin, B.R. Antifibrinolytic Role of a Bee Venom Serine Protease Inhibitor That Acts as a Plasmin Inhibitor. PLoS ONE 2012, 7, e32269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkinson, N.M.; Conyers, C.; Keen, J.; MacNicoll, A.; Smith, I.; Audsley, N.; Weaver, R. Towards a Comprehensive View of the Primary Structure of Venom Proteins from the Parasitoid Wasp Pimpla hypochondriaca. Insect Biochem. Mol. Biol. 2004, 34, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Lee, K.S.; Choo, Y.M.; Kong, D.; Yoon, H.J.; Jin, B.R. Molecular Cloning and Antifibrinolytic Activity of a Serine Protease Inhibitor from Bumblebee (Bombus terrestris) Venom. Toxicon 2013, 63, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP Superfamily: Cysteine-Rich Secretory Proteins, Antigen 5, and Pathogenesis-Related 1 Proteins—Roles in Reproduction, Cancer, and Immune Defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef]
- Blank, S.; Bazon, M.L.; Grosch, J.; Schmidt-Weber, C.B.; Brochetto-Braga, M.R.; Bilò, M.B.; Jakob, T. Antigen 5 Allergens of Hymenoptera Venoms and Their Role in Diagnosis and Therapy of Venom Allergy. Curr. Allergy Asthma Rep. 2020, 20, 58:1–58:13. [Google Scholar] [CrossRef]
- Hoffman, D.R. Reactions to Less Common Species of Fire Ants. J. Allergy Clin. Immunol. 1997, 100, 679–683. [Google Scholar] [CrossRef]
- Hoffman, D.R.; Smith, A.M.; Schmidt, M.; Moffitt, J.E.; Guralnick, M. Allergens in Hymenoptera Venom. XXII. Comparison of Venoms from Two Species of Imported Fire Ants, Solenopsis invicta and richteri. J. Allergy Clin. Immunol. 1990, 85, 988–996. [Google Scholar] [CrossRef]
- Hoffman, D.R. Hymenoptera Venom Allergens. Clin. Rev. Allergy Immunol. 2006, 30, 109–128. [Google Scholar] [CrossRef]
- Jentsch, J. A Procedure for Purification of Myrmica Venom: The Isolation of the Compulsive Component. In Proceedings of the VI Congress of the IUSSI, Bern, Switzerland, September 15 1969; pp. 69–75. [Google Scholar]
- Piek, T. Venoms of the Hymenoptera: Biochemical, Pharmacological and Behavioural Aspects; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 978-1-4832-6370-0. [Google Scholar]
- Mebs, D. Gifttiere: Ein Handbuch für Biologen, Toxikologen, Ärzte, Apotheker; Wissenschaftliche Verlagsgesellschaft: Stuttgart, Germany, 1992; ISBN 978-3-8047-1219-5. [Google Scholar]
- Arevalo, H.A.; Groden, E. European Fire Ant, Red Ant (Suggested Common Names), Myrmica rubra Linnaeus (Insecta: Hymenoptera: Formicidae: Myrmicinae): EENY-410/IN746, 8/2007. EDIS 2007, 2007, 1–5. [Google Scholar] [CrossRef]
- Shiomi, K.; Honma, T.; Ide, M.; Nagashima, Y.; Ishida, M.; Chino, M. An Epidermal Growth Factor-like Toxin and Two Sodium Channel Toxins from the Sea Anemone Stichodactyla gigantea. Toxicon 2003, 41, 229–236. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Fuentes-Silva, D.; King, G.F. Development of a Rational Nomenclature for Naming Peptide and Protein Toxins from Sea Anemones. Toxicon 2012, 60, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; von Reumont, B.M.; Förster, F.; Billion, A.; Timm, T.; Lochnit, G.; Vilcinskas, A.; Lemke, S. An Economic Dilemma between Molecular Weapon Systems May Explain an Arachno-Atypical Venom in Wasp Spiders (Argiope bruennichi). Biomolecules 2020, 10, 978. [Google Scholar] [CrossRef] [PubMed]
- Lüddecke, T.; Herzig, V.; von Reumont, B.M.; Vilcinskas, A. The Biology and Evolution of Spider Venoms. Biol. Rev. 2022, 97, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Undheim, E.A.B.; Grimm, L.L.; Low, C.-F.; Morgenstern, D.; Herzig, V.; Zobel-Thropp, P.; Pineda, S.S.; Habib, R.; Dziemborowicz, S.; Fry, B.G.; et al. Weaponization of a Hormone: Convergent Recruitment of Hyperglycemic Hormone into the Venom of Arthropod Predators. Structure 2015, 23, 1283–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvidson, R.; Kaiser, M.; Lee, S.S.; Urenda, J.-P.; Dail, C.; Mohammed, H.; Nolan, C.; Pan, S.; Stajich, J.E.; Libersat, F.; et al. Parasitoid Jewel Wasp Mounts Multipronged Neurochemical Attack to Hijack a Host Brain. Mol. Cell Proteom. 2019, 18, 99–114. [Google Scholar] [CrossRef] [Green Version]
- Robinson, S.D.; Li, Q.; Bandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.T.; Purcell, A.W.; Norton, R.S.; Safavi-Hemami, H. Hormone-like Peptides in the Venoms of Marine Cone Snails. Gen. Comp. Endocrinol. 2017, 244, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Barnett, A. Exenatide. Expert Opin. Pharmacother. 2007, 8, 2593–2608. [Google Scholar] [CrossRef]
- Osman, M.F.H.; Brander, J. Weitere Beiträge zur Kenntnis der chemischen Zusammensetzung des Giftes von Ameisen aus der Gattung Formica. Z. Nat. B 1961, 16, 749–753. [Google Scholar] [CrossRef]
- Kazandjian, T.D.; Petras, D.; Robinson, S.D.; van Thiel, J.; Greene, H.W.; Arbuckle, K.; Barlow, A.; Carter, D.A.; Wouters, R.M.; Whiteley, G.; et al. Convergent Evolution of Pain-Inducing Defensive Venom Components in Spitting Cobras. Science 2021, 371, 386–390. [Google Scholar] [CrossRef]
- Mouchbahani-Constance, S.; Lesperance, L.S.; Petitjean, H.; Davidova, A.; Macpherson, A.; Prescott, S.A.; Sharif-Naeini, R. Lionfish Venom Elicits Pain Predominantly through the Activation of Nonpeptidergic Nociceptors. PAIN 2018, 159, 2255–2266. [Google Scholar] [CrossRef]
- Walker, A.A.; Robinson, S.D.; Paluzzi, J.-P.V.; Merritt, D.J.; Nixon, S.A.; Schroeder, C.I.; Jin, J.; Goudarzi, M.H.; Kotze, A.C.; Dekan, Z.; et al. Production, Composition, and Mode of Action of the Painful Defensive Venom Produced by a Limacodid Caterpillar, Doratifera vulnerans. Proc. Natl. Acad. Sci. USA 2021, 118, e2023815118. [Google Scholar] [CrossRef] [PubMed]
- Nixon, S.A.; Robinson, S.D.; Agwa, A.J.; Walker, A.A.; Choudhary, S.; Touchard, A.; Undheim, E.A.B.; Robertson, A.; Vetter, I.; Schroeder, C.I.; et al. Multipurpose Peptides: The Venoms of Amazonian Stinging Ants Contain Anthelmintic Ponericins with Diverse Predatory and Defensive Activities. Biochem. Pharmacol. 2021, 192, 114693. [Google Scholar] [CrossRef]
- Fischer, M.L.; Wielsch, N.; Heckel, D.G.; Vilcinskas, A.; Vogel, H. Context-Dependent Venom Deployment and Protein Composition in Two Assassin Bugs. Ecol. Evol. 2020, 10, 9932–9947. [Google Scholar] [CrossRef]
- Walker, A.A.; Mayhew, M.L.; Jin, J.; Herzig, V.; Undheim, E.A.B.; Sombke, A.; Fry, B.G.; Meritt, D.J.; King, G.F. The Assassin Bug Pristhesancus plagipennis Produces Two Distinct Venoms in Separate Gland Lumens. Nat. Commun. 2018, 9, 755. [Google Scholar] [CrossRef] [Green Version]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of Separate Predation- and Defence-Evoked Venoms in Carnivorous Cone Snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, B. Die Ameisen Mittel- und Nordeuropas; lutra Verlags- und Vertriebsgesellschaft: Görlitz/Tauer, Germany, 2007; ISBN 978-3-936412-03-1. [Google Scholar]
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 February 2022).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Haas, B.J. HPC GridRunner. Available online: https://github.com/HpcGridRunner/HpcGridRunner (accessed on 8 February 2021).
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo Transcriptome Assembler and Its Application to RNA-Seq Data. GigaScience 2019, 8, giz100. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Florea, L. Rcorrector: Efficient and Accurate Error Correction for Illumina RNA-Seq Reads. GigaScience 2015, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. In Gene Prediction: Methods and Protocols; Kollmar, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 227–245. ISBN 978-1-4939-9173-0. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome Assembly from Long-Read RNA-Seq Alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- The UniProt Consortium UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef] [PubMed] [Green Version]
- de Castro, E.; Jungo, F. VenomZone. Available online: https://venomzone.expasy.org/ (accessed on 11 January 2021).
- Jungo, F.; Bairoch, A. Tox-Prot, the Toxin Protein Annotation Program of the Swiss-Prot Protein Knowledgebase. Toxicon 2005, 45, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Henikoff, J.G. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R. The InterPro Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2008, 21, 2.7.1–2.7.18. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584:1–e2584:22. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- von Reumont, B.M.; Lüddecke, T.; Timm, T.; Lochnit, G.; Vilcinskas, A.; von Döhren, J.; Nilsson, M.A. Proteo-Transcriptomic Analysis Identifies Potential Novel Toxins Secreted by the Predatory, Prey-Piercing Ribbon Worm Amphiporus lactifloreus. Mar. Drugs 2020, 18, 407. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- King, G.F.; Gentz, M.C.; Escoubas, P.; Nicholson, G.M. A Rational Nomenclature for Naming Peptide Toxins from Spiders and Other Venomous Animals. Toxicon 2008, 52, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
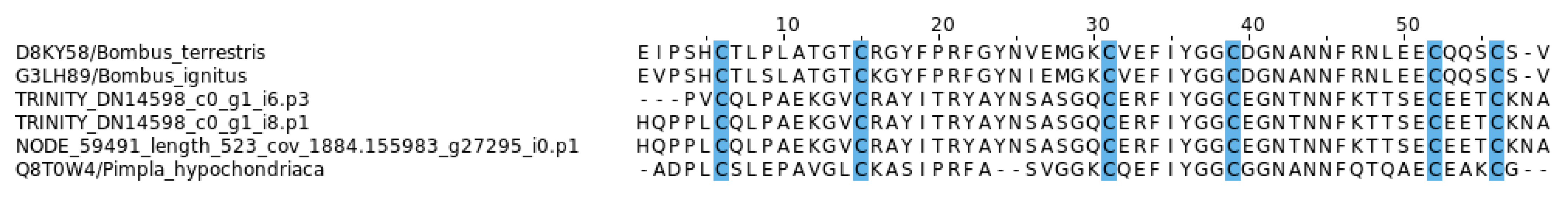

| Species | Toxin | # AA Precursor | # AA Mature Toxin | MW (kDa) | pI (pH) |
|---|---|---|---|---|---|
| M. rubra | U-MYRTX-Mrub1a | 79 | 49 | 5.4 | 5.29 |
| U-MYRTX-Mrub1b | 86 | 56 | 6.0 | 4.80 | |
| U-MYRTX-Mrub1c | 76 | 46 | 5.2 | 4.59 | |
| U-MYRTX-Mrub1d | 81 | 51 | 5.7 | 5.78 | |
| M. ruginodis | U-MYRTX-Mrug1a * | 79 | 49 | 5.4 | 5.41 |
| U-MYRTX-Mrug1b * | 79 | 49 | 5.4 | 5.41 | |
| U-MYRTX-Mrug1c | 86 | 56 | 6.0 | 4.80 | |
| U-MYRTX-Mrug1d | 81 | 51 | 5.6 | 4.72 | |
| U-MYRTX-Mrug1e | 79 | 49 | 5.3 | 4.76 |
| Toxin | BLAST Result ID | Similarity (%) | Clade | EGF-Type |
|---|---|---|---|---|
| ECTX(02)-Rm1a | RXN00400.1 | 48.0 | ECTX-clade | Vertebrate betacellulin-like |
| ECTX(02)-Rm1b | RXN00400.1 | 46.0 | ECTX-clade | Vertebrate betacellulin-like |
| ECTX(02)-Rm1c | XP_032961198.1 | 50.0 | ECTX-clade | Vertebrate betacellulin-like |
| ECTX(02)-Rm1d | XP_042333623.1 | 46.3 | ECTX-clade | Vertebrate epiregulin-like |
| ECTX(02)-Rm1e | XP_036083747.1 | 41.4 | ECTX-clade | Vertebrate epiregulin-like |
| MIITX(02)-Mc1a | XP_041957410.1 | 62.2 | MIICTX-clade | HBEGF-like |
| U-MIITX(02)-Mg1a | XP_038648772.1 | 65.8 | MIICTX-clade | HBEGF-like |
| U-MYRTX-Mrub1a * | XP_041634697.1 | 57.1 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrub1d * | XP_041634697.1 | 57.1 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrug1a * | XP_041634697.1 | 55.3 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrug1b * | XP_041634697.1 | 51.1 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrug1d * | XP_041634697.1 | 57.1 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrug1e * | XP_041634697.1 | 57.1 | MYRTX-clade A | Vertebrate TGF-like |
| U-MYRTX-Mrub1c * | XP_019908008.1 | 58.8 | MYRTX-clade B | Vertebrate epiregulin-like |
| U-MYRTX-Mrub1b * | XP_016916180.1 | 63.0 | MYRTX-clade C | Insect Spitz-like |
| U-MYRTX-Mrug1c * | XP_016916180.1 | 63.0 | MYRTX-clade C | Insect Spitz-like |
| U18-MYRTX-Mri1a | XP_016916180.1 | 63.0 | MYRTX-clade C | Insect Spitz-like |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hurka, S.; Brinkrolf, K.; Özbek, R.; Förster, F.; Billion, A.; Heep, J.; Timm, T.; Lochnit, G.; Vilcinskas, A.; Lüddecke, T. Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis. Toxins 2022, 14, 358. https://doi.org/10.3390/toxins14050358
Hurka S, Brinkrolf K, Özbek R, Förster F, Billion A, Heep J, Timm T, Lochnit G, Vilcinskas A, Lüddecke T. Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis. Toxins. 2022; 14(5):358. https://doi.org/10.3390/toxins14050358
Chicago/Turabian StyleHurka, Sabine, Karina Brinkrolf, Rabia Özbek, Frank Förster, André Billion, John Heep, Thomas Timm, Günter Lochnit, Andreas Vilcinskas, and Tim Lüddecke. 2022. "Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis" Toxins 14, no. 5: 358. https://doi.org/10.3390/toxins14050358
APA StyleHurka, S., Brinkrolf, K., Özbek, R., Förster, F., Billion, A., Heep, J., Timm, T., Lochnit, G., Vilcinskas, A., & Lüddecke, T. (2022). Venomics of the Central European Myrmicine Ants Myrmica rubra and Myrmica ruginodis. Toxins, 14(5), 358. https://doi.org/10.3390/toxins14050358







