ExoU Induces Lung Endothelial Cell Damage and Activates Pro-Inflammatory Caspase-1 during Pseudomonas aeruginosa Infection
Abstract
1. Introduction
2. Results
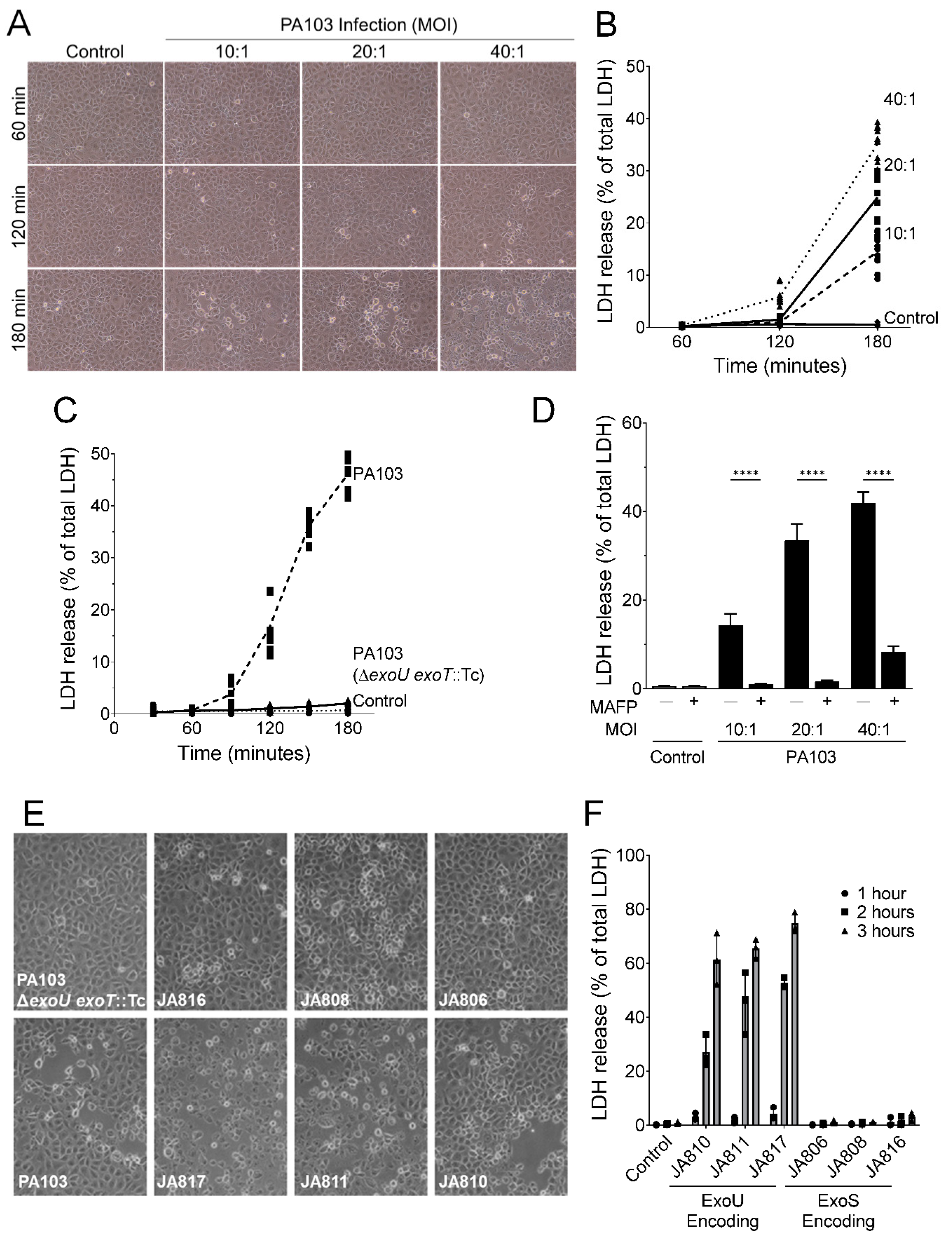
2.1. P. aeruginosa Strains Expressing ExoU Cause PMVEC Lysis and Barrier Disruption in a Time-, Dose-, and PLA2-Dependent Manner
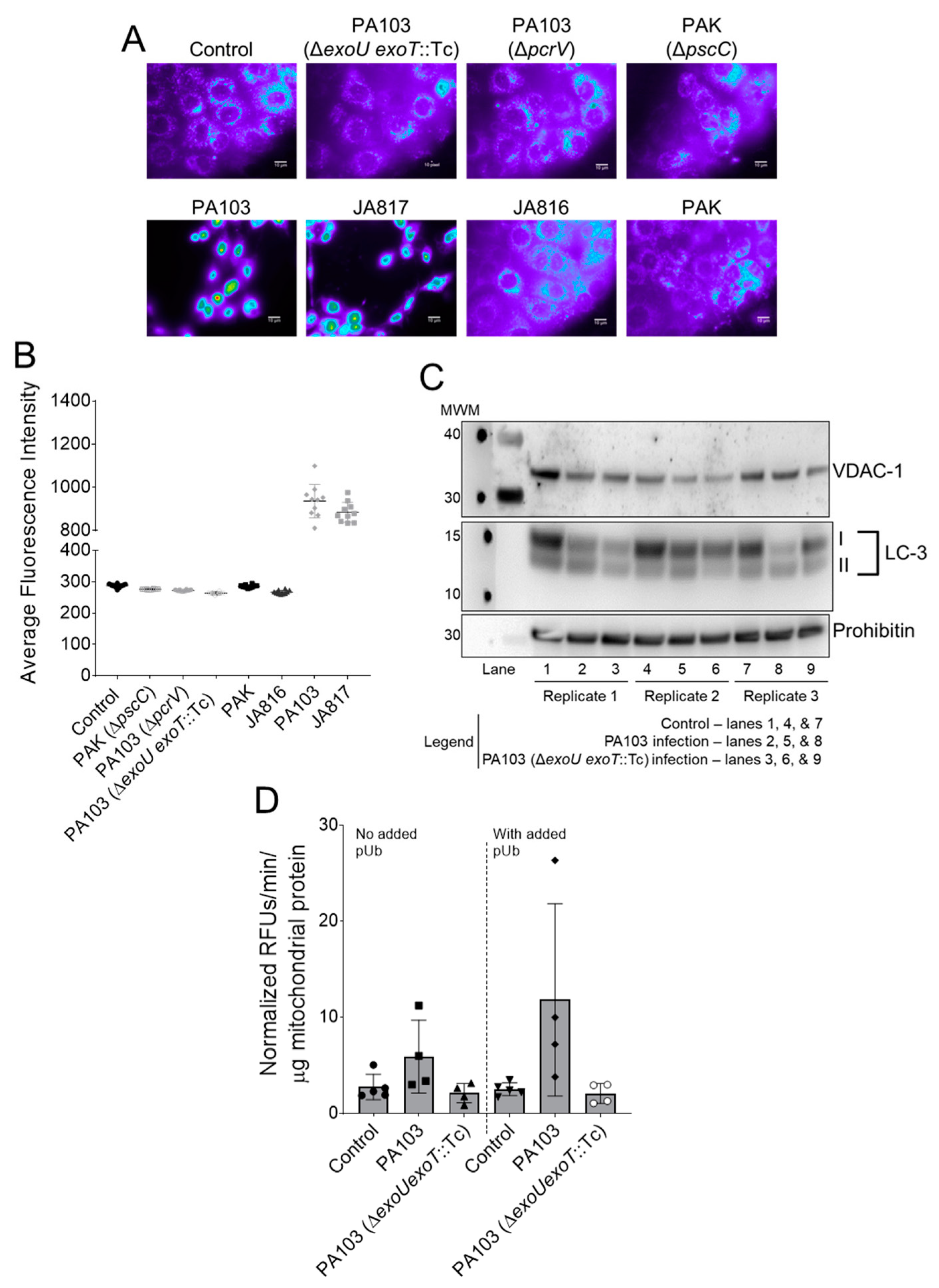
2.2. ExoU Triggers Caspase-1 Activation in PMVECs during P. aeruginosa Infection
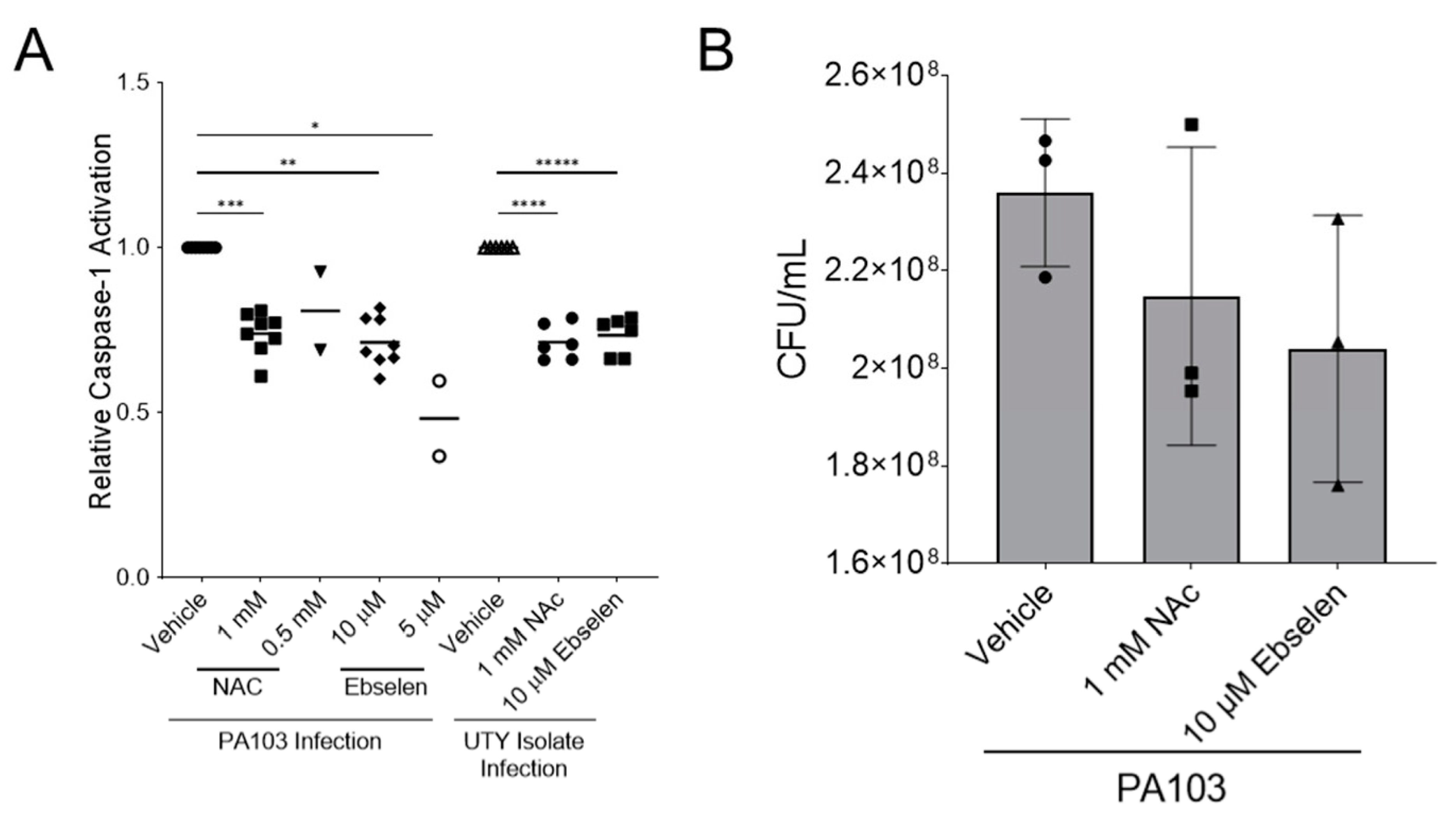
2.3. ExoU-Induced Reactive Oxidative Species Signaling Contributes to Caspase-1 Activation in PMVECs during P. aeruginosa Infection
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagents
5.2. Bacterial Strains and Culture Conditions
5.3. Eukaryotic Cell Culture and Infection Conditions
5.4. Cell Damage and Lysis Measurements
5.5. FLICA and Flow Cytometry
5.6. Reactive Oxygen Species Measurements
5.7. Isolation and Assay of Mitochondria and Mitochondria-Associated Membrane Fractions
5.8. Statistical Analyses of Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feltman, H.; Schulert, G.; Khan, S.; Jain, M.; Peterson, L.; Hauser, A.R. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 2001, 147 Pt 10, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R.; Cobb, E.; Bodi, M.; Mariscal, D.; Valles, J.; Engel, J.N.; Rello, J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 2002, 30, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, K.; Kajikawa, O.; Sawa, T.; Ohara, M.; Gropper, M.A.; Frank, D.W.; Martin, T.R.; Wiener-Kronish, J.P. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 1999, 104, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Roy-Burman, A.; Savel, R.H.; Racine, S.; Swanson, B.L.; Revadigar, N.S.; Fujimoto, J.; Sawa, T.; Frank, D.W.; Wiener-Kronish, J.P. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 2001, 183, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua= (accessed on 27 February 2017).
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Frank, D.W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 1997, 26, 621–629. [Google Scholar] [CrossRef]
- Barbieri, J.T. Pseudomonas aeruginosa exoenzyme S, a bifunctional type-III secreted cytotoxin. Int. J. Med. Microbiol. 2000, 290, 381–387. [Google Scholar] [CrossRef]
- Barbieri, J.T.; Sun, J. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 2004, 152, 79–92. [Google Scholar] [PubMed]
- Finck-Barbancon, V.; Goranson, J.; Zhu, L.; Sawa, T.; Wiener-Kronish, J.P.; Fleiszig, S.M.; Wu, C.; Mende-Mueller, L.; Frank, D.W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997, 25, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Yahr, T.L.; Vallis, A.J.; Hancock, M.K.; Barbieri, J.T.; Frank, D.W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 1998, 95, 13899–13904. [Google Scholar] [CrossRef]
- Anderson, D.M.; Schmalzer, K.M.; Sato, H.; Casey, M.; Terhune, S.S.; Haas, A.L.; Feix, J.B.; Frank, D.W. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 2011, 82, 1454–1467. [Google Scholar] [CrossRef]
- Belyy, A.; Raoux-Barbot, D.; Saveanu, C.; Namane, A.; Ogryzko, V.; Worpenberg, L.; David, V.; Henriot, V.; Fellous, S.; Merrifield, C.; et al. Actin activates Pseudomonas aeruginosa ExoY nucleotidyl cyclase toxin and ExoY-like effector domains from MARTX toxins. Nat. Commun. 2016, 7, 13582. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Coburn, J.; Collier, R.J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 1993, 90, 2320–2324. [Google Scholar] [CrossRef]
- Housley, N.A.; Winkler, H.H.; Audia, J.P. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. J. Bacteriol. 2011, 193, 4634–4642. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Feix, J.B.; Frank, D.W. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry 2006, 45, 10368–10375. [Google Scholar] [CrossRef]
- Allewelt, M.; Coleman, F.T.; Grout, M.; Priebe, G.P.; Pier, G.B. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 2000, 68, 3998–4004. [Google Scholar] [CrossRef] [PubMed]
- El-Solh, A.A.; Hattemer, A.; Hauser, A.R.; Alhajhusain, A.; Vora, H. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit. Care Med. 2012, 40, 1157–1163. [Google Scholar] [CrossRef]
- Schulert, G.S.; Feltman, H.; Rabin, S.D.; Martin, C.G.; Battle, S.E.; Rello, J.; Hauser, A.R. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 2003, 188, 1695–1706. [Google Scholar] [CrossRef]
- Hardy, K.S.; Tessmer, M.H.; Frank, D.W.; Audia, J.P. Perspectives on the Pseudomonas aeruginosa Type III Secretion System Effector ExoU and Its Subversion of the Host Innate Immune Response to Infection. Toxins 2021, 13, 880. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.H.; Hauser, A.R. Pseudomonas aeruginosa cytotoxin ExoU is injected into phagocytic cells during acute pneumonia. Infect. Immun. 2010, 78, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Finck-Barbancon, V.; Frank, D.W. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 2001, 183, 4330–4344. [Google Scholar] [CrossRef][Green Version]
- Howell, H.A.; Logan, L.K.; Hauser, A.R. Type III secretion of ExoU is critical during early Pseudomonas aeruginosa pneumonia. mBio 2013, 4, e00032-13. [Google Scholar] [CrossRef]
- Sato, H.; Frank, D.W. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 2004, 53, 1279–1290. [Google Scholar] [CrossRef]
- Sato, H.; Frank, D.W.; Hillard, C.J.; Feix, J.B.; Pankhaniya, R.R.; Moriyama, K.; Finck-Barbancon, V.; Buchaklian, A.; Lei, M.; Long, R.M.; et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003, 22, 2959–2969. [Google Scholar] [CrossRef]
- Anderson, D.M.; Feix, J.B.; Monroe, A.L.; Peterson, F.C.; Volkman, B.F.; Haas, A.L.; Frank, D.W. Identification of the major ubiquitin-binding domain of the Pseudomonas aeruginosa ExoU A2 phospholipase. J. Biol. Chem. 2013, 288, 26741–26752. [Google Scholar] [CrossRef]
- Pankhaniya, R.R.; Tamura, M.; Allmond, L.R.; Moriyama, K.; Ajayi, T.; Wiener-Kronish, J.P.; Sawa, T. Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit. Care Med. 2004, 32, 2293–2299. [Google Scholar] [CrossRef]
- Shaver, C.M.; Hauser, A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 2004, 72, 6969–6977. [Google Scholar] [CrossRef]
- Diaz, M.H.; Shaver, C.M.; King, J.D.; Musunuri, S.; Kazzaz, J.A.; Hauser, A.R. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect. Immun. 2008, 76, 4414–4421. [Google Scholar] [CrossRef]
- Lins, R.X.; de Assis, M.C.; Mallet de Lima, C.D.; Freitas, C.; Maciel Plotkowski, M.C.; Saliba, A.M. ExoU modulates soluble and membrane-bound ICAM-1 in Pseudomonas aeruginosa-infected endothelial cells. Microbes Infect. 2010, 12, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.A.; Lanter, B.B.; Yonker, L.M.; Eaton, A.D.; Pirzai, W.; Gronert, K.; Bonventre, J.V.; Hurley, B.P. Pseudomonas aeruginosa ExoU augments neutrophil transepithelial migration. PLoS Pathog. 2017, 13, e1006548. [Google Scholar] [CrossRef] [PubMed]
- Plotkowski, M.C.; Brandao, B.A.; de Assis, M.C.; Feliciano, L.F.; Raymond, B.; Freitas, C.; Saliba, A.M.; Zahm, J.M.; Touqui, L.; Bozza, P.T. Lipid body mobilization in the ExoU-induced release of inflammatory mediators by airway epithelial cells. Microb. Pathog. 2008, 45, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Sutterwala, F.S.; Mijares, L.A.; Li, L.; Ogura, Y.; Kazmierczak, B.I.; Flavell, R.A. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 2007, 204, 3235–3245. [Google Scholar] [CrossRef]
- da Cunha, L.G., Jr.; Ferreira, M.F.; de Moraes, J.A.; Reis, P.A.; Castro-Faria-Neto, H.C.; Barja-Fidalgo, C.; Plotkowski, M.C.; Saliba, A.M. ExoU-induced redox imbalance and oxidative stress in airway epithelial cells during Pseudomonas aeruginosa pneumosepsis. Med. Microbiol. Immunol. 2015, 204, 673–680. [Google Scholar] [CrossRef]
- de Lima, C.D.; Calegari-Silva, T.C.; Pereira, R.M.; Santos, S.A.; Lopes, U.G.; Plotkowski, M.C.; Saliba, A.M. ExoU activates NF-kappaB and increases IL-8/KC secretion during Pseudomonas aeruginosa infection. PLoS ONE 2012, 7, e41772. [Google Scholar] [CrossRef]
- Freitas, C.; Assis, M.C.; Saliba, A.M.; Morandi, V.M.; Figueiredo, C.C.; Pereira, M.; Plotkowski, M.C. The infection of microvascular endothelial cells with ExoU-producing Pseudomonas aeruginosa triggers the release of von Willebrand factor and platelet adhesion. Mem. Inst. Oswaldo Cruz 2012, 107, 728–734. [Google Scholar] [CrossRef]
- Saliba, A.M.; de Assis, M.C.; Nishi, R.; Raymond, B.; Marques Ede, A.; Lopes, U.G.; Touqui, L.; Plotkowski, M.C. Implications of oxidative stress in the cytotoxicity of Pseudomonas aeruginosa ExoU. Microbes Infect. 2006, 8, 450–459. [Google Scholar] [CrossRef]
- Saliba, A.M.; Nascimento, D.O.; Silva, M.C.; Assis, M.C.; Gayer, C.R.; Raymond, B.; Coelho, M.G.; Marques, E.A.; Touqui, L.; Albano, R.M.; et al. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell. Microbiol. 2005, 7, 1811–1822. [Google Scholar] [CrossRef]
- Wagener, B.M.; Anjum, N.; Christiaans, S.C.; Banks, M.E.; Parker, J.C.; Threet, A.T.; Walker, R.R.; Isbell, K.D.; Moser, S.A.; Stevens, T.; et al. Exoenzyme Y Contributes to End-Organ Dysfunction Caused by Pseudomonas aeruginosa Pneumonia in Critically Ill Patients: An Exploratory Study. Toxins 2020, 12, 369. [Google Scholar] [CrossRef]
- Bouillot, S.; Attree, I.; Huber, P. Pharmacological activation of Rap1 antagonizes the endothelial barrier disruption induced by exotoxins ExoS and ExoT of Pseudomonas aeruginosa. Infect. Immun. 2015, 83, 1820–1829. [Google Scholar] [CrossRef] [PubMed]
- Renema, P.; Kozhukhar, N.; Pastukh, V.; Spadafora, D.; Paudel, S.S.; Tambe, D.T.; Alexeyev, M.; Frank, D.W.; Stevens, T. Exoenzyme Y induces extracellular active caspase-7 accumulation independent from apoptosis: Modulation of transmissible cytotoxicity. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L380–L390. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.F.; Housley, N.; Koloteva, A.; Zhou, C.; O’Donnell, K.; Audia, J.P. Caspase-1 activation protects lung endothelial barrier function during infection-induced stress. Am. J. Respir. Cell Mol. Biol. 2016, 55, 500–510. [Google Scholar] [CrossRef]
- Renema, P.; Hardy, K.S.; Housley, N.; Dunbar, G.; Annamdevula, N.; Britain, A.; Spadafora, D.; Leavesley, S.; Rich, T.; Audia, J.P.; et al. cAMP signaling primes lung endothelial cells to activate caspase-1 during Pseudomonas aeruginosa infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L1074–L1083. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Strowig, T.; Henao-Mejia, J.; Elinav, E.; Flavell, R. Inflammasomes in health and disease. Nature 2012, 481, 278–286. [Google Scholar] [CrossRef]
- Thornberry, N.A.; Bull, H.G.; Calaycay, J.R.; Chapman, K.T.; Howard, A.D.; Kostura, M.J.; Miller, D.K.; Molineaux, S.M.; Weidner, J.R.; Aunins, J.; et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992, 356, 768–774. [Google Scholar] [CrossRef]
- Lee, V.T.; Pukatzki, S.; Sato, H.; Kikawada, E.; Kazimirova, A.A.; Huang, J.; Li, X.; Arm, J.P.; Frank, D.W.; Lory, S. Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 2007, 75, 1089–1098. [Google Scholar] [CrossRef]
- Morrow, K.A.; Ochoa, C.D.; Balczon, R.; Zhou, C.; Cauthen, L.; Alexeyev, M.; Schmalzer, K.M.; Frank, D.W.; Stevens, T. Pseudomonas aeruginosa exoenzymes U and Y induce a transmissible endothelial proteinopathy. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 310, L337–L353. [Google Scholar] [CrossRef] [PubMed]
- Schmalzer, K.M.; Benson, M.A.; Frank, D.W. Activation of ExoU phospholipase activity requires specific C-terminal regions. J. Bacteriol. 2010, 192, 1801–1812. [Google Scholar] [CrossRef]
- Evavold, C.L.; Kagan, J.C. Inflammasomes: Threat-assessment organelles of the innate immune system. Immunity 2019, 51, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Jabir, M.S.; Hopkins, L.; Ritchie, N.D.; Ullah, I.; Bayes, H.K.; Li, D.; Tourlomousis, P.; Lupton, A.; Puleston, D.; Simon, A.K.; et al. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy 2015, 11, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.I.; Miller, A.N.; Banoth, B.; Iyer, S.S.; Stotland, A.; Weiss, J.P.; Gottlieb, R.A.; Sutterwala, F.S.; Cassel, S.L. Cutting Edge: Mitochondrial assembly of the NLRP3 inflammasome complex is initiated at priming. J. Immunol. 2018, 200, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Merkwirth, C.; Langer, T. Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta 2009, 1793, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Gupta, R.; Blanco, L.P.; Yang, S.; Shteinfer-Kuzmine, A.; Wang, K.; Zhu, J.; Yoon, H.E.; Wang, X.; Kerkhofs, M.; et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 2019, 366, 1531–1536. [Google Scholar] [CrossRef]
- Anderson, D.M.; Sato, H.; Dirck, A.T.; Feix, J.B.; Frank, D.W. Ubiquitin activates patatin-like phospholipases from multiple bacterial species. J. Bacteriol. 2015, 197, 529–541. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abeliovich, H.; Agostinis, P.; Agrawal, D.K.; Aliev, G.; Askew, D.S.; Baba, M.; Baehrecke, E.H.; Bahr, B.A.; Ballabio, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008, 4, 151–175. [Google Scholar] [CrossRef]
- Zhou, R.; Tardivel, A.; Thorens, B.; Choi, I.; Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010, 11, 136–140. [Google Scholar] [CrossRef]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Hauser, A.R.; Engel, J.N. Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 1999, 67, 5530–5537. [Google Scholar] [CrossRef] [PubMed]
- Rabin, S.D.; Veesenmeyer, J.L.; Bieging, K.T.; Hauser, A.R. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 2006, 74, 2552–2561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stirling, F.R.; Cuzick, A.; Kelly, S.M.; Oxley, D.; Evans, T.J. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell. Microbiol. 2006, 8, 1294–1309. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Halavaty, A.S.; Kim, H.; Geissler, B.; Agard, M.; Satchell, K.J.; Cho, W.; Anderson, W.F.; Hauser, A.R. A novel phosphatidylinositol 4,5-bisphosphate binding domain mediates plasma membrane localization of ExoU and other patatin-like phospholipases. J. Biol. Chem. 2015, 290, 2919–2937. [Google Scholar] [CrossRef]
- Tyson, G.H.; Hauser, A.R. Phosphatidylinositol 4,5-bisphosphate is a novel coactivator of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 2013, 81, 2873–2881. [Google Scholar] [CrossRef]
- Veesenmeyer, J.L.; Howell, H.; Halavaty, A.S.; Ahrens, S.; Anderson, W.F.; Hauser, A.R. Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect. Immun. 2010, 78, 3346–3357. [Google Scholar] [CrossRef]
- Broz, P.; Monack, D.M. Measuring inflammasome activation in response to bacterial infection. Methods Mol. Biol. 2013, 1040, 65–84. [Google Scholar]
- Bagayoko, S.; Leon-Icaza, S.A.; Pinilla, M.; Hessel, A.; Santoni, K.; Pericat, D.; Bordignon, P.J.; Moreau, F.; Eren, E.; Boyance, A.; et al. Host phospholipid peroxidation fuels ExoU-dependent cell necrosis and supports Pseudomonas aeruginosa-driven pathology. PLoS Pathog. 2021, 17, e1009927. [Google Scholar] [CrossRef]
- Pendyala, S.; Gorshkova, I.A.; Usatyuk, P.V.; He, D.; Pennathur, A.; Lambeth, J.D.; Thannickal, V.J.; Natarajan, V. Role of Nox4 and Nox2 in hyperoxia-induced reactive oxygen species generation and migration of human lung endothelial cells. Antioxid. Redox Signal. 2009, 11, 747–764. [Google Scholar] [CrossRef]
- Sawa, T.; Yahr, T.L.; Ohara, M.; Kurahashi, K.; Gropper, M.A.; Wiener-Kronish, J.P.; Frank, D.W. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 1999, 5, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Vallis, A.J.; Finck-Barbancon, V.; Yahr, T.L.; Frank, D.W. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect. Immun. 1999, 67, 2040–2044. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.P.; Lindsey, A.S.; Housley, N.A.; Ochoa, C.R.; Zhou, C.; Toba, M.; Oka, M.; Annamdevula, N.S.; Fitzgerald, M.S.; Frank, D.W.; et al. In the absence of effector proteins, the Pseudomonas aeruginosa type three secretion system needle tip complex contributes to lung injury and systemic inflammatory responses. PLoS ONE 2013, 8, e81792. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.A.; Schmalzer, K.M.; Frank, D.W. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA2 activity. Clin. Chim. Acta 2010, 411, 190–197. [Google Scholar] [CrossRef][Green Version]
- Ghasemi, A.; Zahediasl, S. Normality tests for statistical analysis: A guide for non-statisticians. Int. J. Endocrinol. Metab. 2012, 10, 486–489. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Strain | Genotype | Phenotype | Source |
|---|---|---|---|
| PA103 | Wild type | Virulent expressing functional T3SS, ExoU, ExoT | D. W. Frank |
| PA103ΔU | ΔexoU | Attenuated virulent expressing functional T3SS, ExoT | D. W. Frank |
| PA103ΔT | exoT::Tc | Attenuated virulent expressing functional T3SS, ExoU | D. W. Frank |
| PA103ΔUT | ΔexoU exoT::Tc | Attenuated virulent expressing functional T3SS | D. W. Frank |
| PA103ΔpcrV | ΔpcrV | Avirulent lacking T3SS, expressing ExoU, ExoT | D. W. Frank |
| JA806, JA808, JA816 | Wild type | Clinical isolate expressing functional T3SS, ExoS, ExoT, ExoY | B. M. Wagener, J-F Pittet, W. Richter |
| JA810, JA811, JA817 | Wild type | Clinical isolate expressing functional T3SS, ExoU, ExoT, ExoY | B. M. Wagener, J-F Pittet, W. Richter |
| PAK | Wild type | Virulent expressing functional T3SS, ExoS, ExoT, ExoY | W. Richter |
| PAKΔpscC | ΔpscC | Avirulent lacking T3SS, expressing ExoS, ExoT, ExoY | W. Richter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardy, K.S.; Tuckey, A.N.; Renema, P.; Patel, M.; Al-Mehdi, A.-B.; Spadafora, D.; Schlumpf, C.A.; Barrington, R.A.; Alexeyev, M.F.; Stevens, T.; et al. ExoU Induces Lung Endothelial Cell Damage and Activates Pro-Inflammatory Caspase-1 during Pseudomonas aeruginosa Infection. Toxins 2022, 14, 152. https://doi.org/10.3390/toxins14020152
Hardy KS, Tuckey AN, Renema P, Patel M, Al-Mehdi A-B, Spadafora D, Schlumpf CA, Barrington RA, Alexeyev MF, Stevens T, et al. ExoU Induces Lung Endothelial Cell Damage and Activates Pro-Inflammatory Caspase-1 during Pseudomonas aeruginosa Infection. Toxins. 2022; 14(2):152. https://doi.org/10.3390/toxins14020152
Chicago/Turabian StyleHardy, Kierra S., Amanda N. Tuckey, Phoibe Renema, Mita Patel, Abu-Bakr Al-Mehdi, Domenico Spadafora, Cody A. Schlumpf, Robert A. Barrington, Mikhail F. Alexeyev, Troy Stevens, and et al. 2022. "ExoU Induces Lung Endothelial Cell Damage and Activates Pro-Inflammatory Caspase-1 during Pseudomonas aeruginosa Infection" Toxins 14, no. 2: 152. https://doi.org/10.3390/toxins14020152
APA StyleHardy, K. S., Tuckey, A. N., Renema, P., Patel, M., Al-Mehdi, A.-B., Spadafora, D., Schlumpf, C. A., Barrington, R. A., Alexeyev, M. F., Stevens, T., Pittet, J.-F., Wagener, B. M., Simmons, J. D., Alvarez, D. F., & Audia, J. P. (2022). ExoU Induces Lung Endothelial Cell Damage and Activates Pro-Inflammatory Caspase-1 during Pseudomonas aeruginosa Infection. Toxins, 14(2), 152. https://doi.org/10.3390/toxins14020152






