Smp24, a Scorpion-Venom Peptide, Exhibits Potent Antitumor Effects against Hepatoma HepG2 Cells via Multi-Mechanisms In Vivo and In Vitro
Abstract
1. Introduction
2. Results
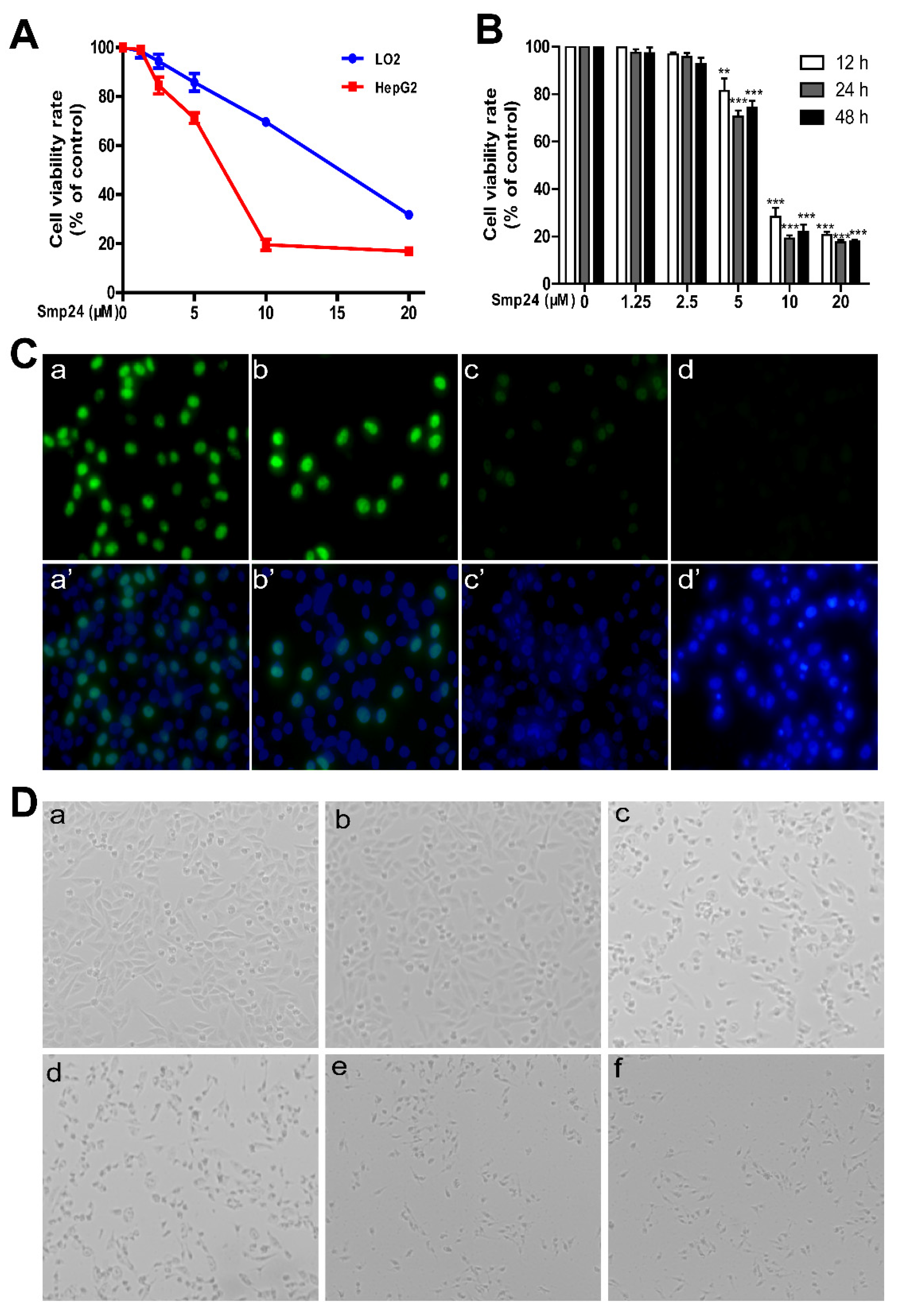
2.1. Smp24 Inhibits the Proliferation of Hepatoma Cells
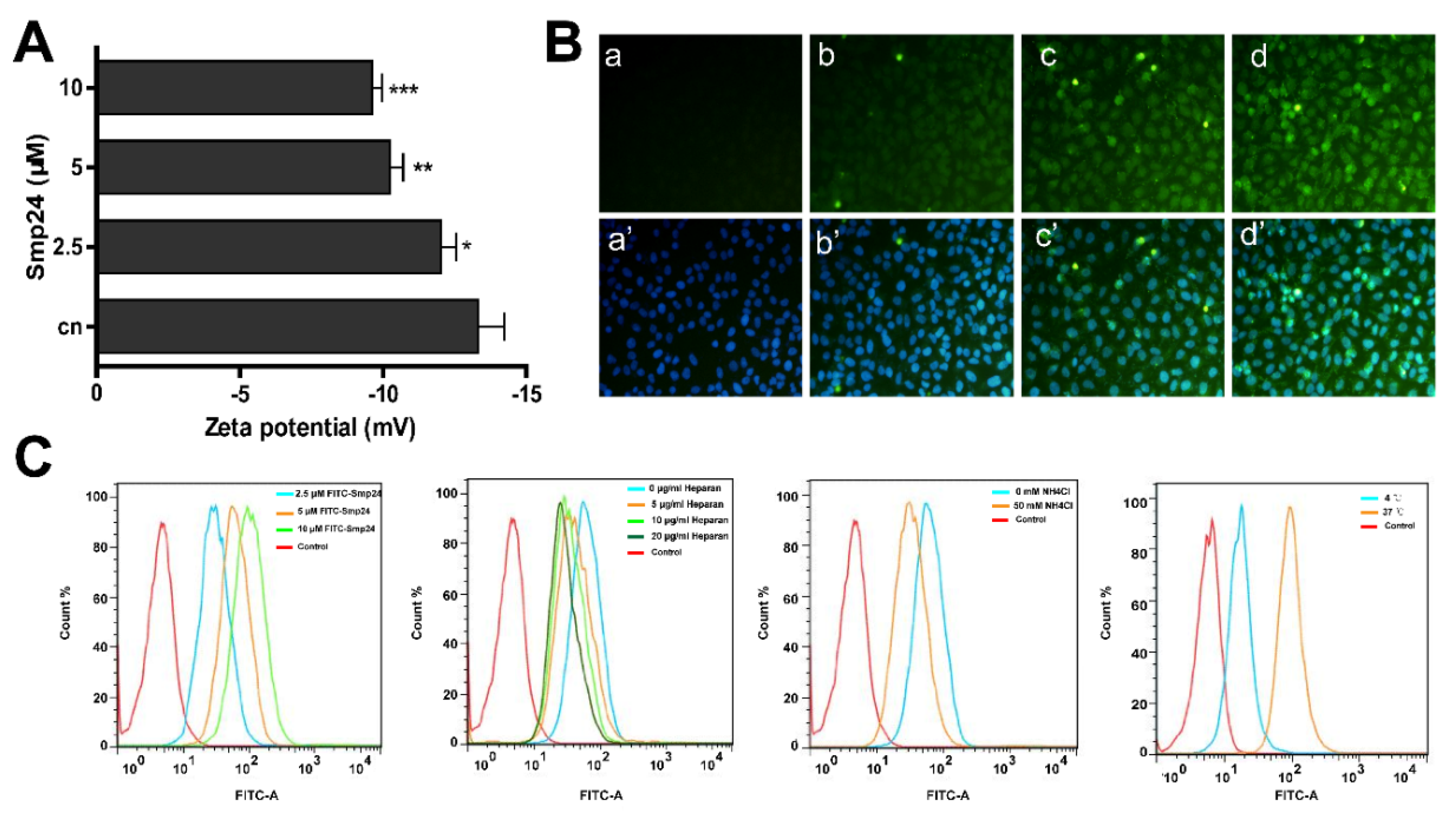
2.2. Smp24 Enters into HepG2 Cells through Endocytosis and Pore Formation
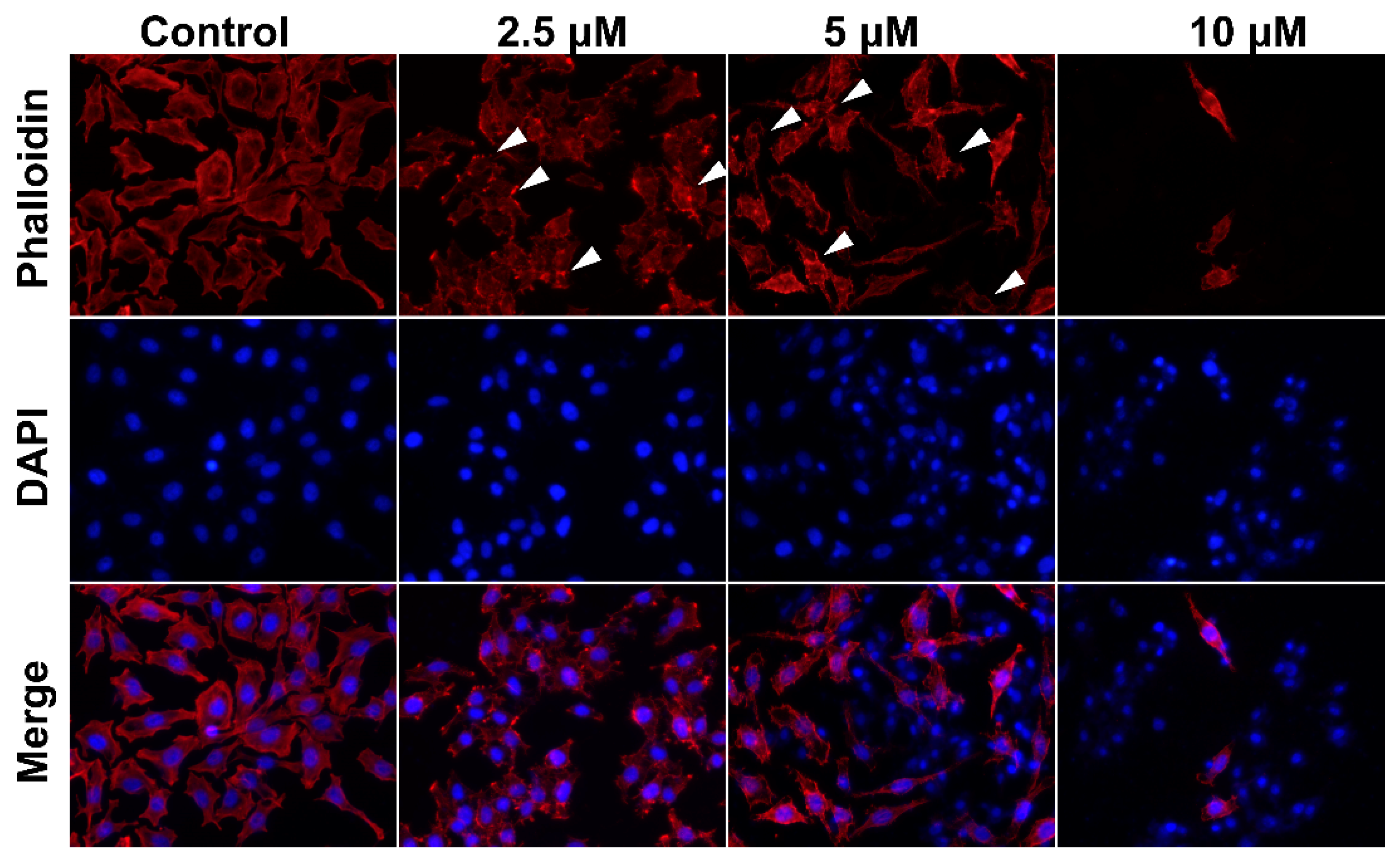
2.3. Smp24 Changes Cell Cytoskeleton Conformation
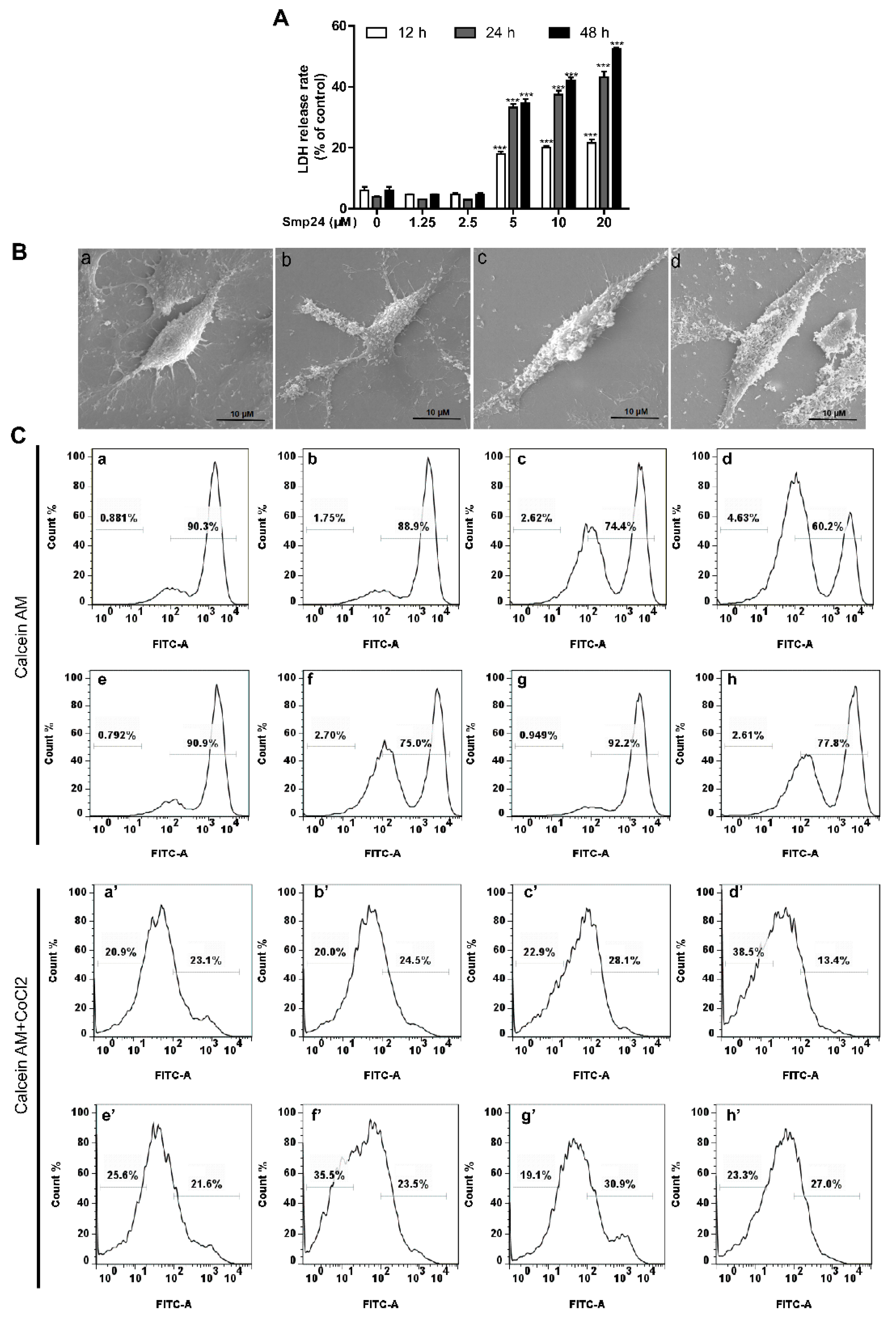
2.4. Smp24 Destroys the Cellular and Mitochondrial Membrane of HepG2 Cells
2.5. Smp24 Declines Mitochondrial Membrane Potential but Promotes ROS Production
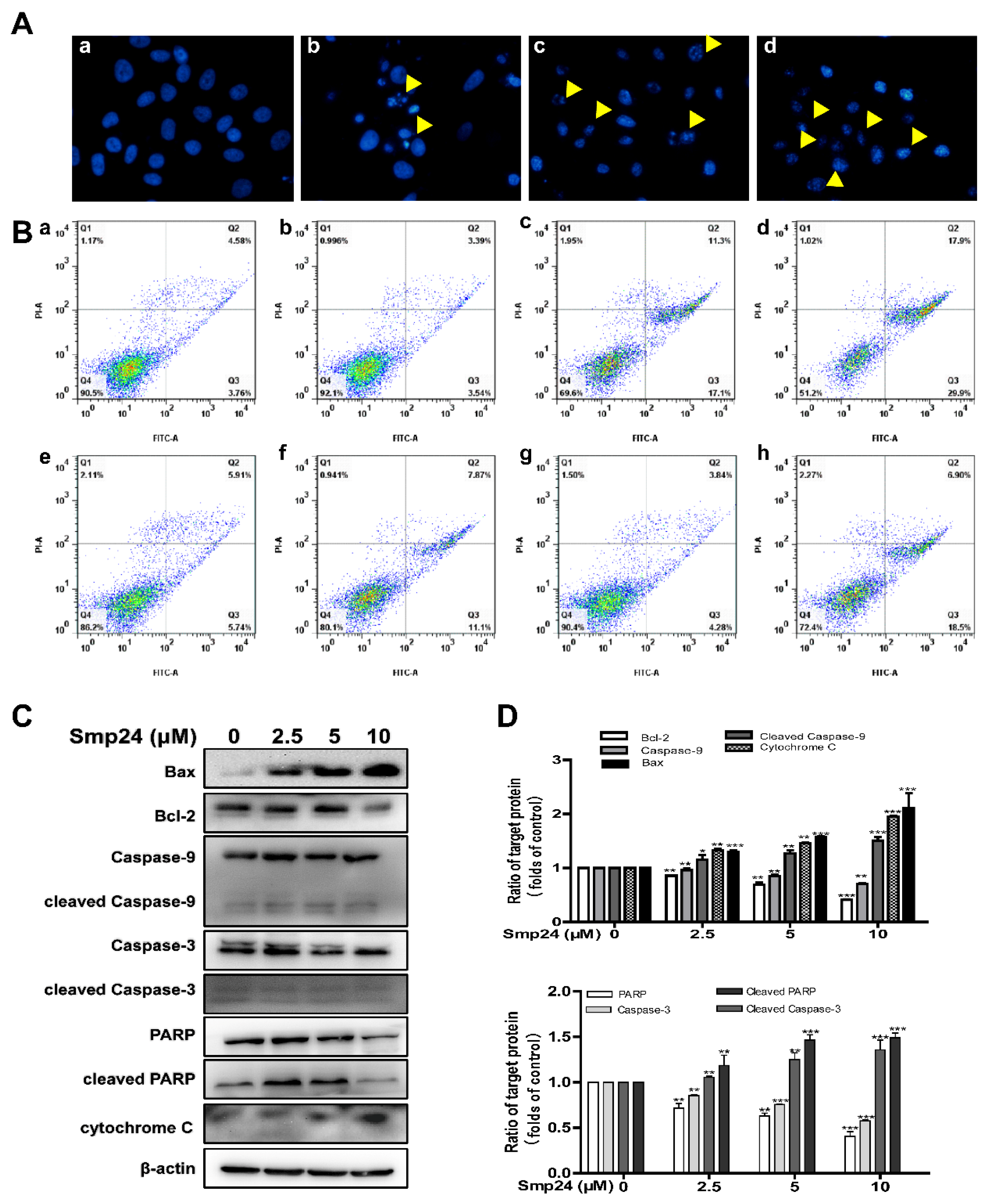
2.6. Smp24 Activates Mitochondrion-Mediated Intrinsic Pathway and Induces Apoptosis in HepG2 Cells
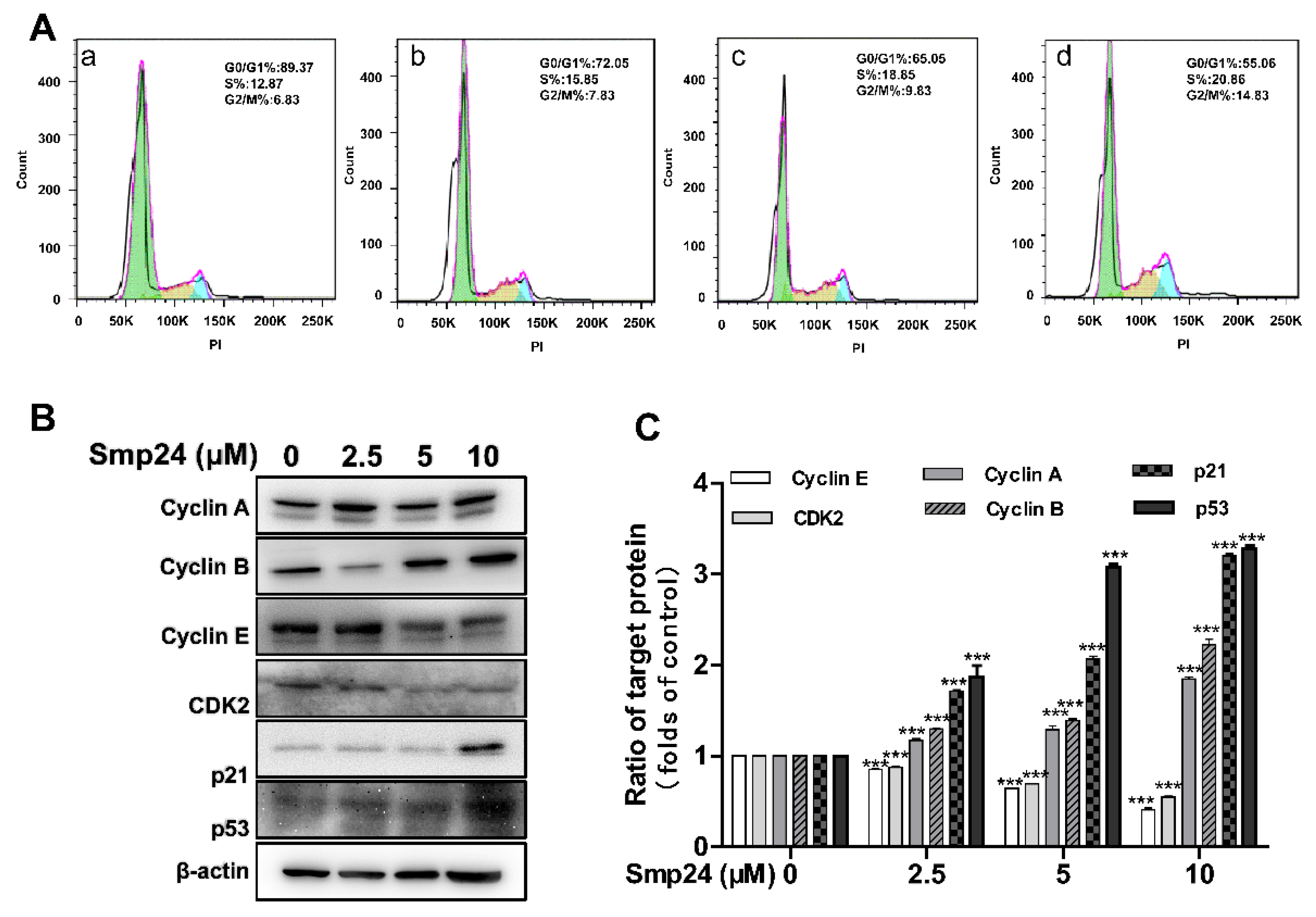
2.7. Smp24 Regulates the Expression of G2/M Phase-Related Proteins and Induces Cell Cycle Arrest
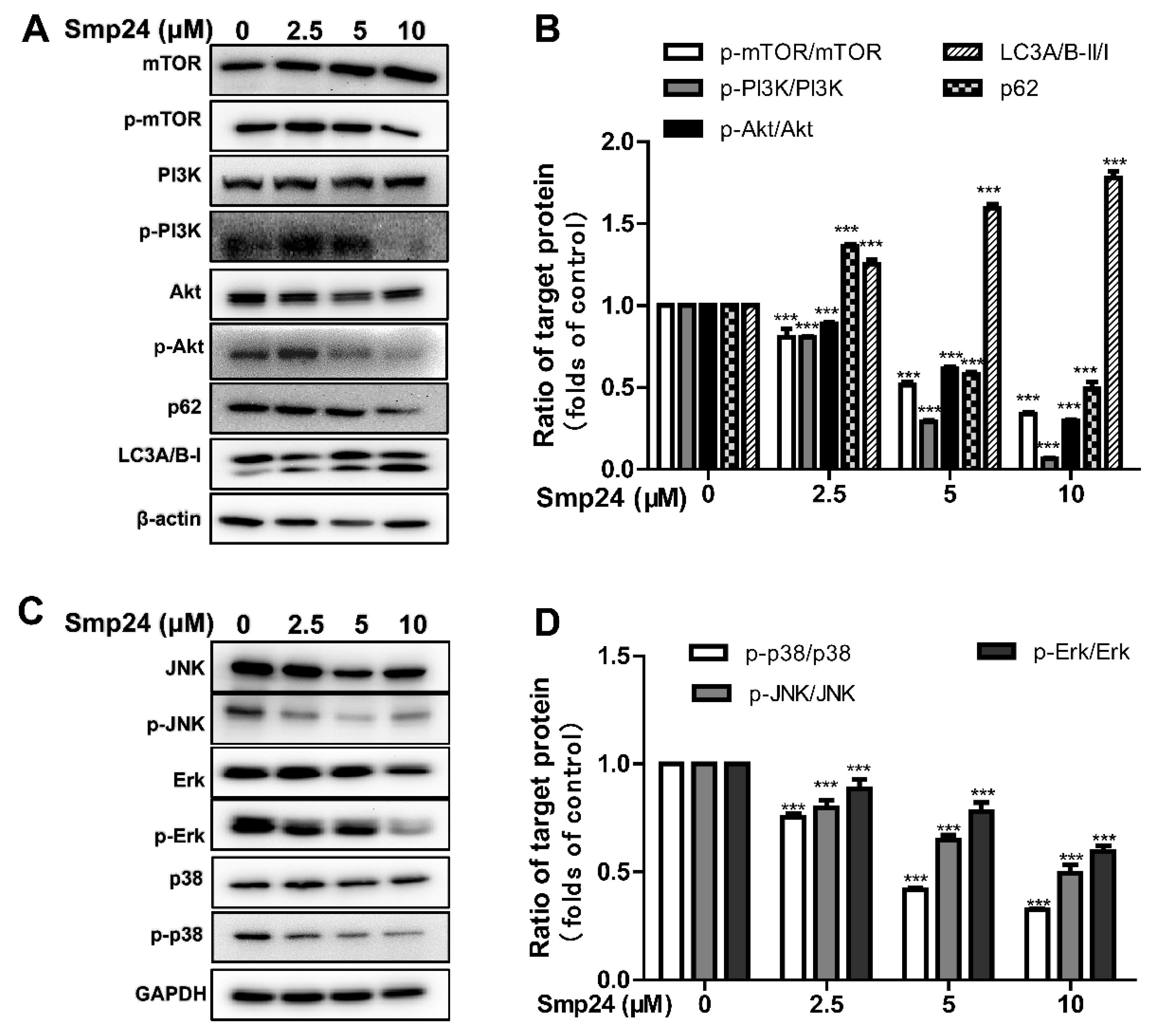
2.8. Smp24 Regulates Autophagy-Related Signaling Pathways
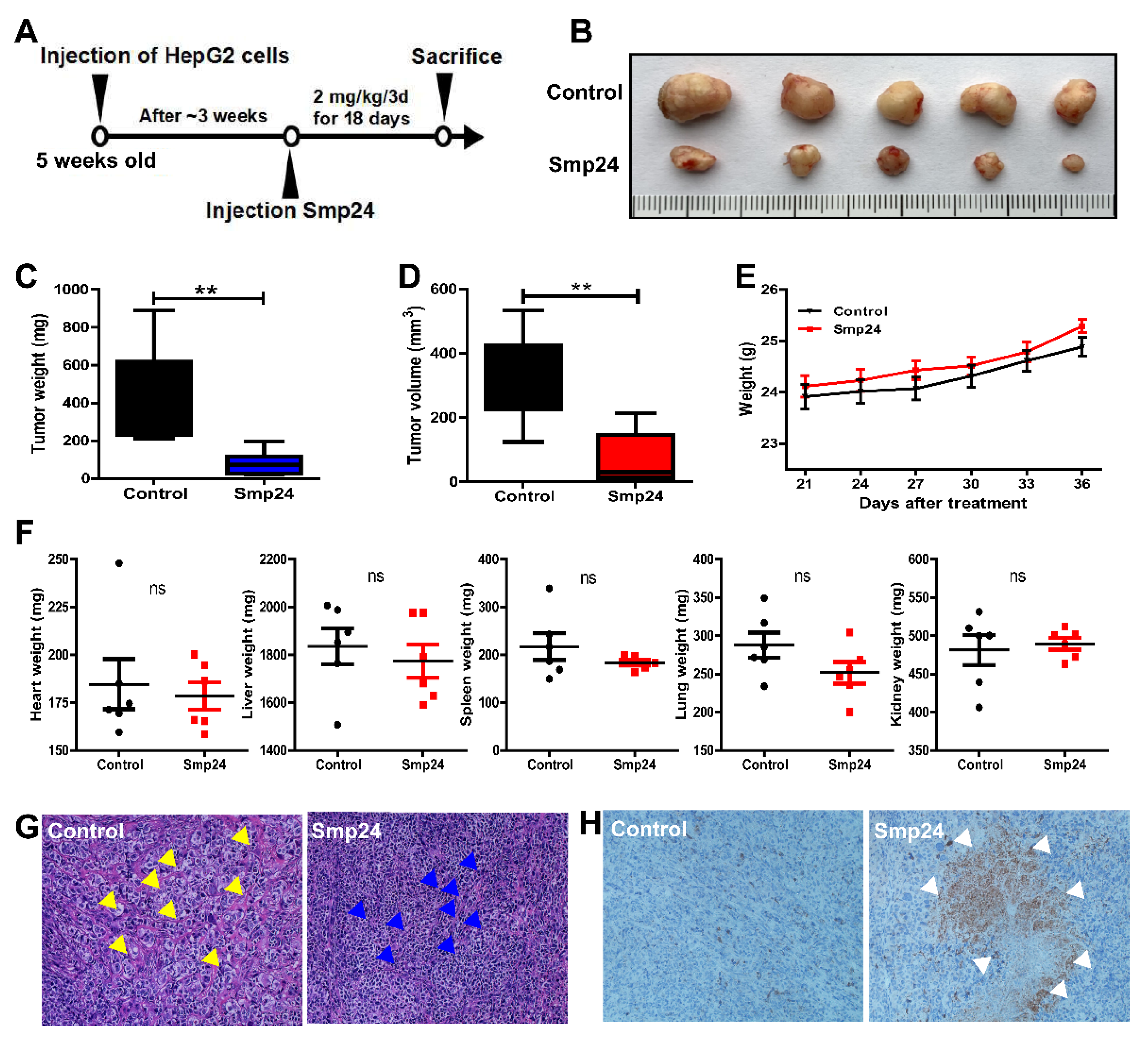
2.9. Smp24 Shows Antitumor Effects In Vivo
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals and Ethics Statement
5.2. Chemicals and Cell Culture
5.3. Cell Viability and Proliferation Analysis
5.4. Peptide Internalization Measurement
5.5. Membrane Integrity Assay
5.6. Cell Morphology Observation
5.7. LDH-Release Assay
5.8. Scanning Electron Microscopy Determination
5.9. Fluorescence Microscopy Analysis
5.10. Cell Cycle and Apoptosis Measurement
5.11. Western Blot Analysis
5.12. Animal Experiments
5.13. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, B.; Lyu, P.; Xi, X.; Ge, L.; Mahadevappa, R.; Shaw, C.; Kwok, H.F. Triggering of cancer cell cycle arrest by a novel scorpion venom-derived peptide-Gonearrestide. J. Cell. Mol. Med. 2018, 22, 4460–4473. [Google Scholar] [CrossRef]
- Shen, B.; Cao, Z.; Li, W.; Sabatier, J.M.; Wu, Y. Treating autoimmune disorders with venom-derived peptides. Expert Opin. Biol. Ther. 2017, 17, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Albalas, Q. Scorpion venom peptides with no disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hmed, B.; Serriayt, H.T.; Mounir, Z.K. Scorpion peptides: Potential use for new drug development. J. Toxicol. 2013, 2013, 958797. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Kuo, H.M.; Tseng, C.C.; Chen, N.F.; Tai, M.H.; Hung, H.C.; Feng, C.W.; Cheng, S.Y.; Huang, S.Y.; Jean, Y.H.; Wen, Z.H. MSP-4, an Antimicrobial Peptide, Induces Apoptosis via Activation of Extrinsic Fas/FasL- and Intrinsic Mitochondria-Mediated Pathways in One Osteosarcoma Cell Line. Mar. Drugs 2018, 16, 8. [Google Scholar] [CrossRef]
- Liu, S.; Aweya, J.J.; Zheng, L.; Zheng, Z.; Huang, H.; Wang, F.; Yao, D.; Ou, T.; Zhang, Y. LvHemB1, a novel cationic antimicrobial peptide derived from the hemocyanin of Litopenaeus vannamei, induces cancer cell death by targeting mitochondrial voltage-dependent anion channel 1. Cell Biol. Toxicol. 2022, 38, 87–110. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Strong, P.N.; Tawfik, M.M.; Miller, K. Characterisation of three alpha-helical antimicrobial peptides from the venom of Scorpio maurus palmatus. Toxicon 2016, 117, 30–36. [Google Scholar] [CrossRef]
- Elrayess, R.A.; Mohallal, M.E.; Mobarak, Y.M.; Ebaid, H.M.; Haywood-Small, S.; Miller, K.; Strong, P.N.; Abdel-Rahman, M.A. Scorpion Venom Antimicrobial Peptides Induce Caspase-1 Dependant Pyroptotic Cell Death. Front. Pharmacol. 2021, 12, 788874. [Google Scholar] [CrossRef]
- Guo, R.; Chen, X.; Nguyen, T.; Chai, J.; Gao, Y.; Wu, J.; Li, J.; Abdel-Rahman, M.A.; Chen, X.; Xu, X. The Strong Anti-Tumor Effect of Smp24 in Lung Adenocarcinoma A549 Cells Depends on Its Induction of Mitochondrial Dysfunctions and ROS Accumulation. Toxins 2022, 14, 590. [Google Scholar] [CrossRef]
- Guo, R.; Liu, J.; Chai, J.; Gao, Y.; Abdel-Rahman, M.A.; Xu, X. Scorpion Peptide Smp24 Exhibits a Potent Antitumor Effect on Human Lung Cancer Cells by Damaging the Membrane and Cytoskeleton In Vivo and In Vitro. Toxins 2022, 14, 483. [Google Scholar] [CrossRef] [PubMed]
- Poon, G.M.; Gariépy, J. Cell-surface proteoglycans as molecular portals for cationic peptide and polymer entry into cells. Biochem. Soc. Trans. 2007, 35, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.J.; McMahon, H.T. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Ann. Rev. Biophys. 2008, 37, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.; Kumavath, R.; Ghosh, P.; Barh, D. Mitochondria as a therapeutic target for aging and neurodegenerative diseases. Curr. Alzheimer Res. 2011, 8, 393–409. [Google Scholar] [CrossRef]
- Chipuk, J.E.; Bouchier-Hayes, L.; Green, D.R. Mitochondrial outer membrane permeabilization during apoptosis: The innocent bystander scenario. Cell Death Differ. 2006, 13, 1396–1402. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, J.; Wallace, D.C.; Jones, D.P. Cytochrome c-mediated apoptosis in cells lacking mitochondrial DNA. Signaling pathway involving release and caspase 3 activation is conserved. J. Biol. Chem. 1999, 274, 29905–29911. [Google Scholar] [CrossRef]
- Xiong, S.; Mu, T.; Wang, G.; Jiang, X. Mitochondria-mediated apoptosis in mammals. Protein Cell 2014, 5, 737–749. [Google Scholar] [CrossRef]
- Hao, X.; Bu, W.; Lv, G.; Xu, L.; Hou, D.; Wang, J.; Liu, X.; Yang, T.; Zhang, X.; Liu, Q.; et al. Disrupted mitochondrial homeostasis coupled with mitotic arrest generates antineoplastic oxidative stress. Oncogene 2022, 41, 427–443. [Google Scholar] [CrossRef]
- Mulcahy, L.J.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ 2020, 27, 843–857. [Google Scholar] [CrossRef]
- Su, M.; Mei, Y.; Sinha, S. Role of the Crosstalk between Autophagy and Apoptosis in Cancer. J. Oncol. 2013, 2013, 102735. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Kalamida, D.; Giatromanolaki, A.; Zois, C.E.; Sivridis, E.; Pouliliou, S.; Mitrakas, A.; Gatter, K.C.; Harris, A.L. Autophagosome Proteins LC3A, LC3B and LC3C Have Distinct Subcellular Distribution Kinetics and Expression in Cancer Cell Lines. PLoS ONE 2015, 10, e0137675. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gao, H.; Su, X. Autophagy-related signaling pathways are involved in cancer. Exp. Ther. Med. 2021, 22, 710. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, L.; Lippman, S.M.; El-Naggar, A.K. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 103–119. [Google Scholar] [CrossRef]
- Kunda, N.K. Antimicrobial peptides as novel therapeutics for non-small cell lung cancer. Drug Discov. Today 2020, 25, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 62, 190–194. [Google Scholar] [CrossRef]
- Yoon, W.H.; Park, H.D.; Lim, K.; Hwang, B.D. Effect of O-glycosylated mucin on invasion and metastasis of HM7 human colon cancer cells. Biochem. Biophys. Res. Commun. 1996, 222, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Y.W.; Zhang, L.; Gong, X.G.; Zhou, Y.; Shang, D.J. Melanoma cell surface-expressed phosphatidylserine as a therapeutic target for cationic anticancer peptide, temporin-1CEa. J. Drug Target. 2016, 24, 548–556. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, C.B.; Kim, J.M.; Jang, S.A.; Park, I.Y.; Kim, M.S.; Cho, J.H.; Kim, S.C. Mechanism of anticancer activity of buforin IIb, a histone H2A-derived peptide. Cancer Lett. 2008, 271, 47–55. [Google Scholar] [CrossRef]
- Fadnes, B.; Rekdal, O.; Uhlin-Hansen, L. The anticancer activity of lytic peptides is inhibited by heparan sulfate on the surface of the tumor cells. BMC Cancer 2009, 9, 183. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar] [CrossRef]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef] [PubMed]
- Kohno, M.; Horibe, T.; Ohara, K.; Ito, S.; Kawakami, K. The membrane-lytic peptides K8L9 and melittin enter cancer cells via receptor endocytosis following subcytotoxic exposure. Chem. Biol. 2014, 21, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, H.; Yang, Y.; Xu, X.; Lv, T.; Zhang, H.; Liu, K.; Zhang, S.; Chen, Y. N-myristoylation of Antimicrobial Peptide CM4 Enhances Its Anticancer Activity by Interacting with Cell Membrane and Targeting Mitochondria in Breast Cancer Cells. Front. Pharmacol. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Gaspar, P.; Holder, M.V.; Aerne, B.L.; Janody, F.; Tapon, N. Zyxin antagonizes the FERM protein expanded to couple F-actin and Yorkie-dependent organ growth. Curr. Biol. 2015, 25, 679–689. [Google Scholar] [CrossRef]
- Illescas, M.; Peñas, A.; Arenas, J.; Martín, M.A.; Ugalde, C. Regulation of Mitochondrial Function by the Actin Cytoskeleton. Front. Cell Dev. Biol. 2021, 9, 95838. [Google Scholar] [CrossRef]
- Gourlay, C.W.; Ayscough, K.R. The actin cytoskeleton: A key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005, 6, 583–589. [Google Scholar] [CrossRef]
- Antico, A.V.; Elguero, M.E.; Poderoso, J.J.; Carreras, M.C. Mitochondrial regulation of cell cycle and proliferation. Antioxid. Redox Signal. 2012, 16, 1150–1180. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Martín-Caballero, J.; Flores, J.M.; García-Palencia, P.; Serrano, M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001, 61, 6234–6238. [Google Scholar]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, J.H.; Jin, L.; Lin, S.M.; Yang, Y.; Sui, Y.X.; Shi, H. Over-expression of the Beclin1 gene upregulates chemosensitivity to anti-cancer drugs by enhancing therapy-induced apoptosis in cervix squamous carcinoma CaSki cells. Cancer Lett. 2010, 294, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Akar, U.; Chaves-Reyez, A.; Barria, M.; Tari, A.; Sanguino, A.; Kondo, Y.; Kondo, S.; Arun, B.; Lopez-Berestein, G.; Ozpolat, B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 2008, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Huang, D.; Zhai, M.; Yang, L.; Peng, S.; Chen, C.; Feng, X.; Weng, Q.; Zhang, B.; Xu, M. Isolation of a novel bio-peptide from walnut residual protein inducing apoptosis and autophagy on cancer cells. BMC Complement. Altern. Med. 2015, 15, 413. [Google Scholar] [CrossRef]
- Ren, S.X.; Shen, J.; Cheng, A.S.; Lu, L.; Chan, R.L.; Li, Z.J.; Wang, X.J.; Wong, C.C.; Zhang, L.; Ng, S.S.; et al. FK-16 derived from the anticancer peptide LL-37 induces caspase-independent apoptosis and autophagic cell death in colon cancer cells. PLoS ONE 2013, 8, e63641. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, A.S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 2019, 59, 125–132. [Google Scholar] [CrossRef]
- Yang, J.; Pi, C.; Wang, G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed. Pharmacother. 2018, 103, 699–707. [Google Scholar] [CrossRef]
- Zhou, L.; Li, S.; Sun, J. Ginkgolic acid induces apoptosis and autophagy of endometrial carcinoma cells via inhibiting PI3K/Akt/mTOR pathway in vivo and in vitro. Hum. Exp. Toxicol. 2021, 40, 2156–2164. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 2015, 14, 48. [Google Scholar] [CrossRef]
- Reddy, D.; Kumavath, R.; Ghosh, P.; Barh, D. Lanatoside C Induces G2/M Cell Cycle Arrest and Suppresses Cancer Cell Growth by Attenuating MAPK, Wnt, JAK-STAT, and PI3K/AKT/mTOR Signaling Pathways. Biomolecules 2019, 9, 792. [Google Scholar] [CrossRef]
- Chai, J.; Yang, W.; Gao, Y.; Guo, R.; Peng, Q.; Abdel-Rahman, M.A.; Xu, X. Antitumor Effects of Scorpion Peptide Smp43 through Mitochondrial Dysfunction and Membrane Disruption on Hepatocellular Carcinoma. J. Nat. Prod. 2021, 84, 3147–3160. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Li, L.; Ma, Y.X.; Li, W.T.; Li, L.; Zhu, H.Z.; Wu, M.H.; Zhou, J.R. Quercetin inhibits growth of hepatocellular carcinoma by apoptosis induction in part via autophagy stimulation in mice. J. Nutr. Biochem. 2019, 69, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.J. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 1995, 80, 179–185. [Google Scholar] [CrossRef]
- Li, C.M.; Haratipour, P.; Lingeman, R.G.; Perry, J.; Gu, L.; Hickey, R.J.; Malkas, L.H. Novel Peptide Therapeutic Approaches for Cancer Treatment. Cells 2021, 10, 2908. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.; Guo, R.; Chai, J.; Wu, J.; Liu, J.; Chen, X.; Abdel-Rahman, M.A.; Xia, H.; Xu, X. Smp24, a Scorpion-Venom Peptide, Exhibits Potent Antitumor Effects against Hepatoma HepG2 Cells via Multi-Mechanisms In Vivo and In Vitro. Toxins 2022, 14, 717. https://doi.org/10.3390/toxins14100717
Nguyen T, Guo R, Chai J, Wu J, Liu J, Chen X, Abdel-Rahman MA, Xia H, Xu X. Smp24, a Scorpion-Venom Peptide, Exhibits Potent Antitumor Effects against Hepatoma HepG2 Cells via Multi-Mechanisms In Vivo and In Vitro. Toxins. 2022; 14(10):717. https://doi.org/10.3390/toxins14100717
Chicago/Turabian StyleNguyen, Tienthanh, Ruiyin Guo, Jinwei Chai, Jiena Wu, Junfang Liu, Xin Chen, Mohamed A. Abdel-Rahman, Hu Xia, and Xueqing Xu. 2022. "Smp24, a Scorpion-Venom Peptide, Exhibits Potent Antitumor Effects against Hepatoma HepG2 Cells via Multi-Mechanisms In Vivo and In Vitro" Toxins 14, no. 10: 717. https://doi.org/10.3390/toxins14100717
APA StyleNguyen, T., Guo, R., Chai, J., Wu, J., Liu, J., Chen, X., Abdel-Rahman, M. A., Xia, H., & Xu, X. (2022). Smp24, a Scorpion-Venom Peptide, Exhibits Potent Antitumor Effects against Hepatoma HepG2 Cells via Multi-Mechanisms In Vivo and In Vitro. Toxins, 14(10), 717. https://doi.org/10.3390/toxins14100717






