Early Warning of Resistance to Bt Toxin Vip3Aa in Helicoverpa zea
Abstract
:1. Introduction
2. Results
2.1. Variation in Susceptibility to Vip3Aa among Lab Strains of H. zea
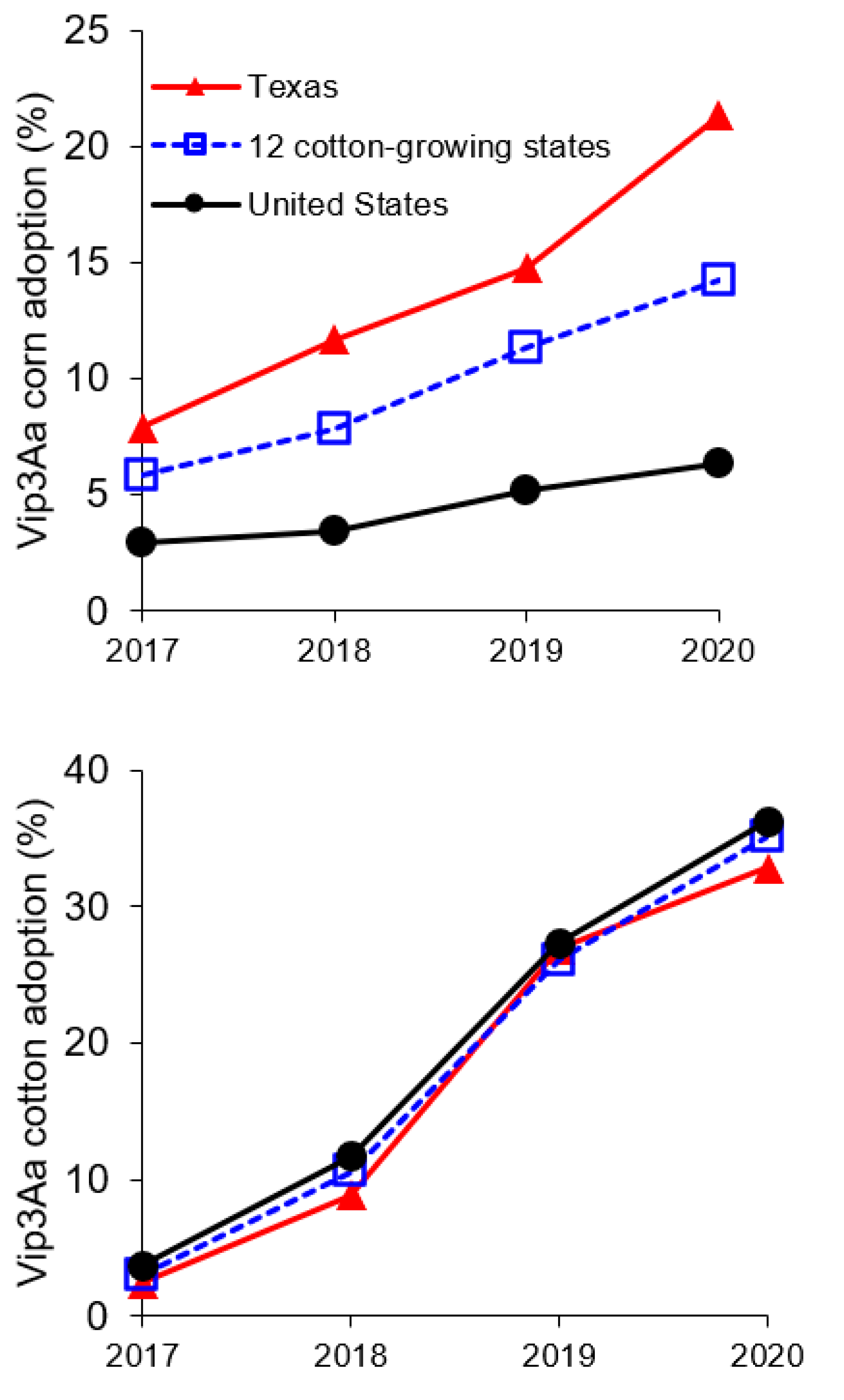
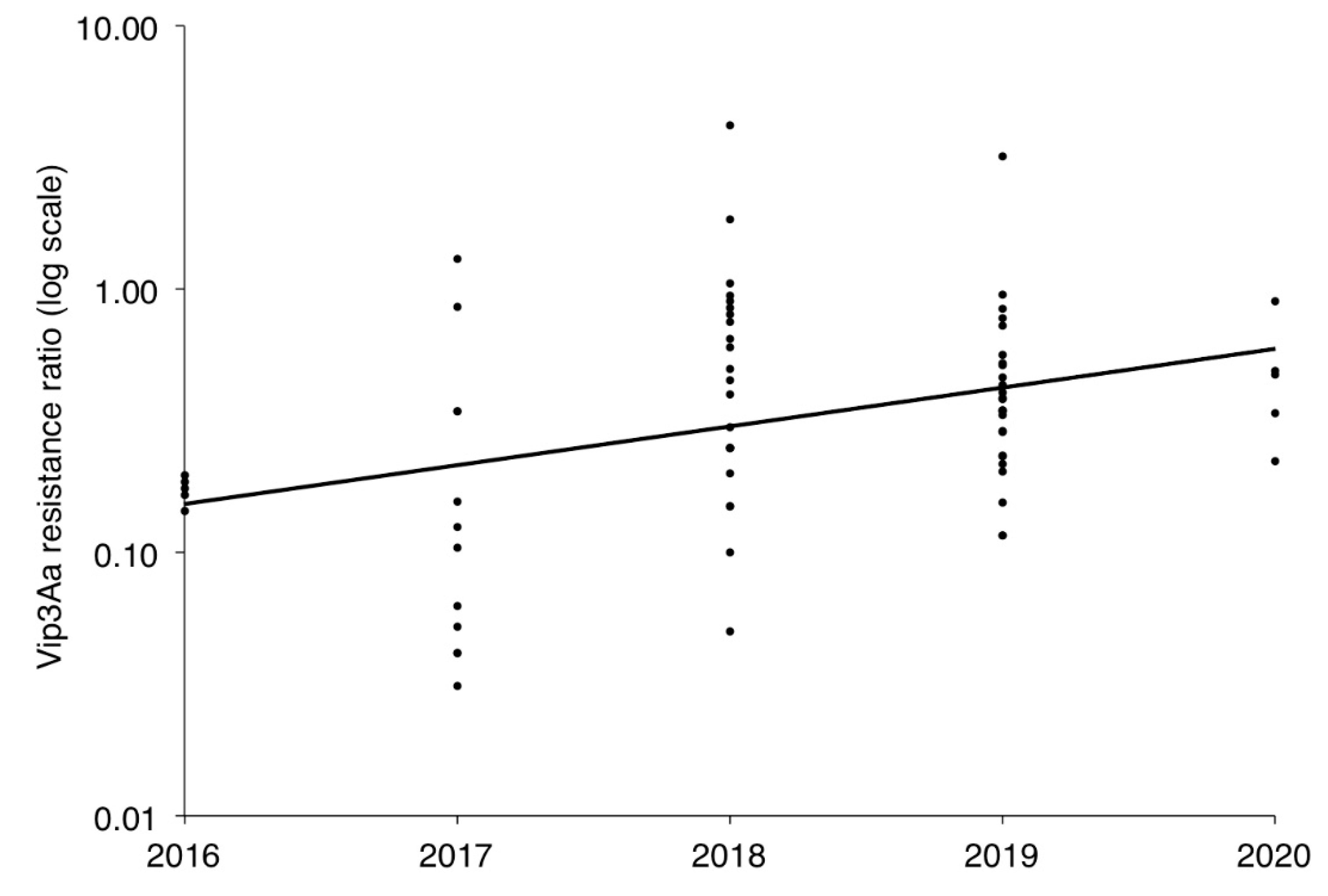
2.2. Resistance to Vip3Aa in Field-Derived Strains of H. zea Relative to Lab Strains
2.3. Resistance to Vip3Aa in H. zea from Vip3Aa Plants Relative to Non-Vip3Aa Plants
3. Discussion
4. Materials and Methods
4.1. Strains of H. zea
4.2. ELISA Tests for Vip3Aa in Putative Vip3Aa Plants
4.3. Diet Overlay Bioassays
4.4. Vip3Aa
4.5. Data Analysis
4.5.1. Diet Bioassay Data
4.5.2. Efficacy of Vip3Aa Corn
4.6. Population Genetic Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchison, W.D.; Burkness, E.C.; Mitchell, P.D.; Moon, R.D.; Leslie, T.W.; Fleischer, S.J.; Abrahamson, M.; Hamilton, K.L.; Steffey, K.L.; Gray, M.E.; et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 2010, 330, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Gould, F.; Amasino, R.M.; Brossard, D.; Buell, C.R.; Dixon, R.A.; Falck-Zepeda, J.B.; Gallo, M.A.; Giller, K.; Glenna, L.; Grin, T.S.; et al. Genetically Engineered Crops: Experiences and Prospects; National Academies of Sciences, Engineering and Medicine; National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Dively, G.P.; Venugopal, P.D.; Bean, D.; Whalen, J.; Holmstrom, K.; Kuhar, T.P.; Doughty, H.B.; Patton, T.; Cissel, W.; Hutchison, W.D. Regional pest suppression associated with widespread Bt maize adoption benefits vegetable growers. Proc. Natl. Acad. Sci. USA 2018, 115, 3320–3325. [Google Scholar] [CrossRef] [Green Version]
- Tabashnik, B.E.; Liesner, L.R.; Ellsworth, P.C.; Unnithan, G.C.; Fabrick, J.A.; Naranjo, S.E.; Li, X.; Dennehy, T.J.; Antilla, L.; Staten, R.T.; et al. Transgenic cotton and sterile insect releases synergize eradication of pink bollworm a century after it invaded the United States. Proc. Natl. Acad. Sci. USA 2021, 118, e2019115118. [Google Scholar] [CrossRef] [PubMed]
- International Service for the Acquisition of Agri-Biotech Applications. Global Status of Commercialized Biotech/GM Crops in 2019: Biotech Crops Drive Socio-Economic Development and Sustainable Environment in the New Frontier; ISAAA Brief No. 55; International Service for the Acquisition of Agri-Biotech Applications: Ithaca, NY, USA, 2019. [Google Scholar]
- Tabashnik, B.E.; Carrière, Y. Global patterns of resistance to Bt crops highlighting pink bollworm in the United States, China, and India. J. Econ. Entomol. 2019, 112, 2513–2523. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Carrière, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nature Biotech. 2017, 35, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Mota-Sanchez, D.; Whalon, M.E.; Hollingworth, R.M.; Carrière, Y. Defining terms for proactive management of resistance to Bt crops and pesticides. J. Econ. Entomol. 2014, 107, 496–507. [Google Scholar] [CrossRef]
- Calles-Torrez, V.; Knodel, J.J.; Boetel, M.A.; French, B.W.; Fuller, B.W.; Ransom, J.K. Field-evolved resistance of northern and western corn rootworm (Coleoptera: Chrysomelidae) populations to corn hybrids expressing single and pyramided Cry3Bb1 and Cry34/35Ab1 Bt proteins in North Dakota. J. Econ. Entomol. 2019, 112, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Farhan, Y.; Schaafsma, A.W. Practical resistance of Ostrinia nubilalis (Lepidoptera: Crambidae) to Cry1F Bacillus thuringiensis maize discovered in Nova Scotia, Canada. Sci. Rep. 2019, 9, 18247. [Google Scholar] [CrossRef] [Green Version]
- Luttrell, R.G.; Jackson, R.E. Helicoverpa zea and Bt cotton in the United States. GM Crops Food 2012, 3, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Luttrell, R.G.; Ali, I.; Allen, K.C.; Young, S.Y., III; Szalanski, A.; Williams, K.; Lorenz, G.; Parker, C.D., Jr.; Blanco, C. Resistance to Bt in Arkansas populations of cotton bollworm. In Proceedings of the 2004 Beltwide Cotton Conferences, San Antonio, TX, USA, 5–9 January 2004; Richter, D.A., Ed.; National Cotton Council of America: Memphis, TN, USA, 2004; pp. 1373–1383. [Google Scholar]
- Tabashnik, B.E.; Gassmann, A.J.; Crowder, D.W.; Carrière, Y. Field-evolved resistance to Bt toxins. Nat. Biotech. 2008, 26, 1074–1076. [Google Scholar] [CrossRef]
- Dively, G.P.; Venugopal, P.D.; Finkenbinder, C. Field-evolved resistance in corn earworm to Cry proteins expressed by transgenic sweet corn. PLoS ONE 2016, 11, e0169115. [Google Scholar] [CrossRef]
- Reisig, D.D.; Huseth, A.S.; Bacheler, J.S.; Aghaee, M.A.; Braswell, L.; Burrack, H.J.; Flanders, K.; Greene, J.K.; Herbert, D.A.; Jacobson, A. Long-term empirical and observational evidence of practical Helicoverpa zea resistance to cotton with pyramided Bt toxins. J. Econ. Entomol. 2018, 111, 1824–1833. [Google Scholar] [CrossRef]
- Bilbo, T.R.; Reay-Jones, F.P.; Reisig, D.D.; Greene, J.K. Susceptibility of corn earworm (Lepidoptera: Noctuidae) to Cry1A.105 and Cry2Ab2 in North and South Carolina. J. Econ. Entomol. 2019, 112, 1845–1857. [Google Scholar] [CrossRef]
- Kaur, G.; Guo, J.; Brown, S.; Head, G.P.; Price, P.A.; Paula-Moraes, S.; Ni, X.; Dimase, M.; Huang, F. Field-evolved resistance of Helicoverpa zea (Boddie) to transgenic maize expressing pyramided Cry1A.105/Cry2Ab2 proteins in northeast Louisiana, the United States. J. Invertebr. Pathol. 2019, 163, 11–20. [Google Scholar] [CrossRef]
- Little, N.S.; Elkins, B.H.; Mullen, R.M.; Perera, O.P.; Parys, K.A.; Allen, K.C.; Boykin, D.L. Differences between two populations of bollworm, Helicoverpa zea (Lepidoptera: Noctuidae), with variable measurements of laboratory susceptibilities to Bt toxins exposed to non-Bt and Bt cottons in large field cages. PLoS ONE 2019, 14, e0212567. [Google Scholar] [CrossRef]
- Yang, F.; González, J.C.; Williams, J.; Cook, D.C.; Gilreath, R.T.; Kerns, D.L. Occurrence and ear damage of Helicoverpa zea on transgenic Bacillus thuringiensis maize in the field in Texas, US and its susceptibility to Vip3A protein. Toxins 2019, 11, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dively, G.P.; Kuhar, T.P.; Taylor, S.; Doughty, H.B.; Holmstrom, K.; Gilrein, D.; Nault, B.A.; Ingerson-Mahar, J.; Whalen, J.; Reisig, D.; et al. Sweet corn sentinel monitoring for lepidopteran field-evolved resistance to Bt toxins. J. Econ. Entomol. 2021, 114, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Oyediran, I.; Rice, M.E.; Brown, S.; Dimase, M.; Lin, S.; Walker, W.; Yu, W.; Niu, Y.; Huang, F. Seed blends of pyramided Cry/Vip maize reduce Helicoverpa zea populations from refuge ears. J. Pest. Sci. 2021, 94, 959–968. [Google Scholar] [CrossRef]
- Niu, Y.; Oyediran, I.; Yu, W.; Lin, S.; Dimase, M.; Brown, S.; Reay-Jones, F.P.F.; Cook, D.; Reisig, D.; Thrash, B.; et al. Populations of Helicoverpa zea (Boddie) in the southeastern United States are commonly resistant to Cry1Ab, but still susceptible to Vip3Aa20 expressed in MIR 162 corn. Toxins 2021, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Reay-Jones, F.P.F. Pest status and management of corn earworm (Lepidoptera: Noctuidae) in field corn in the United States. J. Integr. Pest. Manag. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Tabashnik, B.E.; Van Rensburg, J.B.J.; Carrière, Y. Field-evolved insect resistance to Bt crops: Definition, theory, and data. J. Econ. Entomol. 2009, 102, 2011–2025. [Google Scholar] [CrossRef]
- Yu, J.; Hennessy, D.A.; Wu, F. The impact of Bt corn on aflatoxin-related insurance claims in the United States. Sci. Rep. 2020, 10, 10046. [Google Scholar] [CrossRef]
- US Department of Agriculture; Agricultural Marketing Service. Cotton Varieties Planted 2020 Crop. Available online: https://www.ams.usda.gov/mnreports/cnavar.pdf (accessed on 15 May 2021).
- Byrne, M.J.; Iadanza, M.G.; Perez, M.A.; Maskell, D.P.; George, R.M.; Hesketh, E.L.; Beales, P.A.; Zack, M.D.; Berry, C.; Thompson, R.F. Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane. Nature Comm. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Syed, T.; Askari, M.; Meng, Z.; Li, Y.; Abid, M.A.; Wei, Y.; Guo, S.; Lian, C.; Zhang, R. Current insights on vegetative insecticidal proteins (Vip) as next generation pest killers. Toxins 2020, 12, 522. [Google Scholar] [CrossRef] [PubMed]
- Chakroun, M.; Banyuls, N.; Bel, Y.; Escriche, B.; Ferré, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 329–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabashnik, B.E.; Carrière, Y. Evaluating cross-resistance between Vip and Cry toxins of Bacillus thuringiensis. J. Econ. Entomol. 2020, 113, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, K.J.; Rodrigo-Simón, A.; Ferré, J.; Pusztai-Carey, M.; Sivasupramaniam, S.; Moar, W.J. Production and characterization of Bacillus thuringiensis Cry1Ac-resistant cotton bollworm Helicoverpa zea (Boddie). Appl. Environ. Microbiol. 2008, 74, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, K.L.; Unnithan, G.C.; Degain, B.A.; Wei, J.; Zhang, J.; Li, X.; Tabashnik, B.E.; Carrière, Y. Cross-resistance to toxins used in pyramided Bt crops and resistance to Bt sprays in Helicoverpa zea. J. Invertebr. Pathol. 2015, 132, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Burkness, E.C.; Dively, G.; Patton, T.; Morey, A.C.; Hutchison, W.D. Novel Vip3A Bacillus thuringiensis (Bt) maize approaches high-dose efficacy against Helicoverpa zea (Lepidoptera: Noctuidae) under field conditions: Implications for resistance management. GM Crops 2010, 1, 337–343. [Google Scholar] [CrossRef]
- Yang, F.; Kerns, D.L.; Leonard, B.R.; Oyediran, I.; Burd, T.; Niu, Y.; Huang, F. Performance of Agrisure®VipteraTM 3111 corn against Helicoverpa zea (Lepidoptera: Noctuidae) in seed mixed plantings. Crop. Prot. 2015, 69, 77–82. [Google Scholar] [CrossRef]
- Brown, S.; Walker, W.; Cole, C. Efficacy and field performance of Bt cotton in Louisiana. In Proceedings of the 2019 Beltwide Cotton Conferences, New Orleans, LA, USA, 8–10 January 2019; National Cotton Council of America: Memphis, TN, USA, 2019; pp. 477–480. [Google Scholar]
- Yang, F.; González, J.C.S.; Sword, G.A.; Kerns, D.L. Genetic basis of resistance to the Vip3Aa Bt protein in Helicoverpa zea. Pest. Manag. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S. Introduction to Quantitative Genetics, 3rd ed.; John Wiley & Sons: New York, NY, USA, 1989. [Google Scholar]
- Yang, F.; González, J.C.S.; Little, N.; Reisig, D.; Payne, G.; Dos Santos, R.F.; Jurat-Fuentes, J.L.; Kurtz, R.; Kerns, D.L. First documentation of major Vip3Aa resistance alleles in field populations of Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) in Texas, USA. Sci. Rep. 2020, 10, 5867. [Google Scholar] [CrossRef] [Green Version]
- Leite, N.A.; Pereira, R.M.; Durigan, M.R.; Amado, D.; Fatoretto, J.; Medeiros, F.C.L.; Omoto, C. Susceptibility of Brazilian populations of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) to Vip3Aa20. J. Econ. Entomol. 2018, 111, 399–404. [Google Scholar] [CrossRef]
- Ali, M.I.; Luttrell, R.G. Susceptibility of Helicoverpa zea and Heliothis virescens (Lepidoptera: Noctuidae) to Vip3A insecticidal toxin expressed in VipCotTM cotton. J. Invertebr. Pathol. 2011, 108, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Venette, R.C.; Hutchison, W.D.; Andow, D.A. An in-field screen for early detection and monitoring of insect resistance to Bacillus thuringiensis in transgenic crops. J. Econ. Entomol. 2000, 93, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Roush, R.T.; Miller, G.L. Considerations for design of insecticide resistance monitoring programs. J. Econ. Entomol. 1986, 79, 293–298. [Google Scholar] [CrossRef]
- Reisig, D.D.; Kurtz, R. Bt resistance implications for Helicoverpa zea (Lepidoptera: Noctuidae) insecticide resistance management in the United States. Environ. Entomol. 2018, 47, 1357–1364. [Google Scholar] [CrossRef] [Green Version]
- US Environmental Protection Agency. Resistance in Lepidopteran Pests to Bacillus thuringiensis (Bt) Plant Incorporated Protectants (PIPs) in the United States; July 2018 SAP Meeting Minutes. Available online: https://www.regulations.gov/document?D=EPA-HQ-OPP-2017-0617-0078 (accessed on 15 June 2021).
- Reisig, D.D.; Dively, G.; Gore, J.; DiFonzo, C.; Farhan, Y.; Smith, J. Response to the EPA Draft Proposal to Address Resistance Risks to Lepidopteran Pests of Bt Following the July 2018 FIFRA Scientific Advisory Panel Recommendation Memorandum. Available online: https://www.regulations.gov/comment/EPA-HQ-OPP-2019-0682-0031 (accessed on 15 June 2021).
- Caprio, M.A.; Kurtz, R.; Catchot, A.; Kerns, D.; Reisig, D.; Gore, J.; Reay-Jones, F.P.F. The corn-cotton agroecosystem in the mid-southern United States: What insecticidal event pyramids should be used in each crop to extend Vip3A durability. J. Econ. Entomol. 2019, 112, 2894–2906. [Google Scholar] [CrossRef]
- Bacterial Pesticidal Protein Resource Center. Available online: https://camtech-bpp.ifas.ufl.edu/bestmatchfinder_database/ (accessed on 10 June 2021).
- SAS Institute. SAS/STAT 9.3 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2011. [Google Scholar]
- Tabashnik, B.E.; Cushing, N.L.; Johnson, M.W. Diamondback moth (Lepidoptera: Plutellidae) resistance to insecticides in Hawaii: Intra-island variation and cross-resistance. J. Econ. Entomol. 1987, 80, 1091–1099. [Google Scholar] [CrossRef]
- Payton, M.E.; Greenstone, M.H.; Schenker, N. Overlapping confidence intervals or standard error intervals: What do they mean in terms of statistical significance? J. Insect Sci. 2003, 3, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Brévault, T.; Heuberger, S.; Zhang, M.; Ellers-Kirk, C.; Ni, X.; Masson, L.; Li, X.; Tabashnik, B.E.; Carrière, Y. Potential shortfall of pyramided Bt cotton for resistance management. Proc. Natl. Acad. Sci. USA 2013, 110, 5806–5811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnell, R.; Horn, K.; Blair, E.; Murray, S. 2020 Texas Corn Performance Variety Trials. Texas A&M AgriLife. SCS-2020-11. 2020. Available online: https://varietytesting.tamu.edu/files/corn/2020/2020-CORN-PUBLICATION.PDF (accessed on 23 August 2021).
- Saavoss, M.; Capehart, T.; McBride, W.; Effland, A. Trends in Production Practices and Costs of the U.S. Corn Sector, ERR-2 USDA, Economic Research Service. 2021. Available online: https://www.ers.usda.gov/webdocs/publications/101722/err-294.pdf?v=2087.7 (accessed on 23 August 2021).
- Robinson, E. Calculator Compares Cotton Seed Costs. 2003. FarmProgress. Available online: https://www.farmprogress.com/calculator-compares-cotton-seed-costs (accessed on 23 August 2021).
- U.S. Environmental Protection Agency (EPA). Acres Planted per Day and Seeding Rates of Crops Grown in the United States. 2010. Available online: https://www.epa.gov/sites/default/files/2018-01/documents/seeding-rates-and-acres-planted-per-day-revised-final-030111.pdf (accessed on 23 August 2021).
- USDA National Agricultural Statistics Service (NASS). Crop Production. 2020. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/cropan20.pdf (accessed on 23 August 2021).
- USDA National Agricultural Statistics Service (NASS). Acreage. 2021. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/acrg0621.pdf (accessed on 23 August 2021).
| Host Plant a | Bt Toxins in Host Plant a | Field Site or Lab Strain Name | Pupae b | Larvae c | Slope ± SE | LC50 (95% FL) d | RR vs. BZ e | RR vs. SIMRU f |
|---|---|---|---|---|---|---|---|---|
| 2016: LSU | ||||||||
| Lab diet | None | BZ | / | 957 | 2.8 ± 0.4 | 0.97 (0.85, 1.11) | 1.0 | 2.8 |
| Lab diet | None | SIMRU | / | 958 | 1.9 ± 0.3 | 0.35 (0.22, 0.56) | 0.4 | 1.0 |
| Non-Bt corn | None | Toad Suck, AR | 114 | 956 | 1.6 ± 0.2 | 0.17 (0.13, 0.23) | 0.2 | 0.5 |
| BG2 cotton | Cry1Ac + Cry2Ab | Alexandria, LA | 37 | 945 | 1.8 ± 0.2 | 0.19 (0.15, 0.24) | 0.2 | 0.5 |
| VT2P corn | Cry1A.105 + Cry2Ab | Leland, MS | 182 | 962 | 2.2 ± 0.1 | 0.14 (0.12, 0.16) | 0.1 | 0.4 |
| Grain sorghum | None | Jackson, TN | 118 | 956 | 2.1 ± 0.2 | 0.16 (0.12, 0.21) | 0.2 | 0.5 |
| BG2 cotton | Cry1Ac + Cry2Ab | Jackson, TN | 92 | 943 | 1.8 ± 0.2 | 0.18 (0.13, 0.23) | 0.2 | 0.5 |
| 2017: TAMU | ||||||||
| Lab diet | None | BZ | / | 895 | 2.8 ± 0.3 | 0.96 (0.86, 1.12) | 1.0 | / |
| Grain sorghum | None | Rohwer, AR | 157 | 448 | 5.2 ± 1.2 | 1.25 (1.00, 1.57) | 1.3 | / |
| Non-Bt cotton | None | Alexandria, LA | 131 | 448 | 2.4 ± 0.2 | 0.10 (0.08, 0.12) | 0.1 | / |
| TwinLink cotton | Cry1Ab + Cry2Ae | Alexandria, LA | 107 | 448 | 1.7 ± 0.3 | 0.33 (0.19, 0.61) | 0.3 | / |
| BG2 cotton | Cry1Ac + Cry2Ab | Jonesville, LA | 121 | 448 | 2.0 ± 0.2 | 0.15 (0.12, 0.18) | 0.2 | / |
| Non-Bt corn | None | Winnsboro, LA | 186 | 895 | 2.1 ± 0.4 | 0.33 (0.18, 0.61) | 0.3 | / |
| BG2 cotton | Cry1Ac + Cry2Ab | Benoit, MS | 69 | 448 | 1.3 ± 0.2 | 0.04 (0.03, 0.06) | 0.04 | / |
| BG2 cotton | Cry1Ac + Cry2Ab | Silver City, MS | 75 | 448 | 3.2 ± 0.3 | 0.05 (0.04, 0.06) | 0.1 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Stoneville, MS | 111 | 894 | 1.8 ± 0.2 | 0.06 (0.04, 0.07) | 0.1 | / |
| VT2P & non-Bt corn | Cry1A.105 + Cry2Ab | Starkville, MS | 97 | 448 | 2.6 ± 0.3 | 0.04 (0.03, 0.05) | 0.0 | / |
| Obsession corn | Cry1A.105 + Cry2Ab | Milan, TN | 135 | 448 | 2.3 ± 0.2 | 0.12 (0.10, 0.14) | 0.1 | / |
| WS cotton | Cry1Ab + Cry1Fa | Snook, TX | 77 | 896 | 2.1 ± 0.4 | 0.03 (0.02, 0.04) | 0.03 | / |
| TwinLink cotton | Cry1Ab + Cry2Ae | Wharton, TX | 20 | 895 | 5.7 ± 0.9 | 0.82 (0.73, 0.90) | 0.9 | / |
| Host Plant a | Bt Toxins in Host Plant a | Field Site or Lab Strain Name | Pupae b | Larvae c | Slope ± SE | LC50 (95% FL) d | RR vs. BZ e | RR vs. TM f |
|---|---|---|---|---|---|---|---|---|
| Lab diet | None | BZ | / | 448 | 1.5 ± 0.1 | 0.20 (0.16, 0.26) | 1.0 | 1.3 |
| Lab diet | None | TM | / | 448 | 2.2 ± 0.4 | 0.16 (0.11, 0.25) | 0.8 | 1.0 |
| Intrasect corn | Cry1Ab + Cry1F | Little Rock, AR | 130 | 448 | 2.1 ± 0.2 | 0.05 (0.04, 0.06) | 0.3 | 0.3 |
| and VT2P corn | Cry1A.105 + Cry2Ab2 | |||||||
| Non-Bt corn | None | Pine Bluff, AR | 280 | 448 | 3.1 ± 0.3 | 0.13 (0.11, 0.16) | 0.7 | 0.8 |
| BG2 cotton | Cry1Ac + Cry2Ab | Alexandria, LA | 300 | 448 | 2.1 ± 0.2 | 0.05 (0.04, 0.06) | 0.3 | 0.3 |
| WS3 cotton | Cry1Ac + Cry1Fa + Vip3Aa | Grant, LA | 240 | 448 | 2.7 ± 0.4 | 0.12 (0.10, 0.16) | 0.6 | 0.8 |
| Crimson clover | None | Winnsboro, LA | 300 | 448 | 2.5 ± 0.2 | 0.06 (0.05, 0.07) | 0.3 | 0.4 |
| Non-Bt corn | None | Winnsboro, LA | 300 | 448 | 3.1 ± 0.3 | 0.05 (0.04, 0.06) | 0.3 | 0.3 |
| Soybean | None | Indianola, MS | 120 | 448 | 2.5 ± 0.4 | 0.18 (0.12, 0.25) | 0.9 | 1.1 |
| Crimson clover | None | Natchez, MS | 220 | 448 | 2.0 ± 0.3 | 0.19 (0.12, 0.31) | 1.0 | 1.2 |
| Obsession corn | Cry1A.105 + Cry2Ab | Jackson, TN | 180 | 448 | 2.2 ± 0.3 | 0.21 (0.15, 0.31) | 1.1 | 1.3 |
| BG2 cotton | Cry1Ac + Cry2Ab | Jackson, TN | 150 | 448 | 1.9 ± 0.3 | 0.01 (0.01, 0.02) | 0.1 | 0.1 |
| Non-Bt corn | None | Amarillo, TX | 193 | 448 | 2.2 ± 0.4 | 0.15 (0.13, 0.18) | 0.8 | 0.9 |
| VT3P corn | Cry1A.105 + Cry2Ab | Snook, TX g | 300 | 448 | 2.8 ± 0.3 | 0.04 (0.03, 0.05) | 0.2 | 0.3 |
| WS cotton | Cry1Ac + Cry1Fa | Snook, TX | 200 | 448 | 3.3 ± 1.0 | 0.37 (0.20, 0.71) | 1.9 | 2.3 |
| Leptra corn | Cry1Ab + Cry1Fa + Vip3Aa | Snook, TX h | 100 | 448 | 4.9 ± 1.0 | 0.84 (0.69, 0.97) | 4.2 * | 5.3 * |
| BG2 cotton | Cry1Ac + Cry2Ab | EI Campo, TX | 28 | 448 | 2.8 ± 0.3 | 0.05 (0.04, 0.06) | 0.3 | 0.3 |
| Non-Bt corn | None | Los Indios, TX | 150 | 448 | 2.5 ± 0.2 | 0.10 (0.08, 0.12) | 0.5 | 0.6 |
| Non-Bt corn | None | Lubbock, TX | 272 | 448 | 4.2 ± 0.5 | 0.17 (0.15, 0.20) | 0.9 | 1.1 |
| VT2P corn | Cry1A.105 + Cry2Ab | Muleshoe, TX | 210 | 448 | 1.8 ± 0.2 | 0.03 (0.02, 0.04) | 0.2 | 0.2 |
| Grain sorghum | None | Port Lavaca, TX | 138 | 448 | 2.8 ± 0.3 | 0.09 (0.07, 0.11) | 0.5 | 0.6 |
| STX corn | Cry1A.105 + Cry1Fa + Cry2Ab | Thrall, TX | 87 | 448 | 2.1 ± 0.2 | 0.08 (0.07, 0.10) | 0.4 | 0.5 |
| BG2 cotton | Cry1Ac + Cry2Ab | Wellington, TX | 108 | 448 | 2.5 ± 0.3 | 0.03 (0.03, 0.04) | 0.2 | 0.2 |
| BG2 cotton | Cry1Ac + Cry2Ab | Wharton, TX | 70 | 448 | 2.9 ± 0.6 | 0.02 (0.01, 0.03) | 0.1 | 0.1 |
| VT3P corn | Cry1A.105 + Cry2Ab | Wall, TX | 103 | 448 | 2.3 ± 0.3 | 0.16 (0.12, 0.22) | 0.8 | 1.0 |
| Host Plant a | Bt Toxins in Host Plant a | Field Site or Lab Strain Name | Pupae b | Larvae c | Slope ± SE | LC50 (95% FL) d | RR vs. BZ e | RR vs. TM f |
|---|---|---|---|---|---|---|---|---|
| 2019: TAMU | ||||||||
| Lab diet | None | BZ | / | 448 | 3.7 ± 0.5 | 0.69 (0.56, 0.87) | 1.0 | 4.1 * |
| Lab diet | None | TM g | / | 448 | 2.8 ± 0.3 | 0.17 (0.14, 0.21) | 0.2 | 1.0 |
| VT2P corn | Cry1A.105 + Cry2Ab | Lafayette Co., AR | 120 | 448 | 2.8 ± 0.3 | 0.39 (0.33, 0.47) | 0.6 | 2.3 * |
| VT2P corn | Cry1A.105 + Cry2Ab | Tillar, AR | 115 | 448 | 2.5 ± 0.3 | 0.15 (0.13, 0.19) | 0.2 | 0.9 |
| VT2P corn | Cry1A.105 + Cry2Ab | Alexandria, LA | 278 | 448 | 2.2 ± 0.2 | 0.23 (0.18, 0.28) | 0.3 | 1.4 |
| BG2 cotton | Cry1Ac + Cry2Ab | Alexandria, LA | 180 | 448 | 2.5 ± 0.2 | 0.24 (0.20, 0.29) | 0.3 | 1.4 |
| VT2P corn | Cry1A.105 + Cry2Ab | Winnsboro, LA | 198 | 448 | 2.5 ± 0.2 | 0.14 (0.12, 0.17) | 0.2 | 0.8 |
| Leptra corn | Cry1Ab + Cry1Fa + Vip3Aa | Stoneville, MS | 82 | 448 | 1.6 ± 0.3 | 2.21 (1.27, 4.44) | 3.2 * | 13.0 * |
| VT2P corn | Cry1A.105 + Cry2Ab | Stoneville, MS | 105 | 448 | 2.9 ± 0.3 | 0.08 (0.07, 0.10) | 0.1 | 0.5 |
| VT2P corn | Cry1A.105 + Cry2Ab | Starkville, MS | 285 | 448 | 2.8 ± 0.3 | 0.16 (0.13, 0.19) | 0.2 | 0.9 |
| VT2P corn | Cry1A.105 + Cry2Ab | Jackson, TN | 210 | 448 | 2.5 ± 0.3 | 0.32 (0.24, 0.44) | 0.5 | 1.9 * |
| VT2P corn | Cry1A.105 + Cry2Ab | Hillsboro, TX | 90 | 448 | 3.6 ± 0.4 | 0.30 (0.25, 0.35) | 0.4 | 1.8 * |
| BG2 cotton | Cry1Ac + Cry2Ab | Jackson, TX | 100 | 448 | 3.0 ± 1.3 | 0.20 (0.03, 0.17) | 0.3 | 1.2 |
| Non-Bt corn | None | Lubbock, TX | 172 | 448 | 2.9 ± 0.3 | 0.28 (0.24, 0.34) | 0.4 | 1.6 * |
| BG2 cotton | Cry1Ac + Cry2Ab | Navasota, TX | 117 | 448 | 3.1 ± 0.3 | 0.08 (0.07, 0.10) | 0.1 | 0.5 |
| Leptra corn | Cry1Ab + Cry1Fa + Vip3Aa | Snook, TX | 46 | 448 | 1.8 ± 0.2 | 0.66 (0.49, 0.89) | 1.0 | 3.9 * |
| BG3 cotton | Cry1Ac + Cry2Ab + Vip3Aa | Snook, TX | 123 | 448 | 1.8 ± 0.2 | 0.50 (0.40, 0.63) | 0.7 | 2.9 * |
| Non-Bt corn | None | Wharton, TX | 102 | 448 | 2.1 ± 0.2 | 0.24 (0.19, 0.29) | 0.3 | 1.4 |
| 2019: SIMRU | ||||||||
| Lab diet | None | BZ | / | 512 | 1.7 ± 0.3 | 0.41 (0.26, 0.68) | 1.0 | / |
| Non-Bt corn | None | Pickens, AR | 246 | 512 | 2.3 ± 0.3 | 0.35 (0.26, 0.47) | 0.8 | / |
| Bt corn | Cry1A.105 + Cry2Ab | Leland, MS | 238 | 512 | 1.7 ± 0.2 | 0.22 (0.16, 0.29) | 0.5 | / |
| Crimson clover | None | Grenada, MS | 242 | 512 | 2.6 ± 0.3 | 0.10 (0.08, 0.12) | 0.2 | / |
| Crimson clover | None | Marks, MS | 177 | 512 | 1.5 ± 0.2 | 0.21 (0.13, 0.35) | 0.5 | / |
| Non-Bt corn | None | Mound Bayou, MS | 451 | 512 | 2.4 ± 0.3 | 0.14 0.11, 0.19) | 0.3 | / |
| Crimson clover | None | Olive Branch, MS | 400 | 384 | 2.4 ± 0.4 | 0.12 (0.08, 0.17) | 0.3 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Rolling Fork, MS | 240 | 512 | 2.1 ± 0.2 | 0.16 (0.12, 0.21) | 0.4 | / |
| Non-Bt corn | None | Stoneville, MS | 282 | 512 | 2.4 ± 0.3 | 0.32 (0.24, 0.44) | 0.8 | / |
| TwinLink cotton | Cry1Ab + Cry2Ae | Stoneville, MS | 79 | 512 | 1.9 ± 0.2 | 0.06 (0.04, 0.10) | 0.2 | / |
| Crimson clover | None | Warren County, MS | 201 | 512 | 1.6 ± 0.2 | 0.16 (0.11, 0.22) | 0.4 | / |
| 2020: TAMU | ||||||||
| Lab diet | None | BZ | / | 448 | 3.2 ± 0.4 | 0.11 (0.09, 0.13) | 1.0 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Stoneville, MS | 186 | 448 | 3.9 ± 0.5 | 0.06 (0.05, 0.06) | 0.5 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Pine Bluff, AR | 135 | 448 | 1.3 ± 0.2 | 0.05 (0.03, 0.08) | 0.5 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Alexandria, LA | 93 | 448 | 2.6 ± 0.5 | 0.03 (0.02, 0.03) | 0.2 | / |
| Non-Bt corn | None | Winnsboro, LA | 210 | 448 | 3.7 ± 0.6 | 0.04 (0.03, 0.04) | 0.3 | / |
| VT2P corn | Cry1A.105 + Cry2Ab | Jackson, TN | 178 | 448 | 3.4 ± 0.4 | 0.10 (0.09, 0.12) | 0.9 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Kerns, D.L.; Little, N.S.; Santiago González, J.C.; Tabashnik, B.E. Early Warning of Resistance to Bt Toxin Vip3Aa in Helicoverpa zea. Toxins 2021, 13, 618. https://doi.org/10.3390/toxins13090618
Yang F, Kerns DL, Little NS, Santiago González JC, Tabashnik BE. Early Warning of Resistance to Bt Toxin Vip3Aa in Helicoverpa zea. Toxins. 2021; 13(9):618. https://doi.org/10.3390/toxins13090618
Chicago/Turabian StyleYang, Fei, David L. Kerns, Nathan S. Little, José C. Santiago González, and Bruce E. Tabashnik. 2021. "Early Warning of Resistance to Bt Toxin Vip3Aa in Helicoverpa zea" Toxins 13, no. 9: 618. https://doi.org/10.3390/toxins13090618
APA StyleYang, F., Kerns, D. L., Little, N. S., Santiago González, J. C., & Tabashnik, B. E. (2021). Early Warning of Resistance to Bt Toxin Vip3Aa in Helicoverpa zea. Toxins, 13(9), 618. https://doi.org/10.3390/toxins13090618






