Proteo-Transcriptomic Characterization of Sirex nitobei (Hymenoptera: Siricidae) Venom
Abstract
1. Introduction
2. Results
2.1. Summary and Analysis of Venom Gland Transcriptome Data
2.2. Identification of Venom Proteins by Integrated Transcriptomic and Proteomic Analysis
2.3. Verification of the Expression of the Selected Venom Gland Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Sample Collection and Tissue Dissection
5.2. RNA Extraction
5.3. cDNA Library Construction and Transcriptome Sequencing
5.4. De Novo Assembly and Analysis
5.5. Extraction and SDS-PAGE Analysis of S. nitobei Venom Proteins
5.6. LC-MS/MS Analysis and S. nitobei Venom Protein Identification
5.7. Quantitative RT-PCR (qRT-PCR) Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiff, N.M.; Goulet, H.; Smith, D.R.; Boudreault, C.; Wilson, A.D.; Scheffler, B.E. Siricidae (Hymenoptera: Symphyta: Siricoidea) of the western hemisphere. Can. J. Arthropod Identif. 2012, 21, 1–305. [Google Scholar] [CrossRef]
- Xiao, G.R.; Huang, X.Y.; Zhou, S.Z.; Wu, J.; Zhang, P. Siricidae; Economic Sawfly Fauna of China, Hymenoptera, Symphyta; Tianze Eldonejo: Xi’an, China, 1991; pp. 37–42. [Google Scholar]
- Wang, M.; Wang, L.X.; Fu, N.N.; Gao, C.L.; Ao, T.G.; Ren, L.L.; Luo, Y.Q. Comparison of wing, ovipositor, and Cornus morphologies between Sirex noctilio and Sirex nitobei using geometric morphometrics. Insects 2020, 11, 84. [Google Scholar] [CrossRef]
- Gao, T.; Xu, Q.; Liu, Y.; Zhao, J.Q.; Shi, J. Predicting the potential geographic distribution of Sirex nitobei in China under climate change using maximum entropy model. Forests 2021, 12, 151. [Google Scholar] [CrossRef]
- Goulet, H. Sirex systematics; problems and solutions. In The Sirex Woodwasp and Its Fungal Symbiont: Research and Management of a Worldwide Invasive Pest; Slippers, B., De Groot, P., Wingfield, M.J., Eds.; Springer: Berlin, Germany, 2011; pp. 5–18. [Google Scholar]
- Coutts, M.P.; Dolezal, J.E. Some effects of bark cincturing on the physiology of Pinus radiata, and on Sirex attack. Aust. For. Res. 1966, 2, 17–28. [Google Scholar]
- Coutts, M.P. Formation of polyphenols in small blocks of Pinus radiata sapwood with and without the fungal symbiont of Sirex. Aust. For. Res. 1969, 4, 29–34. [Google Scholar]
- Coutts, M.P.; Dolezal, J.E. Polyphenols and Resin in the Resistance Mechanism of Pinus Radiata Attacked by the Wood Wasp, Sirex Noctilio, and Its Associated Fungus; Forest Research Institute: Canberra, Australia, 1966; pp. 1–19. [Google Scholar]
- Coutts, M.P. Rapid physiological change in Pinus radiata following attack by Sirex noctilio and its associated fungus, Amylostereum sp. Aust. J. Sci. 1968, 30, 275–277. [Google Scholar]
- Coutts, M.P. Mechanism of pathogenicity of Sirex noctilio on Pinus radiata. I. Effects of symbiotic fungus Amylostereum sp. Aust. J. Biol. Sci. 1969, 22, 915–924. [Google Scholar] [CrossRef]
- Coutts, M.P. Mechanism of pathogenicity of Sirex noctilio on Pinus radiata. 2. Effects of S. noctilio mucus. Aust. J. Biol. Sci. 1969, 22, 1153–1161. [Google Scholar] [CrossRef]
- Coutts, M.P. The physiological effects of the mucus secretion of Sirex noctilio on Pinus radiata. Aust. For. Res. 1970, 4, 23–26. [Google Scholar]
- Spradbery, J.P. A comparative study of the phytotoxic effects of siricid woodwasps on conifers. Ann. Appl. Biol. 1973, 75, 309–320. [Google Scholar] [CrossRef]
- Fong, L.K.; Crowden, R.K. Physiological effects of mucus from the wood wasp, Sirex noctilio F., on the foliage of Pinus radiata D. Aust. J. Biol. Sci. 1973, 26, 365–378. [Google Scholar] [CrossRef]
- Goecks, J.; Mortimer, N.T.; Mobley, J.A.; Bowersock, G.J.; Taylor, J.; Schlenke, T.A. Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS ONE 2013, 8, e64125. [Google Scholar] [CrossRef] [PubMed]
- Mathé-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirié, M.; Gatti, J.L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 β-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Fang, Q.; Wang, L.; Liu, J.; Zhu, Y.; Wang, F.; Li, F.; Werren, J.H.; Ye, G.Y. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 2016, 6, 19604. [Google Scholar] [CrossRef] [PubMed]
- Laurino, S.; Grossi, G.; Pucci, P.; Flagiello, A.; Bufo, S.A.; Bianco, G.; Salvia, R.; Vinson, S.B.; Vogel, H.; Falabella, P. Identification of major Toxoneuron nigriceps venom proteins using an integrated transcriptomic/proteomic approach. Insect Biochem. Mol. Biol. 2016, 76, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Y.; Wang, J.Q.; Zhang, Z.B.; Huang, J.M.; Zhu, J.Y. Unraveling the venom components of an encyrtid endoparasitoid wasp Diversinervus elegans. Toxicon 2017, 136, 15–26. [Google Scholar] [CrossRef]
- Teng, Z.W.; Xiong, S.J.; Xu, G.; Gan, S.Y.; Chen, X.; Stanley, D.; Yan, Z.C.; Ye, G.Y.; Fang, Q. Protein discovery: Combined transcriptomic and proteomic analyses of venom from the endoparasitoid Cotesia chilonis (Hymenoptera: Braconidae). Toxins 2017, 9, 135. [Google Scholar] [CrossRef]
- Liu, N.Y.; Xu, Z.W.; Yan, W.; Ren, X.M.; Zhang, Z.Q.; Zhu, J.Y. Venomics reveals novel ion transport peptide-likes (ITPLs) from the parasitoid wasp. Tetrastichus Brontispae. Toxicon 2018, 14, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wang, R.J.; Cheng, Y.; Du, J.; Volovych, O.; Han, L.B.; Li, J.C.; Hu, Y.; Lu, Z.Y.; Lu, Z.Q.; et al. Insights into the venom protein components of Microplitis mediator, an endoparasitoid wasp. Insect Biochem. Mol. Biol. 2019, 105, 33–42. [Google Scholar] [CrossRef]
- Wilson, D.; Daly, N.L. Venomics: A mini-review. High-Throughput 2018, 7, 19. [Google Scholar] [CrossRef]
- Zbek, R.; Wielsch, N.; Vogel, H.; Lochnit, G.; Foerster, F.; Vilcinskas, A.; Reumont, B.M. Proteo-transcriptomic characterization of the venom from the endoparasitoid wasp Pimpla turionellae with aspects on its biology and evolution. Toxins 2019, 11, 12. [Google Scholar]
- Becchimanzi, A.; Avolio, M.; Bostan, H.; Colantuono, C.; Cozzolino, F.; Mancini, D.; Chiusano, M.L.; Pucci, P.; Pennacchio, F. Venomics of the ectoparasitoid wasp Bracon nigricans. BMC Genom. 2020, 21, 34. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, M.; Rotgans, B.A.; Ni, G.; Dean, J.F.D.; Nahrung, H.F.; Cummins, S.F. Proteomic analysis of the venom and venom sac of the woodwasp, Sirex noctilio—Towards understanding its biological impact. J. Proteomic 2016, 146, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Kessler, A.; Baldwin, I.T. Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 2002, 53, 299–328. [Google Scholar] [CrossRef]
- Arimura, G.I.; Ozawa, R.; Shimoda, T.; Nishioka, T.; Takabayashi, J. Herbivory-induced volatiles elicit defence genes in lima bean leaves. Nature 2000, 406, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Eichenseer, H.; Mathews, M.C.; Bi, J.L.; Murphy, J.B.; Felton, G.W. Salivary glucose oxidase: Multifunctional roles for helicoverpa zea. Arch. Insect Biochem. Physiol. 1999, 42, 99–109. [Google Scholar] [CrossRef]
- Ma, R.; Chen, J.L.; Cheng, D.F.; Sun, J.R. Activation of defense mechanism in wheat by polyphenol oxidase from aphid saliva. J. Agric. Food Chem. 2010, 58, 2410–2418. [Google Scholar] [CrossRef]
- Bordeaux, J.M.; Lorenz, W.W.; Dean, J.F.D. Biomarker genes highlight intraspecific and interspecific variations in the responses of Pinus taeda L. and Pinus radiata D. Don to Sirex noctilio F. acid gland secretions. Tree Physiol. 2012, 32, 1302–1312. [Google Scholar] [CrossRef]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Falabella, P.; Riviello, L.; Caccialupi, P.; Rossodivita, T.; Teresa Valente, M.T.; Luisa De Stradis, M.; Tranfaglia, A.; Varricchio, P.; Gigliotti, S.; Graziani, F.; et al. A gamma-glutamyl transpeptidase of Aphidius ervi venom induces apoptosis in the ovaries of host aphids. Insect Biochem. Mol. Biol. 2007, 37, 453–465. [Google Scholar] [CrossRef]
- Aird, S.D.; Aggarwal, S.; Villar-Briones, A.; Tin, M.M.-Y.; Terada, K.; Mikheyev, A.S. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 2015, 16, 647. [Google Scholar] [CrossRef]
- Yoshida, H. Chemistry of lacquer (urushi). Part I. J. Chem. Soc. 1883, 43, 472–486. [Google Scholar] [CrossRef]
- Mayer, A.M.; Staples, R.C. Laccase: New functions for an old enzyme. Phytochemistry 2002, 60, 551–565. [Google Scholar] [CrossRef]
- Santhanam, N.; Vivanco, J.M.; Decker, S.R.; Reardon, K.F. Expression of industrially relevant laccases: Prokaryotic style. Trends Biotechnol. 2011, 29, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.B.; Fu, C.X.; Yun, J.F.; Shao, H.; Wang, X.Q.; Wang, Z.Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Tobimatsu, Y.; Schuetz, M. Lignin polymerization: How do plants manage the chemistry so well? Curr. Opin. Biotechnol. 2019, 56, 75–81. [Google Scholar] [CrossRef]
- Mikolasch, A.; Schauer, F. Fungal laccases as tools for the synthesis of new hybrid molecules and biomaterials. Appl. Microbiol. Biotechnol. 2009, 82, 605–624. [Google Scholar] [CrossRef] [PubMed]
- Polak, J. Structure/Redox potential relationship of simple organic compounds as potential precursors of dyes for laccase-mediated transformation. Biotechnol. Progr. 2012, 28, 93–102. [Google Scholar] [CrossRef]
- Singh, G.; Bhalla, A.; Kaur, P.; Capalash, N.; Sharma, P. Laccase from prokaryotes: A new source for an old enzyme. Rev. Environ. Sci. Biotechnol. 2011, 10, 309–326. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase: A multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Coy, M.R.; Salem, T.Z.; Denton, J.S.; Kovaleva, E.S.; Liu, Z.; Barber, D.S.; Campbell, J.H.; Davis, D.C.; Buchman, G.W.; Boucias, D.G.; et al. Phenol-oxidizing laccases from the termite gut. Insect Biochem. Mol. Biol. 2010, 40, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Fang, Q.; Wang, L.; Ye, G.Y. Molecular cloning and functional studies of two kazal-type serine protease inhibitors specifically expressed by Nasonia vitripennis venom apparatus. Toxins 2015, 7, 2888–2905. [Google Scholar] [CrossRef]
- Laskowski, M., Jr.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Horita, S.; Ishibashi, J.; Nagata, K.; Miyakawa, T.; Yamakawa, M.; Tanokura, M. Isolation, cDNA cloning, and structure-based functional characterization of oryctin, a hemolymph protein from the coconut rhinoceros beetle, Oryctes rhinoceros, as a novel serine protease inhibitor. J. Biol. Chem. 2010, 285, 30150–30158. [Google Scholar] [CrossRef]
- Zheng, Q.L.; Chen, J.; Nie, Z.M.; Lv, Z.B.; Wang, D.; Zhang, Y.Z. Expression, purification and characterization of a three-domain Kazal-type inhibitor from silkworm pupae (Bombyx mori). Comp. Biochem. Physiol. Biochem. Mol. Biol. 2007, 146, 234–240. [Google Scholar] [CrossRef]
- Kim, B.Y.; Lee, K.S.; Zou, F.M.; Wan, H.; Choi, Y.S.; Yoon, H.J.; Kwon, H.W.; Je, Y.H.; Jin, B.R. Antimicrobial activity of a honeybee (Apis cerana) venom Kazal-type serine protease inhibitor. Toxicon 2013, 76, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zen, K.C.; Choi, H.K.; Krishnamachary, N.; Muthukrishnan, S.; Kramer, K.J. Cloning, expression, and hormonal regulation of an insect β-N-acetylglucosaminidase gene. Insect Biochem. Mol. Biol. 1996, 26, 435–444. [Google Scholar] [CrossRef]
- Nagamatsu, Y.; Yanagisawa, I.; Kimoto, M.; Okamoto, E.; Koga, D. Purification of a chitooligosaccharidolytic β-N-acetylglucosaminidase from Bombyx mori larvae during metamorphosis and the nucleotide sequence of its cDNA. Biosci. Biotechnol. Biochem. 1995, 59, 219–225. [Google Scholar] [CrossRef]
- Bade, M.L.; Wyatt, G.R. Metabolic conversions during putation of the cecropia silkworm. Deposition and utilization of nutrient reserves. Biochem. J. 1962, 83, 470–478. [Google Scholar] [CrossRef]
- Kaznowski, C.; Schneiderman, H.A.; Bryant, P.J. Cuticle secretion during larval growth in Drosophila melanogaster. J. Insect Physiol. 1985, 31, 801–813. [Google Scholar] [CrossRef]
- Harju, M.; Kallioinen, H.; Tossavainen, O. Lactose hydrolysis and other conversions in dairy products: Technological aspects. Int. Dairy J. 2012, 22, 104–109. [Google Scholar] [CrossRef]
- Ranwala, A.P.; Suematsu, C.; Masuda, H. The role of β-galactosidases in the modification of cell wall components during muskmelon fruit ripening. Plant. Physiol. 1992, 100, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.S.; Redgwell, R.J.; MacRae, E.A. Kiwifruit β-galactosidase: Isolation and activity against specific fruit cell-wall polysaccharides. Planta 1993, 189, 499–506. [Google Scholar] [CrossRef]
- Peiren, N.; de Graaf, D.C.; Brunain, M.; Bridts, C.H.; Ebo, D.G.; Stevens, W.J.; Jacobs, F.J. Molecular cloning and expression of icarapin, a novel IgE-binding bee venom protein. FEBS Lett. 2006, 580, 4895–4899. [Google Scholar] [CrossRef]
- Grosch, J.; Hilger, C.; Bilò, M.B.; Kler, S.; Schiener, M.; Dittmar, G.; Bernardin, F.; Lesur, A.; Ollert, M.; Schmidt-Weber, C.B.; et al. Shedding light on the venom proteomes of the allergy-relevant hymenoptera Polistes dominula (European paper wasp) and Vespula spp. (yellow jacket). Toxins 2020, 12, 323. [Google Scholar] [CrossRef]
- Jakob, T.; Rauber, M.M.; Perez-Riverol, A.; Spillner, E.; Blank, S. The honeybee venom major allergen Api m 10 (Icarapin) and its role in diagnostics and treatment of hymenoptera venom allergy. Curr. Allergy Asthma Rep. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Torres, A.M.; Wong, H.Y.; Desai, M.; Moochhala, S.; Kuchel, P.W.; Kini, R.M. Identification of a novel family of proteins in snake venoms: Purification and structural characterization of nawaprin from Naja nigricollis snake venom. J. Biol. Chem. 2003, 278, 40097–40104. [Google Scholar] [CrossRef]
- Wiedow, O.; Schroder, J.M.; Gregory, H.; Young, J.A.; Christophers, E. Elafin: An elastase-specific inhibitor of human skin. Purification, characterization and complete amino acid sequence. J. Biol. Chem. 1990, 265, 14791–14795. [Google Scholar] [CrossRef]
- Hagiwara, K.; Kikuchi, T.; Endo, Y.; Hu, Q.; Usui, K.; Takahashi, M.; Shibata, N.; Kusakabe, T.; Xin, H.; Hoshi, S.; et al. Mouse SWAM1 and SWAM2 are antibacterial proteins composed of a single whey acidic protein motif. J. Immunol. 2003, 170, 1973–1979. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Kuroki, J.; Ito, O.; Kuwada, M.; Tachibana, S. Novel peptide inhibitor (SPAI) of Na+, K+-ATPase from porcine intestine. Biochem. Biophys. Res. Commun. 1989, 164, 496–502. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y.; Lin, W.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of ceratophrys calcarata contains antimicrobial function. Gene 2012, 514, 99–104. [Google Scholar] [CrossRef]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Talbot, P.H.B. The Sirex-Amylostereum-Pinus association. Annu. Rev. Phytopathol. 1977, 15, 41–54. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Jungo, F.; Bougueleret, L.; Xenarios, I.; Poux, S. The UniProtKB/Swiss-Prot Tox-Prot program: A central hub of integrated venom protein data. Toxicon 2012, 60, 551–557. [Google Scholar] [CrossRef]
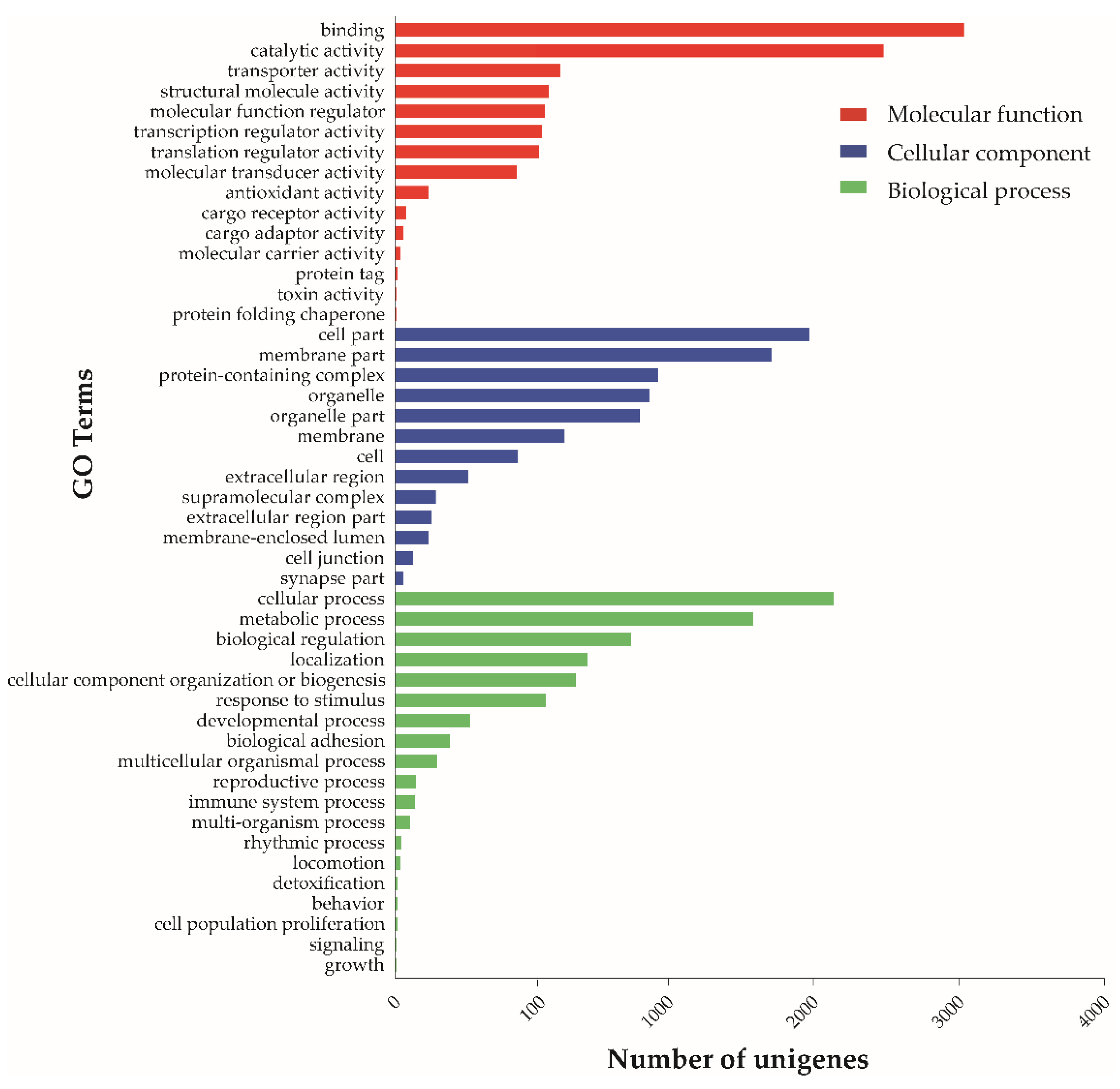
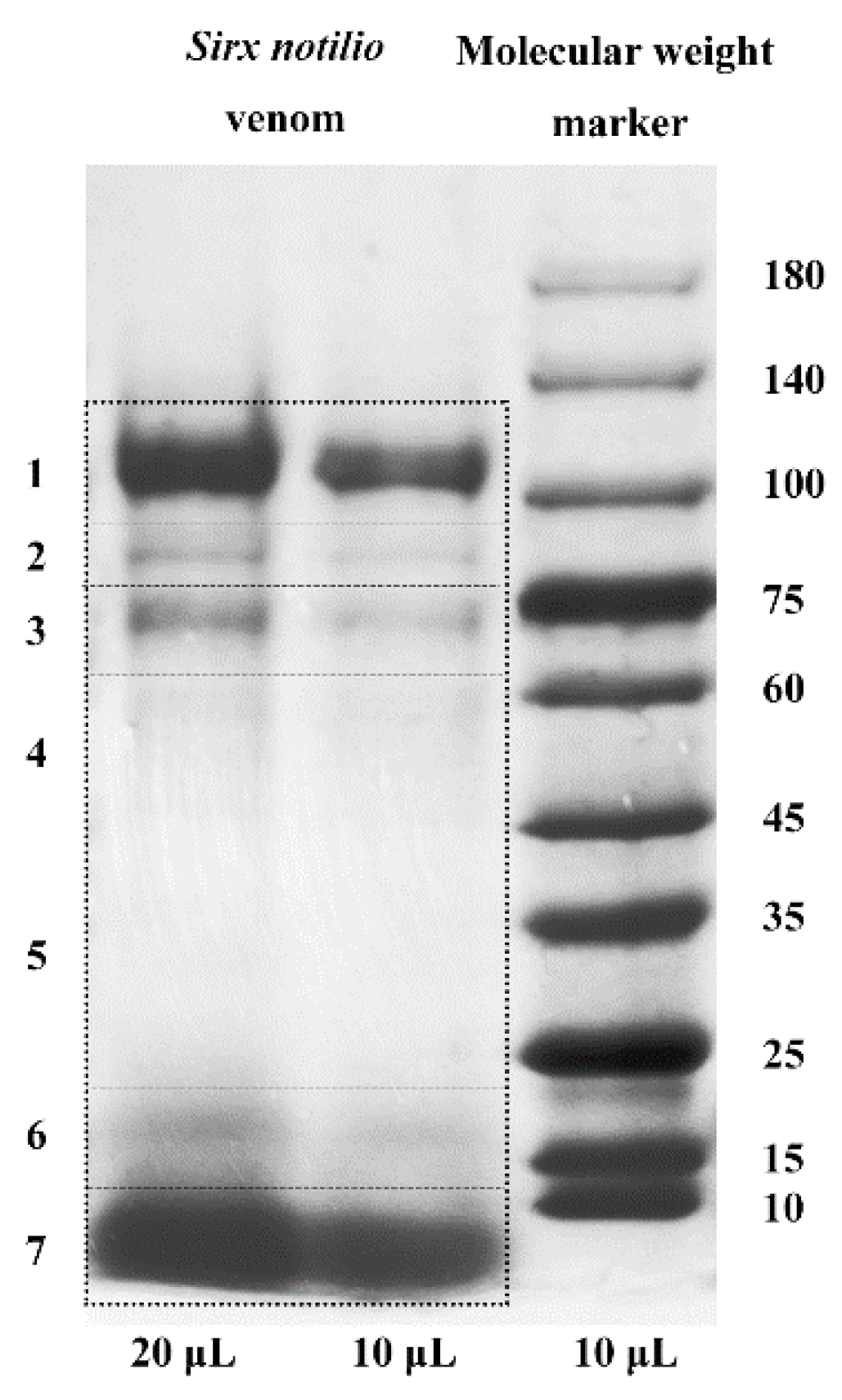
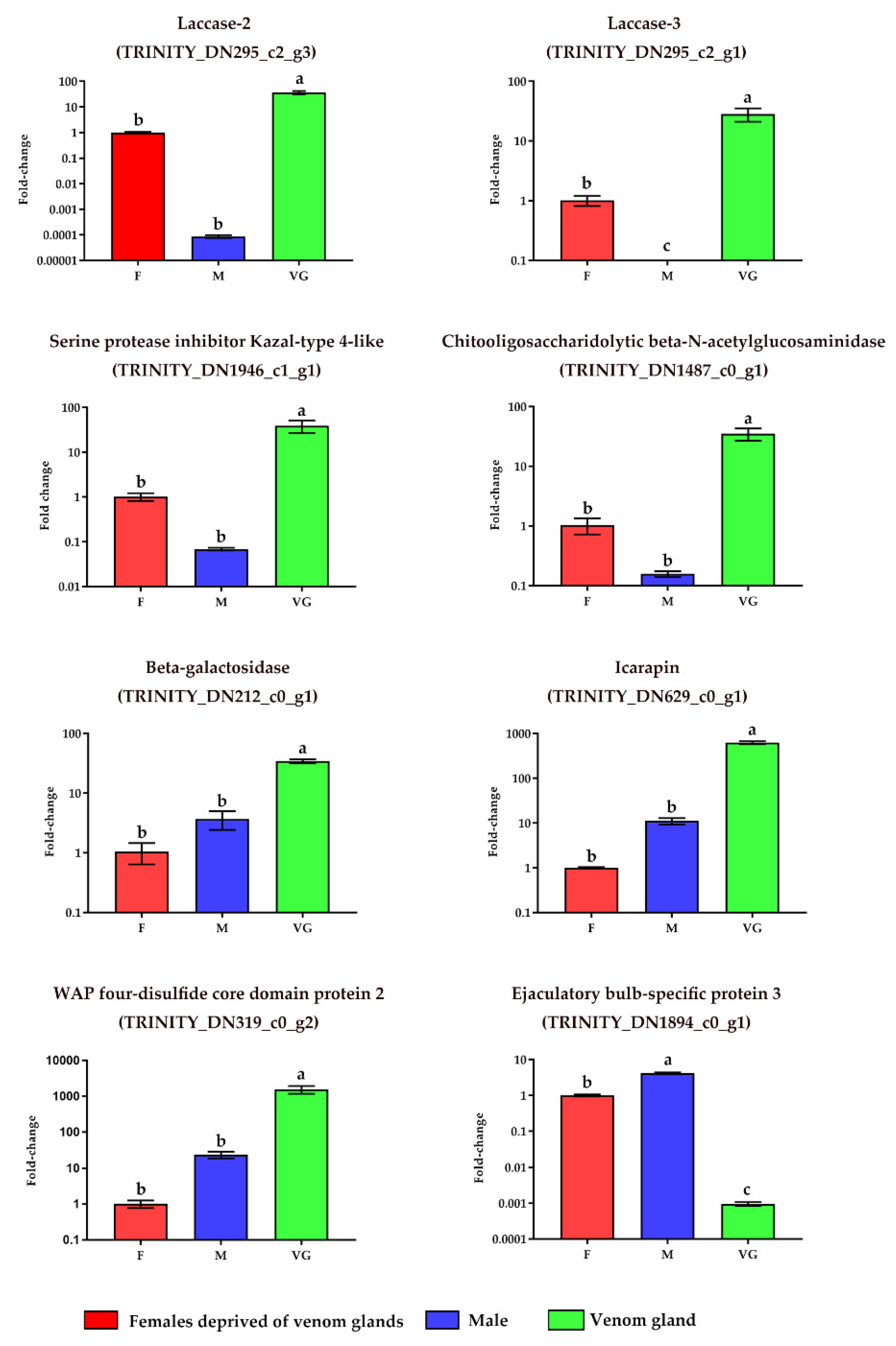
| Sequencing and Assembly Parameters | Value |
|---|---|
| Total number of raw reads | 138,974,566 |
| Total number of clean reads | 137,607,450 |
| Average of GC Content (%) | 43.4 |
| Average of Q20 (%) | 97.54 |
| Average of Q30 (%) | 93.87 |
| Total number of transcripts/unigenes | 18,410/15,036 |
| Average length of transcripts/unigenes (bp) | 1171.30/1085.37 |
| N50 length of transcripts/unigenes (bp) | 1982/1857 |
| Unigene ID | TPM | Best Hit in SwissProt Database | Protein Family | Peptides | Mass |
|---|---|---|---|---|---|
| TRINITY_DN185_c0_g2 | 725.73 | No hits found | None | 12 | 10,259 |
| TRINITY_DN295_c2_g1 | 562.87 | No hits found | Cupredoxin | 53 | 71,463 |
| TRINITY_DN1946_c1_g1 | 342.52 | No hits found | Serine protease inhibitor Kazal-type 4-like | 2 | 11,479 |
| TRINITY_DN1487_c0_g1 | 205.83 | Chitooligosaccharidolytic beta-N-acetylglucosaminidase P49010 Bombyx mori | Beta-hexosaminidase | 87 | 68,318 |
| TRINITY_DN212_c0_g1 | 140.43 | Beta-galactosidase P23780 Mus musculus | Glycoside hydrolase, family 35- Beta-galactosidase 1-like | 54 | 72,755 |
| TRINITY_DN2737_c0_g2 | 42.43 | Ferritin subunit P41822 Aedes aegypti | Beta-hexosaminidase | 7 | 24,849 |
| TRINITY_DN1504_c0_g2 | 40.96 | Basement membrane-specific heparan sulfate proteoglycan core protein P98160 Homo sapiens | None | 3 | 11,856 |
| TRINITY_DN629_c0_g1 | 33.07 | Icarapin Q5EF78 Apis mellifera carnica | None | 9 | 23,713 |
| TRINITY_DN319_c0_g2 | 32.62 | WAP four-disulfide core domain protein 2 Q8CHN3 Rattus norvegicus | WAP superfamily | 4 | 12,716 |
| TRINITY_DN10373_c0_g1 | 29.39 | Probable salivary secreted peptide D8KY57 Bombus ignitus | Transcription activator MBF2 | 4 | 12,877 |
| TRINITY_DN1894_c0_g1 | 24.37 | Ejaculatory bulb-specific protein 3 Q9W1C9 Drosophila melanogaster | Insect odorant-binding protein A10/Ejaculatory bulb-specific protein 3 | 13 | 14,079 |
| TRINITY_DN2893_c1_g2 | 23.82 | Carboxypeptidase Q Q6GQ29 Xenopus laevis | Carboxypeptidase Q | 34 | 54,699 |
| TRINITY_DN2064_c0_g2 | 22.69 | Peptidyl-prolyl cis-trans isomerase FKBP14 Q9NWM8 Homo sapiens | None | 12 | 25,268 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Ren, L.; Wang, M.; Wang, Z.; Fu, N.; Wang, H.; Wang, X.; Ao, T.; Du, W.; Zheng, Z.; et al. Proteo-Transcriptomic Characterization of Sirex nitobei (Hymenoptera: Siricidae) Venom. Toxins 2021, 13, 562. https://doi.org/10.3390/toxins13080562
Gao C, Ren L, Wang M, Wang Z, Fu N, Wang H, Wang X, Ao T, Du W, Zheng Z, et al. Proteo-Transcriptomic Characterization of Sirex nitobei (Hymenoptera: Siricidae) Venom. Toxins. 2021; 13(8):562. https://doi.org/10.3390/toxins13080562
Chicago/Turabian StyleGao, Chenglong, Lili Ren, Ming Wang, Zhengtong Wang, Ningning Fu, Huiying Wang, Xiaochen Wang, Tegen Ao, Wensheng Du, Zijin Zheng, and et al. 2021. "Proteo-Transcriptomic Characterization of Sirex nitobei (Hymenoptera: Siricidae) Venom" Toxins 13, no. 8: 562. https://doi.org/10.3390/toxins13080562
APA StyleGao, C., Ren, L., Wang, M., Wang, Z., Fu, N., Wang, H., Wang, X., Ao, T., Du, W., Zheng, Z., Li, H., & Shi, J. (2021). Proteo-Transcriptomic Characterization of Sirex nitobei (Hymenoptera: Siricidae) Venom. Toxins, 13(8), 562. https://doi.org/10.3390/toxins13080562






