Anticoagulant Activity of Naja nigricollis Venom Is Mediated by Phospholipase A2 Toxins and Inhibited by Varespladib
Abstract
1. Introduction
2. Results
2.1. Anticoagulant Effects of Crude N. nigricollis Venom and Quantification of Inhibition by Small Molecule Toxin Inhibitors
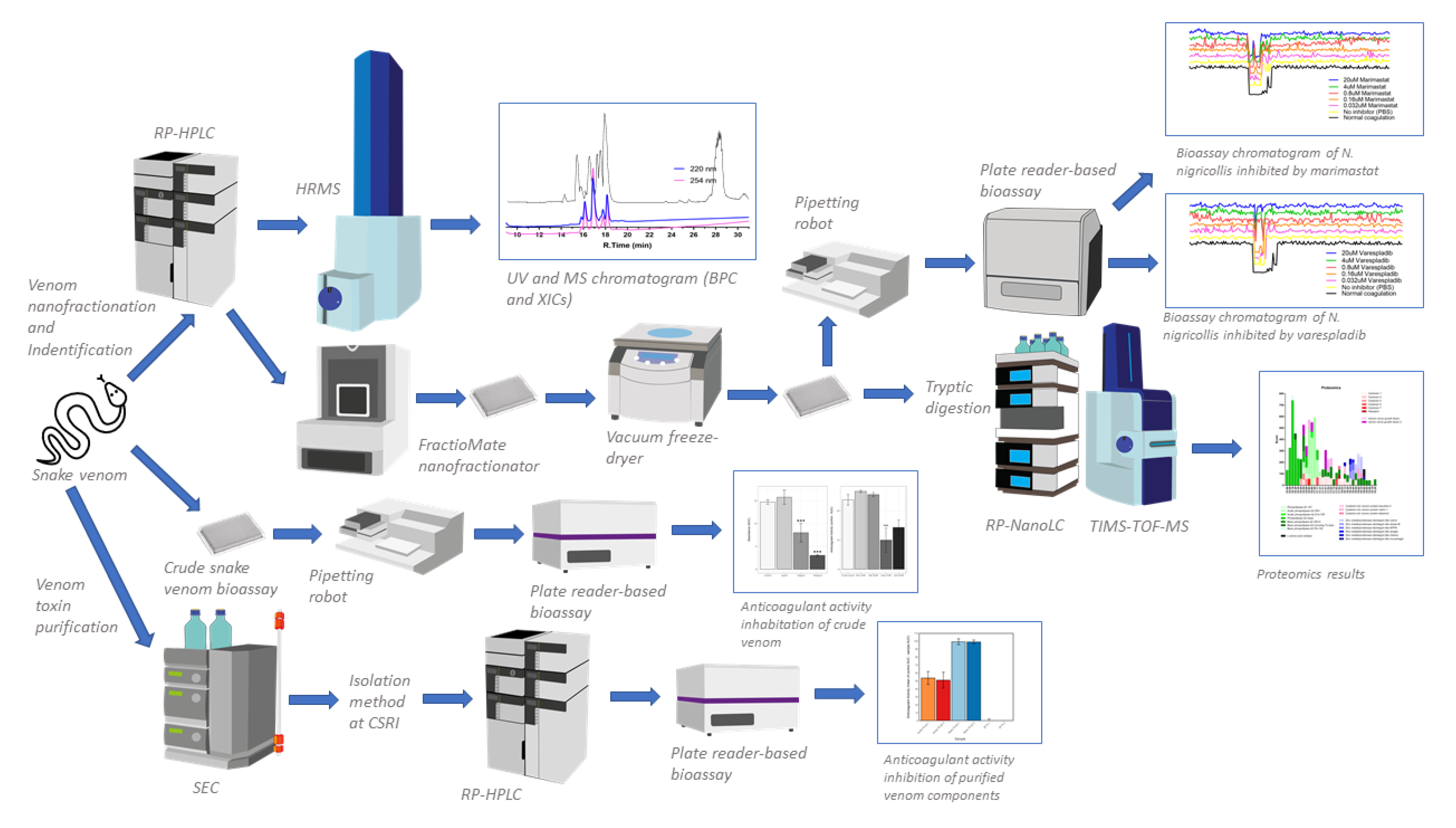
2.2. Identification of Anticoagulant Venom Toxins Via Nanofractionation, Bioactivity Testing and Proteomics
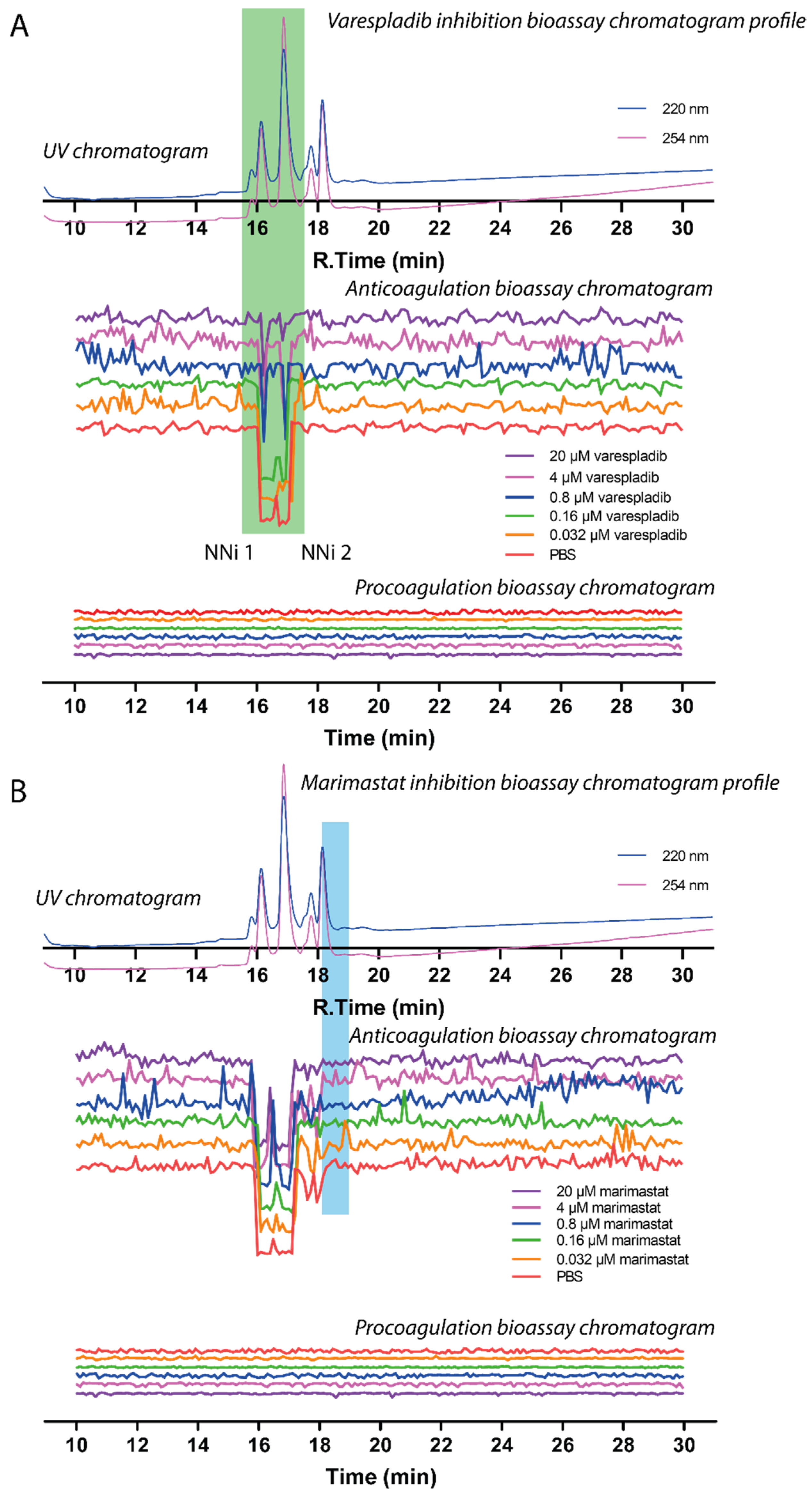
2.3. Assessing Inhibition of Nanofractionated Toxins by Varespladib and Marimastat
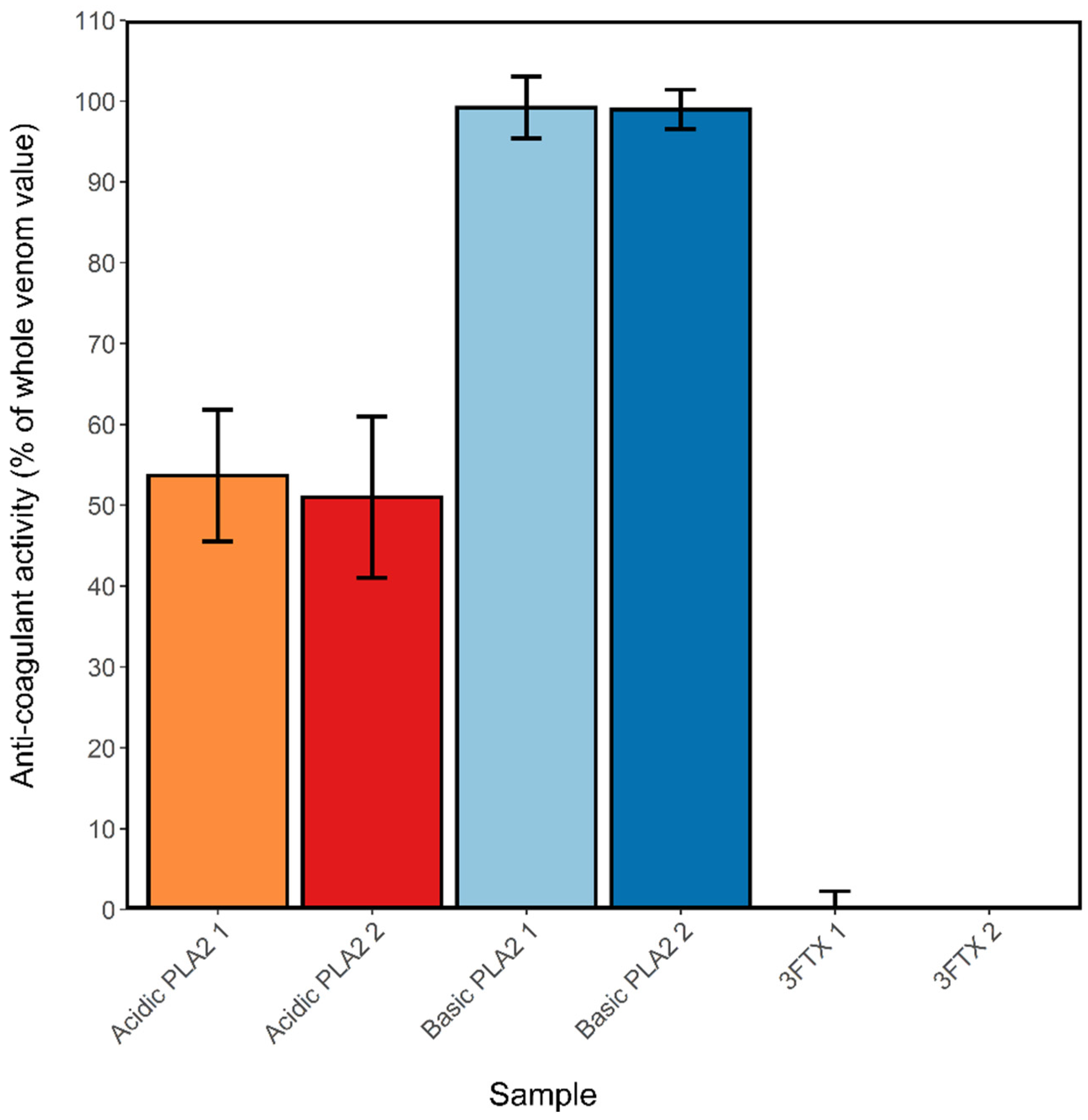
2.4. Anticoagulant Effects of Isolated and Purified N. nigricollis Toxins
3. Discussion
4. Materials and Methods
4.1. Chemical and Biological Reagents
4.2. Coagulopathic Activity of Crude N. nigricollis Venom and Assessment of Inhibition with Varespladib and Marimastat
4.3. HPLC-MS-Nanofractionation and Simultaneous Bioassay of N. nigricollis Venom
4.4. Assessment of Inhibition of Nanofractionated Anticoagulant Toxins with Varespladib and Marimastat
4.5. In-Solution Tryptic Digestions from Nanofractionated 384-Well Plates
4.6. Proteomics of Coagulopathic Venom Toxins
4.7. Chromatographic Isolation of N. nigricollis Venom Components
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kasturiratne, A.; Wickremasinghe, A.R.; De Silva, N.; Gunawardena, N.K.; Pathmeswaran, A.; Premaratna, R.; Savioli, L.; Lalloo, D.G.; De Silva, H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008, 5, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017, 3, 17063. [Google Scholar] [CrossRef] [PubMed]
- Chippaux, J.P. Snake-bites: Appraisal of the global situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.; Rathuwithana, A.C.; Kasturiratne, A.; Lalloo, D.G.; de Silva, H.J. Underestimation of snakebite mortality by hospital statistics in the Monaragala District of Sri Lanka. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 693–695. [Google Scholar] [CrossRef]
- Longbottom, J.; Shearer, F.M.; Devine, M.; Alcoba, G.; Chappuis, F.; Weiss, D.J.; Ray, S.E.; Ray, N.; Warrell, D.A.; Bill, F.; et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet 2018, 6736, 1–12. [Google Scholar] [CrossRef]
- Warrell, D.A. Clinical Toxicology of Snakebite in Asia. In Handbook of Clinical Toxicology of Animal Venoms and Poisons; Meier, J., White, J., Eds.; Informa Healthcare USA Inc.: New York, NY, USA, 2008; pp. 496–588. ISBN 978-0-8493-4489-3. [Google Scholar]
- Warrell, D.A. Clinical Toxinology of Snakebite in Africa and the Middle East/Arabian Penninsula. In Handbook of Clinical Toxicology of Animal Venoms and Poisons; Meier, J., White, J., Eds.; Informa Healthcare USA Inc.: New York, NY, USA, 2008; pp. 433–492. ISBN 978-0-8493-4489-3. [Google Scholar]
- Blaylock, R.S.; Lichtman, A.R.; Potgieter, P.D. Clinical manifestations of Cape cobra (Naja nivea) bites. A report of 2 cases. S. Afr. Med. J. 1985, 68, 342–344. [Google Scholar]
- Warrell, D.A.; Barnes, H.J.; Piburn, M.F. Neurotoxic effects of bites by the Egyptian cobra (Naja haje) in Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1976, 70, 78–79. [Google Scholar] [CrossRef]
- Zouari, N.; Choyakh, F. Les effets neurotoxiques du venin de cobra (Naja haje haje) sur la jonction neuromusculaire. Étude électroclinique de deux cas en Tunisie. Neurophysiol. Clin. 1995, 25, 59–65. [Google Scholar] [CrossRef]
- Strover, H.M. Observations on two cases of snake-bite by Naja nigricollis ss mossambica. Cent. Afr. J. Med. 1973, 19, 12–13. [Google Scholar]
- Tilbury, C.R. Observations on the bite of the Mozambique spitting cobra (Naja mossambica mossambica). S. Afr. Med. J. 1982, 61, 308–313. [Google Scholar]
- Warrell, D.A.; Greenwood, B.M.; Davidson, N.M.; Ormerod, L.D.; Prentice, C.R. Necrosis, haemorrhage and complement depletion following bites by the spitting cobra (Naja nigricollis). Q. J. Med. 1973, 45, 1–22. [Google Scholar]
- Wüster, W.; Crookes, S.; Ineich, I.; Mane, Y.; Pook, C.E.; Trape, J.F.; Broadley, D.G. The phylogeny of cobras inferred from mitochondrial DNA sequences: Evolution of venom spitting and the phylogeography of the African spitting cobras (Serpentes: Elapidae: Naja nigricollis complex). Mol. Phylogenet. Evol. 2007, 45, 437–453. [Google Scholar] [CrossRef]
- Pugh, R.N.H.; Theakston, D.G. Incidence and Mortality of Snake Bite in Savanna Nigeria. Lancet 1980, 316, 1181–1183. [Google Scholar] [CrossRef]
- Pugh, R.N.H.; Theakston, R.D.G.; Reid, H.A.; Bhar, I.S. Malumfashi Endemic Diseases Research Project, XIII. Ann. Trop. Med. Parasitol. 1980, 74, 523–530. [Google Scholar] [CrossRef]
- Habib, A.G.; Gebi, U.I.; Onyemelukwe, G.C. Snake bite in Nigeria. Afr. J. Med. Med. Sci. 2001, 30, 171–178. [Google Scholar]
- Rivel, M.; Solano, D.; Herrera, M.; Vargas, M.; Villalta, M.; Segura, Á.; Arias, A.S.; León, G.; Gutiérrez, J.M. Pathogenesis of dermonecrosis induced by venom of the spitting cobra, Naja nigricollis: An experimental study in mice. Toxicon 2016, 119, 171–179. [Google Scholar] [CrossRef]
- Méndez, I.; Gutiérrez, J.M.; Angulo, Y.; Calvete, J.J.; Lomonte, B. Comparative study of the cytolytic activity of snake venoms from African spitting cobras (Naja spp., Elapidae) and its neutralization by a polyspecific antivenom. Toxicon 2011, 58, 558–564. [Google Scholar] [CrossRef]
- Dutta, S.; Sinha, A.; Dasgupta, S.; Mukherjee, A.K. Binding of a Naja naja venom acidic phospholipase A 2 cognate complex to membrane-bound vimentin of rat L6 cells: Implications in cobra venom-induced cytotoxicity. Biochim. Biophys. Acta Biomembr. 2019, 1861, 958–977. [Google Scholar] [CrossRef]
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef]
- McCleary, R.J.R.; Kini, R.M. Snake bites and hemostasis/thrombosis. Thromb. Res. 2013, 132, 642–646. [Google Scholar] [CrossRef]
- Sundell, I.B.; Rånby, M.; Zuzel, M.; Robinson, K.A.; Theakston, R.D.G. In vitro procoagulant and anticoagulant properties of Naja naja naja venom. Toxicon 2003, 42, 239–247. [Google Scholar] [CrossRef]
- Dutta, S.; Gogoi, D.; Mukherjee, A.K. Anticoagulant mechanism and platelet deaggregation property of a non-cytotoxic, acidic phospholipase A2 purified from Indian cobra (Naja naja) venom: Inhibition of anticoagulant activity by low molecular weight heparin. Biochimie 2015, 110, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Osipov, A.V.; Filkin, S.Y.; Makarova, Y.V.; Tsetlin, V.I.; Utkin, Y.N. A new type of thrombin inhibitor, noncytotoxic phospholipase A2, from the Naja haje cobra venom. Toxicon 2010, 55, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-S.; Fletcher, J.E.; Smith, L.A. Factors influencing the hemolysis of human erythrocytes by cardiotoxins from Naja naja kaouthia and Naja naja atra venoms and a phospholipase A2with cardiotoxin-like activities from Bungarus fasciatus venom. Toxicon 1989, 27, 247–257. [Google Scholar] [CrossRef]
- Banerjee, Y.; Mizuguchi, J.; Iwanaga, S.; Kini, R.M. Hemextin AB complex, a unique anticoagulant protein complex from Hemachatus haemachatus (African Ringhals cobra) venom that inhibits clot initiation and factor VIIa activity. J. Biol. Chem. 2005, 280, 42601–42611. [Google Scholar] [CrossRef]
- Louw, A.I.; Visser, L. The synergism of cardiotoxin and phospholipase A2 in hemolysis. BBA Biomembr. 1978, 512, 163–171. [Google Scholar] [CrossRef]
- Kazandjian, T.D.; Petras, D.; Robinson, S.D.; van Thiel, J.; Greene, H.W.; Arbuckle, K.; Barlow, A.; Carter, D.A.; Wouters, R.M.; Whiteley, G.; et al. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science 2021, 371, 386–390. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Kini, R.M. Serine proteases affecting blood coagulation and fibrinolysis from snake venoms. Pathophysiol. Haemost. Thromb. 2006, 34, 200–204. [Google Scholar] [CrossRef]
- Phillips, D.J.; Swenson, S.D.; Markland, F.S.J. Thrombin-Like Snake Venom Serine Proteinases. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 139–154. ISBN 978-0-8493-9165-1. [Google Scholar]
- Jagadeesha, D.K.; Shashidharamurthy, R.; Girish, K.S.; Kemparaju, K. A non-toxic anticoagulant metalloprotease: Purification and characterization from Indian cobra (Naja naja naja) venom. Toxicon 2002, 40, 667–675. [Google Scholar] [CrossRef]
- Bittenbinder, M.A.; Zdenek, C.N.; Op Den Brouw, B.; Youngman, N.J.; Dobson, J.S.; Naude, A.; Vonk, F.J.; Fry, B.G. Coagulotoxic cobras: Clinical implications of strong anticoagulant actions of african spitting naja venoms that are not neutralised by antivenom but are by LY315920 (varespladib). Toxins 2018, 10, 516. [Google Scholar] [CrossRef]
- Petras, D.; Sanz, L.; Segura, Á.; Herrera, M.; Villalta, M.; Solano, D.; Vargas, M.; León, G.; Warrell, D.A.; Theakston, R.D.G.; et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the Pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011, 10, 1266–1280. [Google Scholar] [CrossRef]
- Evans, H.J. Cleavage of the Aa-chain of fibrinogen and the A-polymer of fibrin by the venom of the spitting cobra (Naja nigricollis). Biochem. Biophys. Acta 1981, 660, 219–226. [Google Scholar]
- Mackay, N.; Ferguson, J.C.; Mcnicol, G.P. Effects of three cobra venoms on blood coagulation, platelet aggregation, and fibrinolysis. J. Clin. Pathol. 1969, 22, 304–311. [Google Scholar] [CrossRef]
- Stefansson, S.; Kini, R.M.; Evans, H.J. The Basic Phospholipase A2 from Naja nigricollis Venom Inhibits the Prothrombinase Complex by a Novel Nonenzymatic Mechanism. Biochemistry 1990, 29, 7742–7746. [Google Scholar] [CrossRef]
- Kini, R.M.; Evans, H.J. The role of enzymatic activity in inhibition of the extrinsic tenase complex by phospholipase A2 isoenzymes from Naja nigricollis venom. Toxicon 1995, 33, 1585–1590. [Google Scholar] [CrossRef]
- Stefansson, S.; Kini, R.M.; Evans, H.J. The inhibition of clotting complexes from the extrinsic coagulation cascade by the phospholipase A2 isoenzymes from Naja nigricollis venom. Thromb. Res. 1989, 55, 481–491. [Google Scholar] [CrossRef]
- Kerns, R.T.; Kini, R.M.; Stefansson, S.; Evans, H.J. Targeting of venom phospholipases: The strongly anticoagulant phospholipase A2 from Naja nigricollis venom binds to coagulation factor Xa to inhibit the prothrombinase complex. Arch. Biochem. Biophys. 1999, 369, 107–113. [Google Scholar] [CrossRef]
- Kini, R.M.; Evans, H.J. Mechanism of platelet effects of cardiotoxins from Naja nigricollis crawshawii (spitting cobra) snake venom. Thromb. Res. 1988, 52, 185–195. [Google Scholar] [CrossRef]
- Still, K.B.M.; Slagboom, J.; Kidwai, S.; Xie, C.; Zhao, Y.; Eisses, B.; Jiang, Y.; Vonk, F.J.; Somsen, G.W.; Casewell, N.R.; et al. Development of high-throughput screening assays for profiling snake venom Phospholipase A2 activity after high-resolution chromatographic fractionation. Toxicon 2020, 178, 61–68. [Google Scholar] [CrossRef]
- Ainsworth, S.; Petras, D.; Engmark, M.; Süssmuth, R.D.; Whiteley, G.; Albulescu, L.O.; Kazandjian, T.D.; Wagstaff, S.C.; Rowley, P.; Wüster, W.; et al. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteom. 2018, 172, 173–189. [Google Scholar] [CrossRef]
- Davidson, T.M.; Eisner, J. Colective Review: United States coral snakes. Wilderness Environ. Med. 1996, 1, 38–45. [Google Scholar] [CrossRef]
- Slagboom, J.; Mladić, M.; Xie, C.; Kazandjian, T.D.; Vonk, F.; Somsen, G.W.; Casewell, N.R.; Kool, J. High throughput screening and identification of coagulopathic snake venom proteins and peptides using nanofractionation and proteomics approaches. PLoS Negl. Trop. Dis. 2020, 14, e0007802. [Google Scholar] [CrossRef]
- Bittenbinder, M.A.; Dobson, J.S.; Zdenek, C.N.; op den Brouw, B.; Naude, A.; Vonk, F.J.; Fry, B.G. Differential destructive (non-clotting) fibrinogenolytic activity in Afro-Asian elapid snake venoms and the links to defensive hooding behavior. Toxicol. Vitr. 2019, 60, 330–335. [Google Scholar] [CrossRef]
- Kini, R.M. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon 2005, 45, 1147–1161. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, C.; Li, Y.; Bao, J. Toxicon Inhibition of platelet aggregation and blood coagulation by a P-III class metalloproteinase purified from Naja atra venom. Toxicon 2020, 187, 223–231. [Google Scholar] [CrossRef]
- Olaoba, O.T.; dos Santos, P.K.; Selistre-de-Araujo, H.S.; Ferreira de Souza, D.H. Snake Venom Metalloproteinases (SVMPs): A structure-function update. Toxicon X 2020, 7, 100052. [Google Scholar] [CrossRef]
- Xie, C.; Albulescu, L.-O.; Still, K.; Slagboom, J.; Zhao, Y.; Jiang, Z.; Somsen, G.; Vonk, F.; Casewell, N.; Kool, J. Varespladib inhibits the phospholipase A2 and coagulopathic activities of venom components from haemotoxic snakes. Biomedicines 2020, 8, 165. [Google Scholar] [CrossRef]
- Youngman, N.J.; Walker, A.; Naude, A.; Coster, K.; Sundman, E.; Fry, B.G. Varespladib (LY315920) neutralises phospholipase A2 mediated prothrombinase-inhibition induced by Bitis snake venoms. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 236, 108818. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Zhang, D.; Xiao, H.; Xiong, S.; Huang, C. Exploration of the inhibitory potential of varespladib for snakebite envenomation. Molecules 2018, 23, 391. [Google Scholar] [CrossRef]
- Liu, C.C.; Wu, C.J.; Hsiao, Y.C.; Yang, Y.H.; Liu, K.L.; Huang, G.J.; Hsieh, C.H.; Chen, C.K.; Liaw, G.W. Snake venom proteome of Protobothrops mucrosquamatus in Taiwan: Delaying venom-induced lethality in a rodent model by inhibition of phospholipase A2 activity with varespladib. J. Proteom. 2021, 234, 104084. [Google Scholar] [CrossRef] [PubMed]
- Salvador, G.H.M.; Gomes, A.A.S.; Bryan-Quirós, W.; Fernández, J.; Lewin, M.R.; Gutiérrez, J.M.; Lomonte, B.; Fontes, M.R.M. Structural basis for phospholipase A2-like toxin inhibition by the synthetic compound Varespladib (LY315920). Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lewin, M.; Samuel, S.; Merkel, J.; Bickler, P. Varespladib (LY315920) appears to be a potent, broad-spectrum, inhibitor of snake venom phospholipase A2 and a possible pre-referral treatment for envenomation. Toxins 2016, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Lewin, M.R.; Gilliam, L.L.; Gilliam, J.; Samuel, S.P.; Bulfone, T.C.; Bickler, P.E.; Gutiérrez, J.M. Delayed LY333013 (oral) and LY315920 (intravenous) reverse severe neurotoxicity and rescue juvenile pigs from lethal doses of Micrurus fulvius (eastern coral snake) venom. Toxins 2018, 10, 479. [Google Scholar] [CrossRef]
- Lewin, M.R.; María Gutiérrez, J.; Samuel, S.P.; Herrera, M.; Bryan-Quirós, W.; Lomonte, B.; Bickler, P.E.; Bulfone, T.C.; Williams, D.J. Delayed oral LY333013 rescues mice from highly neurotoxic, lethal doses of papuan taipan (Oxyuranus scutellatus) venom. Toxins 2018, 10, 380. [Google Scholar] [CrossRef]
- Zinenko, O.; Tovstukha, I.; Korniyenko, Y. PLA2 inhibitor varespladib as an alternative to the antivenom treatment for bites from nikolsky’s viper Vipera berus nikolskii. Toxins 2020, 12, 356. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Lewin, M.R.; Williams, D.J.; Lomonte, B. Varespladib (LY315920) and methyl varespladib (LY333013) abrogate or delay lethality induced by presynaptically acting neurotoxic snake venoms. Toxins 2020, 12, 131. [Google Scholar] [CrossRef]
- Neumann, C.; Slagboom, J.; Somsen, G.W.; Vonk, F.; Casewell, N.R.; Cardoso, C.L.; Kool, J. Development of a generic high-throughput screening assay for profiling snake venom protease activity after high-resolution chromatographic fractionation. Toxicon 2020, 178, 61–68. [Google Scholar] [CrossRef]
- Still, K.B.M.; Nandlal, R.S.S.; Slagboom, J.; Somsen, G.W.; Casewell, N.R.; Kool, J. Multipurpose HTS coagulation analysis: Assay development and assessment of coagulopathic snake venoms. Toxins 2017, 9, 382. [Google Scholar] [CrossRef]
- Kazandjian, T.D.; Robinson, S.D.; Greene, H.W.; Carter, D.A.; Wouters, R.M.; Wagstaff, S.C.; Arias, A.S.; Albulescu, L.-O.; McCabe, C.V.; da Silva, R.R.; et al. Convergent Evolution of Pain-Inducing Defensive Venom Components in Spitting Cobras. bioRxiv 2020. [Google Scholar] [CrossRef]

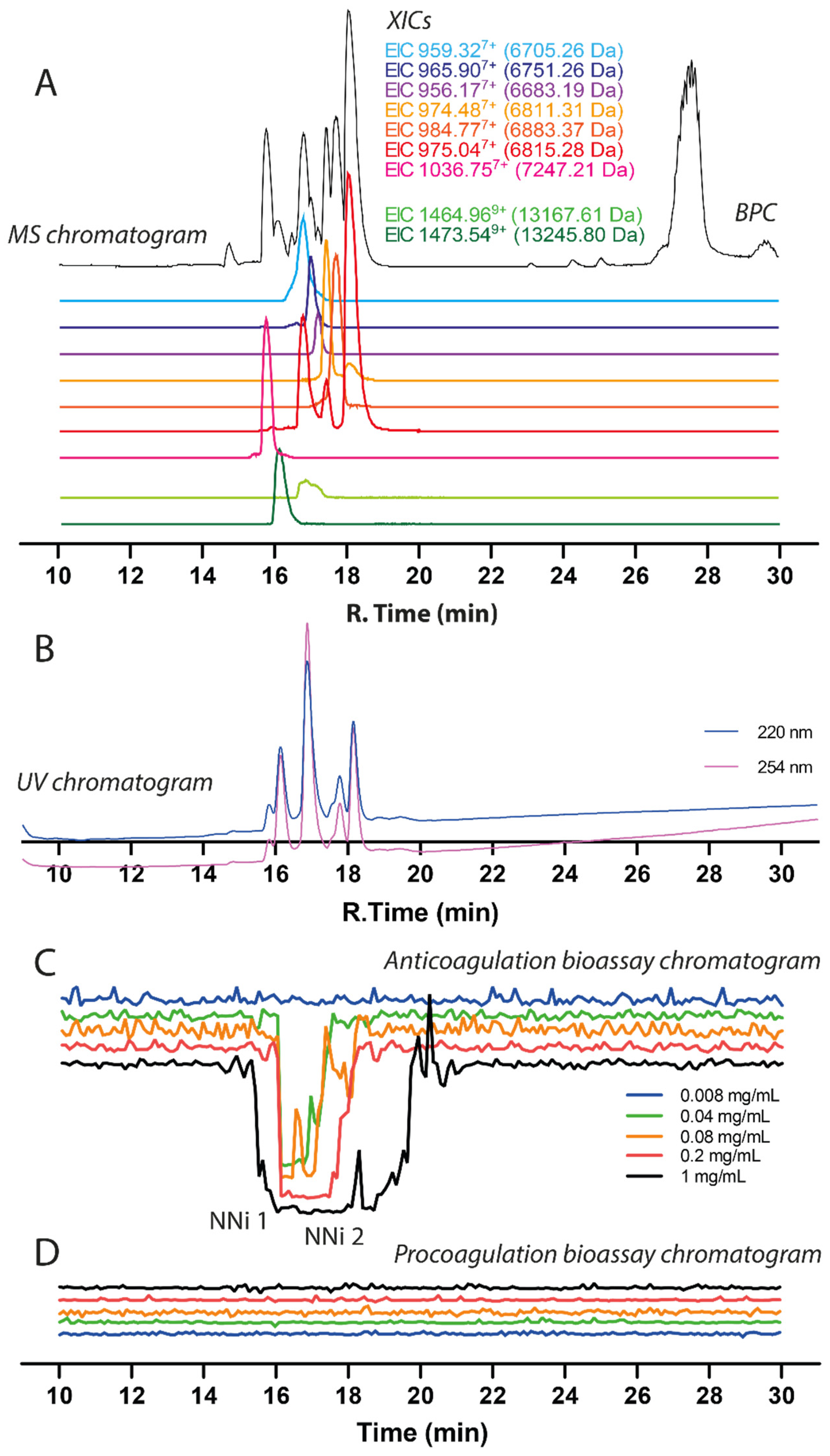
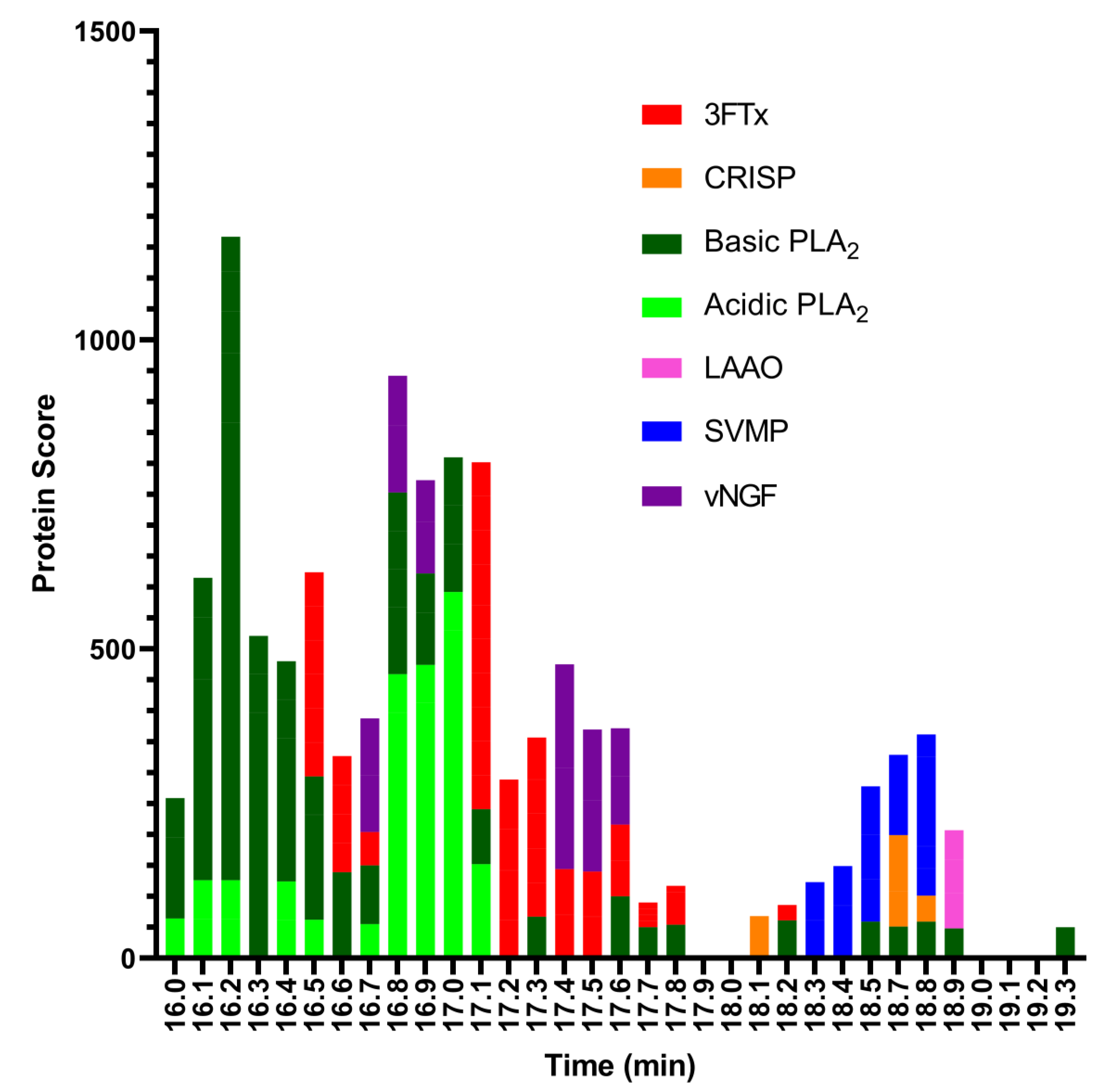
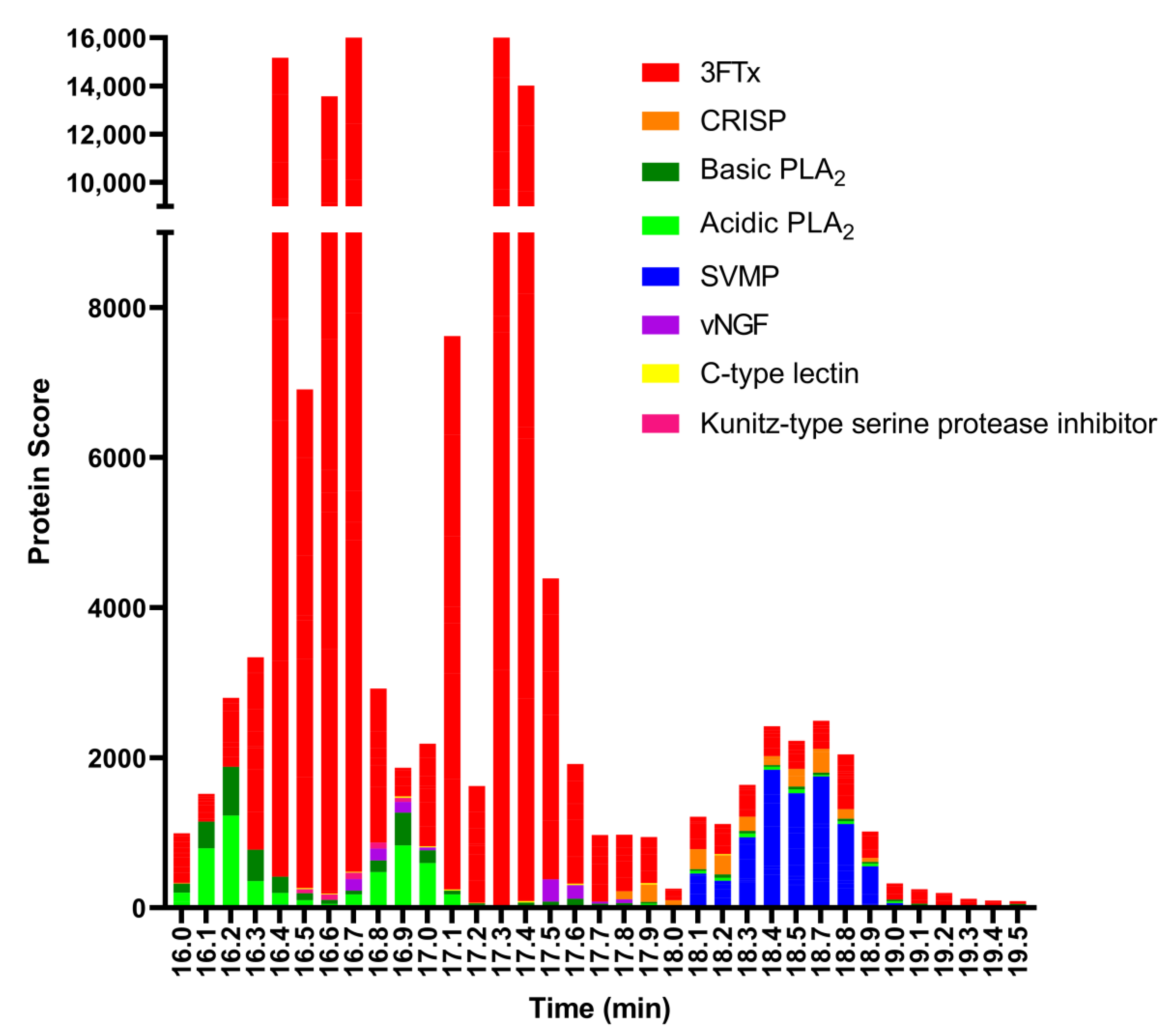
| Retention Time (min) | m/z Value (Charge) | MS Accurate Mass (Da) | Mascot Protein Hits (Protein Identification) | Mascot Exact Mass (Da) | Species-Specific Database Hits | Toxin Class |
|---|---|---|---|---|---|---|
| 15.7 | 1036.75(+7) | 7247.21 | Three-finger toxin | N.A. | N.A. | 3FTx |
| 13,217 | Basic Phospholipase A2 (B1) ** | N.A. | N.A. | PLA2 | ||
| 16.1 | 1473.54 (+9) | 13,245.80 | PA2B4_NAJNG (Naja nigricollis) Phospholipase A2 Basic (B2) ** | 13,244.90 | T0927_T1551_T1904_T2072 | PLA2 |
| 13,184 | Acidic Phospholipase A2 (A1) ** | N.A. | N.A. | PLA2 | ||
| 13,256 | Acidic Phospholiase A2 (A2) ** | N.A. | N.A. | PLA2 | ||
| 16.8 | 1464.96 (+9) | 13,167.61 | PA2A1_NAJMO (Naja mossambica) Acidic Phospholipase A2 CM-I | 13,195.75 | T1049_T1649 | PLA2 |
| 16.8 | 959.32(+7) | 6705.26 | 3SA4_NAJMO (Naja mossambica) Cytotoxin 4 | 6702.34 | T1903_R_6_8306_350 | 3FTx |
| 17.0 | 965.90(+7) | 6751.26 | 3SA9_NAJNA (Naja naja) Cytotoxin 9 (3FTx group 2) ** | 6750.36 * | T0802_R_6_4763_L_609 | 3FTx |
| 17.2 | 956.17(+7) | 6683.19 | 3SAN_NAJNG (Naja nigricollis) Naniproin | 6682.41 | T2051_R_3_6355_L_312 | 3FTx |
| 17.4 | 974.48(+7) | 6811.31 | 3SA5_NAJAT (Naja atra) Cytotoxin 5 | 6810.35 * | T2073_R_2_4857_L_300 | 3FTx |
| 17.7 | 984.77(+7) | 6883.37 | Three-finger toxin (3FTx group 2) ** | N.A. | N.A. | 3FTx |
| 18.0 | 975.04(+7) | 6815.28 | 3SA1_NAJPA (Naja pallida) Cytotoxin 1 (3FTx group 2) ** | 6814.31 | T0939_T1746_R_11_8132_L_555 | 3FTx |
| 6817 | Three-finger toxin (3FTx group 1) ** | N.A. | N.A | 3FTx |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazandjian, T.D.; Arrahman, A.; Still, K.B.M.; Somsen, G.W.; Vonk, F.J.; Casewell, N.R.; Wilkinson, M.C.; Kool, J. Anticoagulant Activity of Naja nigricollis Venom Is Mediated by Phospholipase A2 Toxins and Inhibited by Varespladib. Toxins 2021, 13, 302. https://doi.org/10.3390/toxins13050302
Kazandjian TD, Arrahman A, Still KBM, Somsen GW, Vonk FJ, Casewell NR, Wilkinson MC, Kool J. Anticoagulant Activity of Naja nigricollis Venom Is Mediated by Phospholipase A2 Toxins and Inhibited by Varespladib. Toxins. 2021; 13(5):302. https://doi.org/10.3390/toxins13050302
Chicago/Turabian StyleKazandjian, Taline D., Arif Arrahman, Kristina B. M. Still, Govert W. Somsen, Freek J. Vonk, Nicholas R. Casewell, Mark C. Wilkinson, and Jeroen Kool. 2021. "Anticoagulant Activity of Naja nigricollis Venom Is Mediated by Phospholipase A2 Toxins and Inhibited by Varespladib" Toxins 13, no. 5: 302. https://doi.org/10.3390/toxins13050302
APA StyleKazandjian, T. D., Arrahman, A., Still, K. B. M., Somsen, G. W., Vonk, F. J., Casewell, N. R., Wilkinson, M. C., & Kool, J. (2021). Anticoagulant Activity of Naja nigricollis Venom Is Mediated by Phospholipase A2 Toxins and Inhibited by Varespladib. Toxins, 13(5), 302. https://doi.org/10.3390/toxins13050302






