Characterization and Diversity of Microcystins Produced by Cyanobacteria from the Curonian Lagoon (SE Baltic Sea)
Abstract
1. Introduction
2. Results
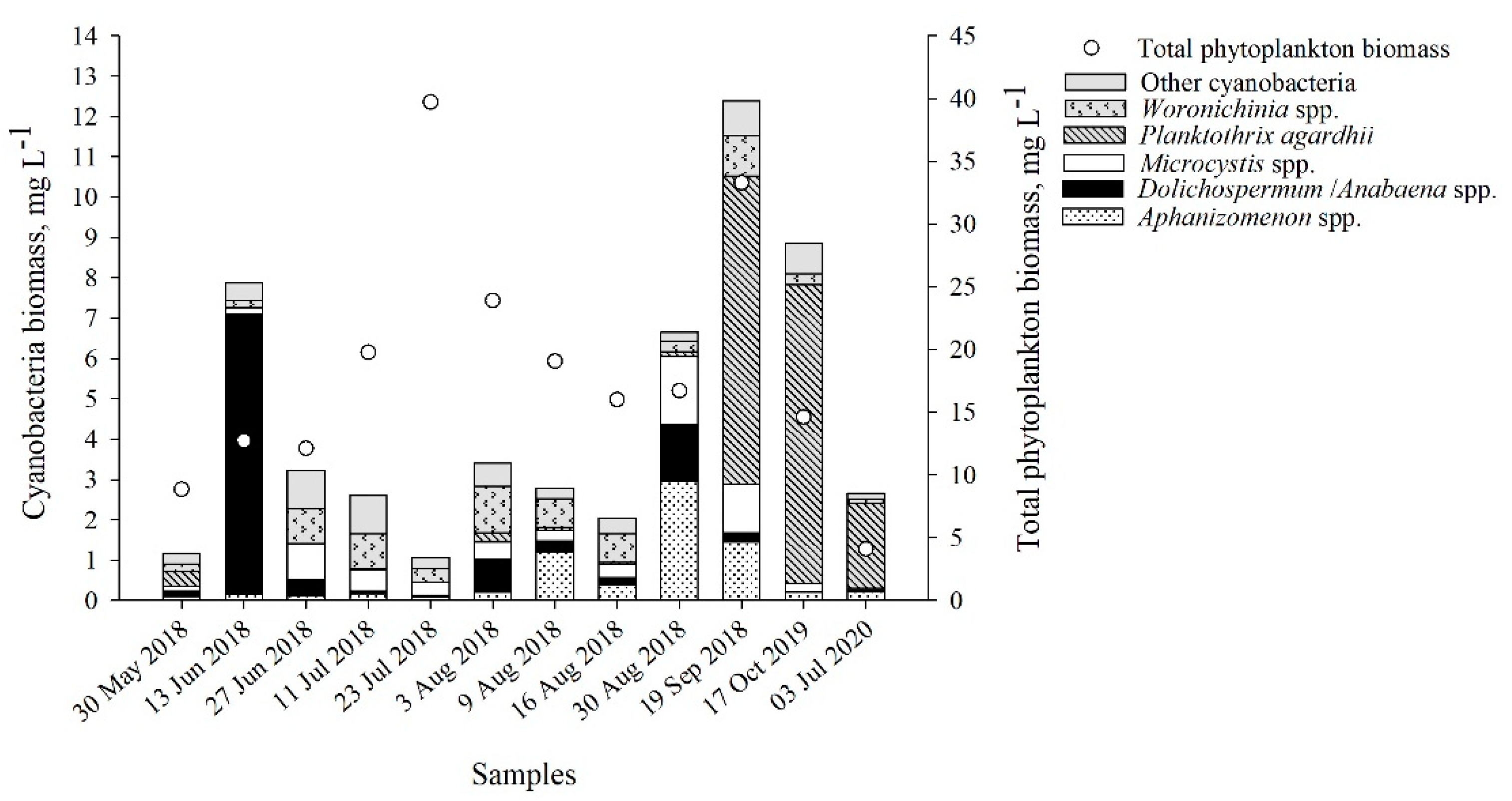
2.1. Cyanobacteria Community
2.2. Microcystins Diversity
2.3. Genetic Analysis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Water Samples
5.2. Phytoplankton Analysis
5.3. DNR Extraction and PCR
5.4. Microcystins Analysis
5.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watson, S.B.; Whitton, B.A.; Higgins, S.N.; Paerl, H.W.; Brooks, B.W.; Wehr, J.D. Harmful Algal Blooms. In Freshwater Algae of North America; Academic Press: Cambridge, MA, USA, 2015; pp. 873–920. [Google Scholar] [CrossRef]
- Svirčev, Z.; Lalić, D.; Bojadžija Savić, G.; Tokodi, N.; Drobac Backović, D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global Geographical and Historical Overview of Cyanotoxin Distribution and Cyanobacterial Poisonings. Arch. Toxicol. 2019, 93, 2429–2481. [Google Scholar] [CrossRef]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural Products from Cyanobacteria: Focus on Beneficial Activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a Comprehensive Public Database of Secondary Metabolites from Cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef]
- Janssen, E.M.L. Cyanobacterial Peptides beyond Microcystins—A Review on Co-Occurrence, Toxicity, and Challenges for Risk Assessment. Water Res. 2019, 151, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Miles, O.C.; Stirling, D. Toxin Mass List, Version 15b [Data Set]. Available online: https://doi.org/10.13140/RG.2.2.15794.50889 (accessed on 17 November 2021).
- Chorus, I.; Welker, M. Toxic Cyanobacteria in Water, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Machado, J.; Azevedo, J.; Vasconcelos, V.; Campos, A.; Machado, J.; Azevedo, J.; Campos, A.; Vasconcelos, V. Cyanobacterial Toxins and Corresponding Chemical Variants. In Microbial Toxins. Toxinology; Gopalakrishnakone, P., Stiles, B., Alape-Giron, A., Dubreuil, J., Mandal, M., Eds.; Springer: Dordrecht, The Netherlands, 2018; pp. 441–464. [Google Scholar]
- Lone, Y.; Koiri, R.K.; Bhide, M. An Overview of the Toxic Effect of Potential Human Carcinogen Microcystin-LR on Testis. Toxicol. Rep. 2015, 2, 289–296. [Google Scholar] [CrossRef]
- Hinojosa, M.G.; Gutiérrez-Praena, D.; Prieto, A.I.; Guzmán-Guillén, R.; Jos, A.; Cameán, A.M. Neurotoxicity Induced by Microcystins and Cylindrospermopsin: A Review. Sci. Total Environ. 2019, 668, 547–565. [Google Scholar] [CrossRef]
- Kubickova, B.; Babica, P.; Hilscherová, K.; Šindlerová, L. Effects of Cyanobacterial Toxins on the Human Gastrointestinal Tract and the Mucosal Innate Immune System. Environ. Sci. Eur. 2019, 31, 1–27. [Google Scholar] [CrossRef]
- World Health Organization. Cyanobacterial Toxins: Microcystins Background Document for Development of WHO Guidelines for Drinking-Water Quality and Guidelines for Safe Recreational Water Environments; World Health Organization: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/handle/10665/338066 (accessed on 17 November 2021).
- Meriluoto, J.; Blaha, L.; Bojadzija, G.; Bormans, M.; Brient, L.; Codd, G.A.; Drobac, D.; Faassen, E.J.; Fastner, J.; Hiskia, A.; et al. Toxic Cyanobacteria and Cyanotoxins in European Waters—Recent Progress Achieved through the CYANOCOST Action and Challenges for Further Research. Adv. Oceanogr. Limnol. 2017, 8, 22–35. [Google Scholar] [CrossRef]
- Botes, D.P.; Tuinman, A.A.; Wessels, P.L.; Viljoen, C.C.; Kruger, H.; Williams, D.H.; Santikarn, S.; Smith, R.J.; Hammond, S.J. The Structure of Cyanoginosin-LA, a Cyclic Heptapeptide Toxin from the Cyanobacterium Microcystis aeruginosa. J. Chem. Soc. Perkin Trans. 1984, 1, 2311–2318. [Google Scholar] [CrossRef]
- Santikarn, S.; Williams, D.H.; Smith, R.J.; Hammond, S.J.; Botes, D.P.; Tuinman, A.; Wessels, P.L.; Viljoen, C.C.; Kruger, H. A Partial Structure for the Toxin BE-4 from the Blue-Green Algae, Microcystis aeruginosa. J. Chem. Soc. Chem. Commun. 1983, 12, 652–654. [Google Scholar] [CrossRef]
- Rouhiainen, L.; Vakkilainen, T.; Siemer, B.L.; Buikema, W.; Haselkorn, R.; Sivonen, K. Genes Coding for Hepatotoxic Heptapeptides (Microcystins) in the Cyanobacterium Anabaena Strain 90. Appl. Environ. Microbiol. 2004, 70, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Tillett, D.; Dittmann, E.; Erhard, M.; von Döhren, H.; Börner, T.; Neilan, B.A. Structural Organization of Microcystin Biosynthesis in Microcystis aeruginosa PCC7806: An Integrated Peptide-Polyketide Synthetase System. Chem. Biol. 2000, 7, 753–764. [Google Scholar] [CrossRef]
- Christiansen, G.; Fastner, J.; Erhard, M.; Börner, T.; Dittmann, E. Microcystin Biosynthesis in Planktothrix: Genes, Evolution, and Manipulation. J. Bacteriol. 2003, 185, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Rantala, A.; Rajaniemi-Wacklin, P.; Lyra, C.; Lepistö, L.; Rintala, J.; Mankiewicz-Boczek, J.; Sivonen, K. Detection of Microcystin-Producing Cyanobacteria in Finnish Lakes with Genus-Specific Microcystin Synthetase Gene E (McyE) PCR and Associations with Environmental Factors. Appl. Environ. Microbiol. 2006, 72, 6101–6110. [Google Scholar] [CrossRef] [PubMed]
- Sipari, H.; Rantala-Ylinen, A.; Jokela, J.; Oksanen, I.; Sivonen, K. Development of a Chip Assay and Quantitative PCR for Detecting Microcystin Synthetase e Gene Expressions. Appl. Environ. Microbiol. 2010, 76, 3797–3805. [Google Scholar] [CrossRef]
- Ngwa, F.F.; Madramootoo, C.A.; Jabaji, S. Comparison of Cyanobacterial Microcystin Synthetase (Mcy) E Gene Transcript Levels, Mcy E Gene Copies, and Biomass as Indicators of Microcystin Risk under Laboratory and Field Conditions. Microbiol. Open 2014, 3, 411–425. [Google Scholar] [CrossRef]
- Vareli, K.; Briasoulis, E.; Pilidis, G.; Sainis, I. Molecular confirmation of Planktothrix rubescens as the cause of intense, microcystin-Synthesizing cyanobacterial bloom in Lake Ziros, Greece. Harmful Algae 2009, 8, 447–453. [Google Scholar] [CrossRef]
- Pilkaitytė, R.; Overlingė, D.; Gasiūnaitė, Z.R.; Mazur-Marzec, H. Spatial and Temporal Diversity of Cyanometabolites in the Eutrophic Curonian Lagoon (SE Baltic Sea). Water 2021, 13, 1760. [Google Scholar] [CrossRef]
- Overlingė, D.; Kataržytė, M.; Vaičiūtė, D.; Gyraite, G.; Gečaitė, I.; Jonikaitė, E.; Mazur-Marzec, H. Are There Concerns Regarding CHAB in Coastal Bathing Waters Affected by Freshwater-Brackish Continuum? Mar. Pollut. Bull. 2020, 159, 111500. [Google Scholar] [CrossRef]
- Overlingė, D.; Toruńska-sitarz, A.; Cegłowska, M.; Błaszczyk, A.; Szubert, K.; Pilkaitytė, R.; Mazur-marzec, H. Phytoplankton of the Curonian Lagoon as a New Interesting Source for Bioactive Natural Products. Special Impact on Cyanobacterial Metabolites. Biomolecules 2021, 11, 1139. [Google Scholar] [CrossRef]
- Smirnova, M.M. Presence of Microcystins in the Littoral Zone of the Curonian Lagoon by the Data of Immunochromatographic Analysis in 2017. Mar. Biol. J. 2019, 4, 109–111. [Google Scholar] [CrossRef]
- Paldavičienė, A.; Mazur-Marzec, H.; Razinkovas, A. Toxic Cyanobacteria Blooms in the Lithuanian Part of the Curonian Lagoon. Oceanologia 2009, 51, 203–216. [Google Scholar] [CrossRef]
- Vaičiūtė, D.; Bučas, M.; Bresciani, M.; Dabulevičienė, T.; Gintauskas, J.; Mėžinė, J.; Tiškus, E.; Umgiesser, G.; Morkūnas, J.; de Santi, F.; et al. Hot Moments and Hotspots of Cyanobacteria Hyperblooms in the Curonian Lagoon (SE Baltic Sea) Revealed via Remote Sensing-Based Retrospective Analysis. Sci. Total Environ. 2021, 769, 145053. [Google Scholar] [CrossRef]
- Olenina, I. Long-Term Changes in the Kuršių Marios Lagoon: Eutrophication and Phytoplankton Response. Ekologija 1998, 1, 56–65. [Google Scholar]
- Belykh, O.I.; Dmitrieva, O.A.; Gladkikh, A.S.; Sorokovikova, E.G. Identification of Toxigenic Cyanobacteria of the Genus Microcystis in the Curonian Lagoon (Baltic Sea). Oceanology 2013, 53, 71–79. [Google Scholar] [CrossRef]
- Semenova, A.S.; Sidelev, S.I.; Dmitrieva, O.A. Experimental Investigation of Natural Populations of Daphnia galeata G.O. Sars from the Curonian Lagoon Feeding on Potentially Toxigenic Cyanobacteria. Biol. Bull. 2017, 44, 538–546. [Google Scholar] [CrossRef]
- Lange, E.K. Structure and Spatial Distribution of Winter Phytoplankton of the Curonian Lagoon (Baltic Sea). Ekologija 2011, 57, 121–127. [Google Scholar] [CrossRef][Green Version]
- Šulčius, S.; Pilkaityte, R.; Mazur-Marzec, H.; KasperovičienE, J.; Ezhova, E.; Błaszczyk, A.; Paškauskas, R. Increased Risk of Exposure to Microcystins in the Scum of the Filamentous Cyanobacterium Aphanizomenon flos-aquae Accumulated on the Western Shoreline of the Curonian Lagoon. Mar. Pollut. Bull. 2015, 99, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Österholm, J.; Popin, R.V.; Fewer, D.P.; Sivonen, K. Phylogenomic Analysis of Secondary Metabolism in the Toxic Cyanobacterial Genera Anabaena, Dolichospermum and Aphanizomenon. Toxins 2020, 12, 248. [Google Scholar] [CrossRef]
- Stoyneva-Gärtner, M.; Stefanova, K.; Descy, J.P.; Uzunov, B.; Radkova, M.; Pavlova, V.; Mitreva, M.; Gärtner, G. Microcystis aeruginosa and M. wesenbergii Were the Primary Planktonic Microcystin Producers in Several Bulgarian Waterbodies (August 2019). Appl. Sci. 2020, 11, 357. [Google Scholar] [CrossRef]
- Oberholster, P.; Botha, A.-M.; Grobbelaar, J. Microcystis Aeruginosa: Source of Toxic Microcystins in Drinking Water. Afr. J. Biotechnol. 2004, 3, 159–168. [Google Scholar] [CrossRef]
- Bober, B.; Lechowski, Z.; Bialczyk, J. Determination of Some Cyanopeptides Synthesized by Woronichinia naegeliana (Chroococcales, Cyanophyceae). Phycol. Res. 2011, 59, 286–294. [Google Scholar] [CrossRef]
- Samdal, I.A.; Strand, D.A.; Ballot, A.; Rusch, J.C.; Haande, S.; Løvberg, K.L.E.; Miles, C.O.; Vralstad, T. Microcystins in European Noble Crayfish Astacus Astacus in Lake Steinsfjorden, a Planktothrix-Dominated Lake. Toxins 2020, 12, 298. [Google Scholar] [CrossRef]
- Bormans, M.; Savar, V.; Legrand, B.; Mineaud, E.; Robert, E.; Lance, E.; Amzil, Z. Cyanobacteria and Cyanotoxins in Estuarine Water and Sediment. Aquat. Ecol. 2020, 54, 625–640. [Google Scholar] [CrossRef]
- Chernova, E.; Sidelev, S.; Russkikh, I.; Voyakina, E.; Zhakovskaya, Z. First Observation of Microcystin- and Anatoxin-a-Producing Cyanobacteria in the Easternmost Part of the Gulf of Finland (the Baltic Sea). Toxicon 2019, 157, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Toporowska, M.; Mazur-Marzec, H.; Pawlik-Skowrońska, B. The Effects of Cyanobacterial Bloom Extracts on the Biomass, Chl-a, MC and Other Oligopeptides Contents in a Natural Planktothrix agardhii Population. Int. J. Environ. Res. Public Health 2020, 17, 2881. [Google Scholar] [CrossRef]
- Sivonen, K.; Jones, G.J. Cyanobacterial Toxins. In Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring, and Management; Chorus, I., Bartram, J., Eds.; E & FN Spon: London, UK, 1999. [Google Scholar]
- Puddick, J.; Prinsep, M.R.; Wood, S.A.; Kaufononga, S.A.F.; Cary, S.C.; Hamilton, D.P. High Levels of Structural Diversity Observed in Microcystins from Microcystis CAWBG11 and Characterization of Six New Microcystin Congeners. Mar. Drugs 2014, 12, 5372–5395. [Google Scholar] [CrossRef] [PubMed]
- Major, Y.; Kifle, D.; Spoof, L.; Meriluoto, J. Cyanobacteria and Microcystins in Koka Reservoir (Ethiopia). Environ. Sci. Pollut. Res. 2018, 25, 26861–26873. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.D.; Dhanji-Rapkova, M.; O’Neill, A.; Coates, L.; Lewis, A.; Lewis, K. Analysis of Microcystins in Cyanobacterial Blooms from Freshwater Bodies in England. Toxins 2018, 10, 39. [Google Scholar] [CrossRef]
- Fastner, J.; Erhard, M.; Carmichael, W.W.; Sun, F.; Rinehart, K.L.; Rönicke, H.; Chorus, I. Characterization and Diversity of microcystins in Natural Blooms and Strains of the Genera and Planktothrix from German Freshwaters. Arch. Hydrobiol. 1999, 145, 147–163. [Google Scholar] [CrossRef]
- Kurmayer, R.; Christiansen, G.; Fastner, J.; Börner, T. Abundance of Active and Inactive Microcystin Genotypes in Populations of the Toxic Cyanobacterium Planktothrix spp. Environ. Microbiol. 2004, 6, 831–841. [Google Scholar] [CrossRef]
- Fewer, D.P.; Halinen, K.; Sipari, H.; Bernardová, K.; Mänttäri, M.; Eronen, E.; Sivonen, K. Non-Autonomous Transposable Elements Associated with Inactivation of Microcystin Gene Clusters in Strains of the Genus Anabaena Isolated from the Baltic Sea. Environ. Microbiol. Rep. 2011, 3, 189–194. [Google Scholar] [CrossRef]
- Wood, S.A.; Rueckert, A.; Hamilton, D.P.; Cary, S.C.; Dietrich, D.R. Switching Toxin Production on and off: Intermittent Microcystin Synthesis in a Microcystis Bloom. Environ. Microbiol. Rep. 2011, 3, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Mikalsen, B.; Boison, G.; Skulberg, O.M.; Fastner, J.; Davies, W.; Gabrielsen, T.M.; Rudi, K.; Jakobsen, K.S. Natural Variation in the Microcystin Synthetase Operon McyABC and Impact on Microcystin Production in Microcystis Strains. J. Bacteriol. 2003, 185, 2774–2785. [Google Scholar] [CrossRef]
- Bouaïcha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Yasmine Benayache, N.; Nguyen-Quang, T.; Chadli Bendjedid, U.; Taref, E. Structural Diversity, Characterization and Toxicology of Microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef] [PubMed]
- Spoof, L.; Catherine, A. Tables of Microcystins and Nodularins. In Hanbook of Cyanobacterial Monitoring and Cyanotoxin Analysis; Meriluoto, J., Spoof, L., Codd, A.G., Eds.; John Wiley & Sons, Ltd.: West Sussex, UK, 2017; pp. 526–537. [Google Scholar]
- Miles, C.O.; Melanson, J.E.; Ballot, A. Sulfide Oxidations for LC-MS Analysis of Methionine-Containing Microcystins in Dolichospermum flos-aquae NIVA-CYA 656. Environ. Sci. Technol. 2014, 48, 13307–13315. [Google Scholar] [CrossRef]
- Yilmaz, M.; Foss, A.J.; Miles, C.O.; Özen, M.; Demir, N.; Balcı, M.; Beach, D.G. Comprehensive Multi-Technique Approach Reveals the High Diversity of Microcystins in Field Collections and an Associated Isolate of Microcystis aeruginosa from a Turkish Lake. Toxicon Off. J. Int. Soc. Toxinol. 2019, 167, 87–100. [Google Scholar] [CrossRef]
- Ballot, A.; Sandvik, M.; Rundberget, T.; Botha, C.J.; Miles, C.O.; Ballot, A.; Sandvik, M.; Rundberget, T.; Botha, C.J.; Miles, C.O. Diversity of Cyanobacteria and Cyanotoxins in Hartbeespoort Dam, South Africa. Mar. Freshw. Res. 2013, 65, 175–189. [Google Scholar] [CrossRef]
- Johansson, E.; Legrand, C.; Björnerås, C.; Godhe, A.; Mazur-Marzec, H.; Säll, T.; Rengefors, K. High Diversity of Microcystin Chemotypes within a Summer Bloom of the Cyanobacterium Microcystis botrys. Toxins 2019, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, M.; Kobos, J.; Toruńska-Sitarz, A.; Mazur-Marzec, H. Non-Ribosomal Peptides Produced by Planktothrix agardhii from Siemianówka Dam Reservoir SDR (Northeast Poland). Arch. Microbiol. 2014, 196, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Rohrlack, T.; Skulberg, R.; Skulberg, O.M. Distribution of Oligopeptide Chemotypes of the Cyanobacterium Planktothrix and Their Persistence in Selected Lakes in Fennoscandia. J. Phycol. 2009, 45, 1259–1265. [Google Scholar] [CrossRef]
- Xie, L.; Rediske, R.R.; Gillett, N.D.; O’Keefe, J.P.; Scull, B.; Xue, Q. The Impact of Environmental Parameters on Microcystin Production in Dialysis Bag Experiments. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Neilan, B.A.; Pearson, L.A.; Muenchhoff, J.; Moffitt, M.C.; Dittmann, E. Environmental Conditions That Influence Toxin Biosynthesis in Cyanobacteria. Environ. Microbiol. 2013, 15, 1239–1253. [Google Scholar] [CrossRef]
- Shishido, T.K.; Kaasalainen, U.; Fewer, D.P.; Rouhiainen, L.; Jokela, J.; Wahlsten, M.; Fiore, M.F.; Yunes, J.S.; Rikkinen, J.; Sivonen, K. Convergent Evolution of [D-Leucine1] Microcystin-LR in Taxonomically Disparate Cyanobacteria. BMC Evol. Biol. 2013, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Fewer, D.P.; Rouhiainen, L.; Jokela, J.; Wahlsten, M.; Laakso, K.; Wang, H.; Sivonen, K. Recurrent Adenylation Domain Replacement in the Microcystin Synthetase Gene Cluster. BMC Evol. Biol. 2007, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, G.; Yoshida, W.Y.; Blom, J.F.; Portmann, C.; Gademann, K.; Hemscheidt, T.; Kurmayer, R. Isolation and Structure Determination of Two Microcystins and Sequence Comparison of the McyABC Adenylation Domains in Planktothrix Species. J. Nat. Prod. 2008, 71, 1881–1886. [Google Scholar] [CrossRef]
- Miles, C.O.; Sandvik, M.; Nonga, H.E.; Rundberget, T.; Wilkins, A.L.; Rise, F.; Ballot, A. Thiol Derivatization for LC-MS Identification of Microcystins in Complex Matrices. Environ. Sci. Technol. 2012, 46, 8937–8944. [Google Scholar] [CrossRef] [PubMed]
- Bateman, K.P.; Thibault, P.; Douglas, D.J.; White, R.L. Mass Spectral Analyses of Microcystins from Toxic Cyanobacteria Using On-Line Chromatographic and Electrophoretic Separations. J. Chromatogr. A 1995, 712, 253–268. [Google Scholar] [CrossRef]
- Del Campo, F.F.; Ouahid, Y. Identification of Microcystins from Three Collection Strains of Microcystis aeruginosa. Environ. Pollut. 2010, 158, 2906–2914. [Google Scholar] [CrossRef]
- Diehnelt, C.W.; Dugan, N.R.; Peterman, S.M.; Budde, W.L. Identification of Microcystin Toxins from a Strain of Microcystis aeruginosa by Liquid Chromatography Introduction into a Hybrid Linear Ion Trap-Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem. 2006, 78, 501–512. [Google Scholar] [CrossRef]
- Namikoshi, M.; Rinehart, K.L.; Sakai, R.; Stotts, R.R.; Dahlem, A.M.; Beasley, V.R.; Carmichael, W.W.; Evans, W.R. Identification of 12 Hepatotoxins from a Homer Lake Bloom of the Cyanobacteria Microcystis aeruginosa, Microcystis viridis, and Microcystis wesenbergii: Nine New Microcystins. J. Org. Chem. 1992, 57, 866–872. [Google Scholar] [CrossRef]
- Namikoshi, M.; Sun, F.; Choi, B.W.; Rinehart, K.L.; Carmichael, W.W.; Evans, W.R.; Beasley, V.R. Seven More Microcystins from Homer Lake Cells: Application of the General Method for Structure Assignment of Peptides Containing α, β-Dehydroamino Acid Unit(s). J. Org. Chem. 1995, 60, 3671–3679. [Google Scholar] [CrossRef]
- Fewer, D.P.; Tooming-Klunderud, A.; Jokela, J.; Wahlsten, M.; Rouhiainen, L.; Kristensen, T.; Rohrlack, T.; Jakobsen, K.S.; Sivonen, K. Natural Occurrence of Microcystin Synthetase Deletion Mutants Capable of Producing Microcystins in Strains of the Genus Anabaena (Cyanobacteria). Microbiology 2008, 154, 1007–1014. [Google Scholar] [CrossRef]
- LeBlanc, P.; Merkley, N.; Thomas, K.; Lewis, N.I.; Békri, K.; Renaud, S.L.B.; Pick, F.R.; McCarron, P.; Miles, C.O.; Quilliam, M.A. Isolation and Characterization of [D-Leu1]Microcystin-LY from Microcystis aeruginosa CPCC-464. Toxins 2020, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Pick, F.R. Blooming Algae: A Canadian Perspective on the Rise of Toxic Cyanobacteria. Can. J. Fish. Aquat. Sci. 2016, 73, 1149–1158. [Google Scholar] [CrossRef]
- Bortoli, S.; Volmer, D.A. Account: Characterization and Identification of Microcystins by Mass Spectrometry. Eur. J. Mass Spectrom. 2014, 20, 1–19. [Google Scholar] [CrossRef]
- Utermöhl, H. Zur Vervollkommnung Der Quantitativen Phytoplankton-Methodik. SIL Commun. 1958, 9, 1–38. [Google Scholar] [CrossRef]
- Starmach, K. Dinophyceae—Dinofity. Raphidophyceae—Rafidofity. In Flora slodkowodna Polski 5; Starmach, K., Sieminska, J., Eds.; PWN: Warszawa, Poland; Krakow, Poland, 1974. [Google Scholar]
- Tikkanen, T.; Willen, T. Vaxtplanktonflora; Statens Naturvardsverk: Solna, Sweden, 1992. [Google Scholar]
- Komárek, J.; Anganostidis, K. Cyanoprokaryota. In Süßwasserflora von Mitteleuropa; Büdel, B., Gärtner, G., Krienitz, L., Schagerl, M., Eds.; Elsevier: München, Germany, 2008; Volume 19/1. [Google Scholar]
- Komárek, J. Cyanoprokaryota: 3rd Part: Heterocystous Genera. In Süßwasserflora von Mitteleuropa; Büdel, B., Gärtner, G., Krienitz, L., Schagerl, M., Eds.; Springer Spektrum: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- HELCOM. Manual of Marine Monitoring in the Combine Programme of HELCOM. Annex C-6: Guidelines Concerning Phytoplankton Species Composition, Abundance and Biomass. Available online: https://helcom.fi/media/publications/Manual-for-Marine-Monitoring-in-the-COMBINE-Programme-of-HELCOM.pdf (accessed on 11 June 2021).
- Mazur-Marzec, H.; Browarczyk-Matusiak, G.; Forycka, K.; Kobos, J.; Pliński, M. Morphological, Genetic, Chemical and Ecophysiological Characterisation of Two Microcystis aeruginosa Isolates from the Vistula Lagoon, Southern Baltic. Oceanologia 2010, 52, 127–146. [Google Scholar] [CrossRef]
- Cegłowska, M.; Szubert, K.; Wieczerzak, E.; Kosakowska, A.; Mazur-Marzec, H. Eighteen New Aeruginosamide Variants Produced by the Baltic Cyanobacterium Limnoraphis CCNP1324. Mar. Drugs 2020, 18, 446. [Google Scholar] [CrossRef]
- Rantala, A.; Fewer, D.P.; Hisbergues, M.; Rouhiainen, L.; Vaitomaa, J.; Börner, T.; Sivonen, K. Phylogenetic Evidence for the Early Evolution of Microcystin Synthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 568–573. [Google Scholar] [CrossRef]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative Real-Time PCR for Determination of Microcystin Synthetase E Copy Numbers for Microcystis and Anabaena in Lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mazur-Marzec, H.; Błaszczyk, A.; Felczykowska, A.; Hohlfeld, N.; Kobos, J.; Toruńska-Sitarz, A.; Devi, P.; Montalvão, S.; D’souza, L.; Tammela, P.; et al. Baltic Cyanobacteria—A Source of Biologically Active Compounds. Eur. J. Phycol. 2015, 50, 343–360. [Google Scholar] [CrossRef]
- Bray, R.J.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]



| MC Variants | m/z | Sampling Dates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 May 2018 | 13 Jun 2018 | 27 Jun 2018 | 11 Jul 2018 | 23 Jul 2018 | 3 Aug 2018 | 9 Aug 2018 | 16 Aug 2018 | 30 Aug 2018 | 19 Sep 2018 | 17 Oct 2019 | 3 Jul 2020 | ||
| [Ser1]MC-HtyR or MC-Y(OMe)R | 1075 | + | + | ||||||||||
| MC-WR or [Ser1]MC-HarR | 1068 | + | + | ||||||||||
| MC-X1R | 1057 | + | + | ||||||||||
| MC-? | 1054 | + | |||||||||||
| MC-(H4)YR | 1049 | + | + | ||||||||||
| MC-YR | 1045 | + | + | + | + | + | |||||||
| MC-HphR | 1043 | + | + | ||||||||||
| MC-RR | 1038 (519) | + | + | + | + | + | + | + | + | + | + | + | + |
| [Asp3]MC-YR or [Asp3]MC-M(O2)R | 1031 | + | + | + | + | ||||||||
| [Asp3]MC-RY | 1031 | + | + | ||||||||||
| MC-FR | 1029 | + | + | ||||||||||
| MC-LW | 1025 | + | |||||||||||
| [Dha7]MC-RR | 1024 (512) | + | + | + | + | + | + | + | |||||
| MC-HilR | 1009 | + | + | ||||||||||
| MC-LY | 1002 | + | |||||||||||
| MC-LR | 995 | + | + | + | + | + | + | + | + | + | + | + | |
| [Asp3]MC-LY | 988 | + | + | ||||||||||
| MC-LF | 986 | + | + | + | + | ||||||||
| [Dha7]MC-LR | 981 | + | + | + | + | ||||||||
| [Asp3]MC-LR | 981 | + | + | + | |||||||||
| Samples | General mcyE Primers | Dolichospermum Specific Primers | Microcystis Specific Primers | Planktothrix Specific Primers |
|---|---|---|---|---|
| 30 May 2018 | + | OK500398 | OK500409 | OK500421 |
| 13 Jun 2018 | + | OK500399 | OK500410 | OK500422 |
| 27 Jun 2018 | + | OK500400 | OK500411 | OK500423 |
| 11 Jul 2018 | + | OK500401 | OK500412 | OK500424 |
| 27 Jul 2018 | + | OK500402 | OK500413 | OK500425 |
| 3 Aug 2018 | + | OK500403 | OK500414 | OK500426 |
| 9 Aug 2018 | + | OK500404 | OK500415 | OK500427 |
| 16 Aug 2018 | + | OK500405 | OK500416 | OK500428 |
| 30 Aug 2018 | + | OK500406 | OK500417 | OK500429 |
| 19 Sep 2018 | + | OK500407 | OK500418 | OK500430 |
| 17 Oct 2019 | + | − | OK500419 | OK500431 |
| 3 Jul 2020 | + | OK500408 | OK500420 | OK500432 |
| M. aeruginosa CCNP1102 | + | − | OK500396 | − |
| P. aghardii CCNP1325 | + | − | − | OK500397 |
| Limnoraphis sp. CCNP1324 | − | − | − | − |
| MilliQ water | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Overlingė, D.; Toruńska-Sitarz, A.; Kataržytė, M.; Pilkaitytė, R.; Gyraitė, G.; Mazur-Marzec, H. Characterization and Diversity of Microcystins Produced by Cyanobacteria from the Curonian Lagoon (SE Baltic Sea). Toxins 2021, 13, 838. https://doi.org/10.3390/toxins13120838
Overlingė D, Toruńska-Sitarz A, Kataržytė M, Pilkaitytė R, Gyraitė G, Mazur-Marzec H. Characterization and Diversity of Microcystins Produced by Cyanobacteria from the Curonian Lagoon (SE Baltic Sea). Toxins. 2021; 13(12):838. https://doi.org/10.3390/toxins13120838
Chicago/Turabian StyleOverlingė, Donata, Anna Toruńska-Sitarz, Marija Kataržytė, Renata Pilkaitytė, Greta Gyraitė, and Hanna Mazur-Marzec. 2021. "Characterization and Diversity of Microcystins Produced by Cyanobacteria from the Curonian Lagoon (SE Baltic Sea)" Toxins 13, no. 12: 838. https://doi.org/10.3390/toxins13120838
APA StyleOverlingė, D., Toruńska-Sitarz, A., Kataržytė, M., Pilkaitytė, R., Gyraitė, G., & Mazur-Marzec, H. (2021). Characterization and Diversity of Microcystins Produced by Cyanobacteria from the Curonian Lagoon (SE Baltic Sea). Toxins, 13(12), 838. https://doi.org/10.3390/toxins13120838






