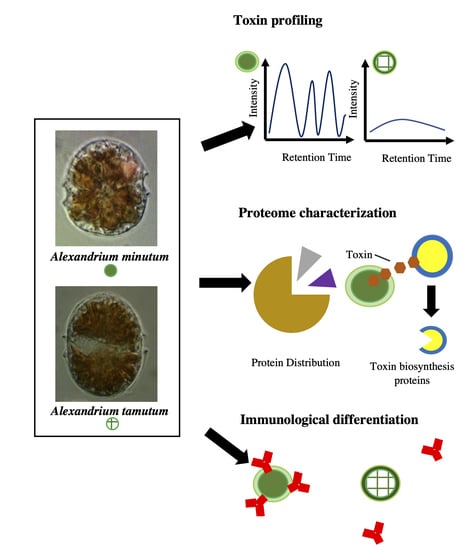
Differentiating Two Closely Related Alexandrium Species Using Comparative Quantitative Proteomics
Abstract
1. Introduction
2. Results
2.1. Toxin Analysis
2.2. Immunological Analysis
2.3. Gel-Free Proteomics Approach
2.4. Gel-Based Proteomics Approach
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cultivation and Cell Collection
5.2. Toxin Analysis
5.3. Immunostaining Using Confocal Laser Scanning Microscopy
5.4. Gel-Free Proteomics
5.4.1. Protein Extraction for iTRAQ-Method
5.4.2. Protein Labelling and Strong Cation Exchange Chromatography
5.4.3. Mass Spectrometry Analysis, Protein Identification and Annotation
5.5. Gel-Based Proteomics
5.5.1. Sample Extraction and Preparation for 2D-DIGE
5.5.2. 2D-DIGE Labeling and 2D Runs
5.5.3. Mass Spectrometry Analysis, Protein Identification and Annotation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zingone, A.; Enevoldsen, H.O. The diversity of harmful algal blooms: A challenge for science and management. Ocean Coast. Manag. 2000, 43, 725–748. [Google Scholar] [CrossRef]
- Subong, B.J.J.; Benico, G.A.; Sulit, A.K.L.; Mendoza, C.O.; Cruz, L.J.; Azanza, R.V.; Jimenez, E.C. Toxicity and protein expression of Alexandrium species collected in the Philippine waters. Philipp. J. Sci. 2017, 146, 425–436. [Google Scholar]
- Anderson, D.M.; Alpermann, T.J.; Cembella, A.D.; Collos, Y.; Masseret, E.; Montresor, M. The globally distributed genus Alexandrium: Multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 2012, 14, 10–35. [Google Scholar] [CrossRef] [PubMed]
- Garthwaite, I. Keeping shellfish safe to eat: A brief review of shellfish toxins, and methods for their detection. Trends Food Sci. Technol. 2000, 11, 235–244. [Google Scholar] [CrossRef]
- Lilly, E.L.; Halanych, K.M.; Anderson, D.M. Phylogeny, biogeography, and species boundaries within the Alexandrium minutum group. Harmful Algae 2005, 4, 1004–1020. [Google Scholar] [CrossRef]
- Montresor, M.; John, U.; Beran, A.; Medlin, L.K. Alexandrium tamutum sp. Nov.(dinophyceae): A new nontoxic species in the genus Alexandrium. J. Phycol. 2004, 40, 398–411. [Google Scholar] [CrossRef]
- Balech, E. The Genus Alexandrium Halim (Dinoflagellata); Sherkin Island Marine Station: Cork, Ireland, 1995; p. 452. [Google Scholar]
- Montresor, M.; Zingone, A.; Sarno, D. Dinoflagellate cyst production at a coastal Mediterranean site. J. Plankton Res. 1998, 20, 2291–2312. [Google Scholar] [CrossRef]
- Penna, A.; Fraga, S.; Maso, M.; Giacobbe, M.G.; Bravo, I.; Garcés, E.; Vila, M. Phylogenetic relationships among the Mediterranean Alexandrium (Dinophyceae) species based on sequences of 5.8 S gene and internal transcript spacers of the rRNA operon. Eur. J. Phycol. 2008, 43, 163–178. [Google Scholar] [CrossRef]
- Kon, N.F.; Hii, K.S.; Tan, T.H.; Yek, L.H.; Lim, P.T.; Leaw, C.P. First record of marine dinoflagellate, Alexandrium tamutum (Dinophyceae) from Malaysia. Malay. J. Sci. 2013, 32, 77–84. [Google Scholar] [CrossRef][Green Version]
- Hallegraeff, G.M.; Bolch, C.J. Transport of toxic dinoflagellate cysts via ships’ ballast water. Mar. Pollut. Bull. 1991, 22, 27–30. [Google Scholar] [CrossRef]
- de Salas, M.F.; van Emmerik, M.J.; Hallegraeff, G.M.; Negri, A.P.; Vaillancourt, R.E.; Bolch, C.J. Toxic Australian Alexandrium Dinoflagellates: Introduced or Indigenous. In Harmful Algal Blooms; Hallegraeff, G.M., Blackburn, S.I., Bolch, C.J., Lewis, R.J., Eds.; UNESCO: Paris, France, 2001; pp. 214–217. [Google Scholar]
- Hwang, D.F.; Lu, Y.H. Influence of environmental and nutritional factors on growth, toxicity, and toxin profile of dinoflagellate Alexandrium minutum. Toxicon 2000, 38, 1491–1503. [Google Scholar] [CrossRef]
- Yoshida, M.; Ogata, T.; Van Thuoc, C.; Matsuoka, K.; Fukuyo, Y.; Hoi, N.C.; Kodama, M. The first finding of toxic dinoflagellate Alexandrium minutum in Vietnam. Fish. Sci. 2000, 66, 177–179. [Google Scholar] [CrossRef]
- Honsell, G. First Report of Alexandrium Minutum in Northern Adriatic Waters (Mediterranean Sea). In Toxic Phytoplankton Blooms in the Sea; Smayda, T.J., Shimizu, Y., Eds.; Amsterdam, The Netherlands: Elsevier, 1993; pp. 127–132. [Google Scholar]
- Honsell, G.; Poletti, R.; Pompei, M.; Sidari, L.; Milandri, A.; Casadei, C.; Viviani, R. Alexandrium minutum Halim and PSP Contamination in the Northern Adriatic Sea (Mediterranean Sea). In Harmful and Toxic Algal Blooms; Yasumoto, T., Oshima, Y., Fukuyo, Y., Eds.; UNESCO: Sendai, Japan, 1996; pp. 77–80. [Google Scholar]
- Touzet, N.; Franco, J.M.; Raine, R. Characterization of nontoxic and toxin-producing strains of Alexandrium minutum (Dinophyceae) in Irish coastal waters. Appl. Environ. Microbiol. 2007, 73, 3333–3342. [Google Scholar] [CrossRef] [PubMed]
- Toebe, K.; Alpermann, T.J.; Tillmann, U.; Krock, B.; Cembella, A.; John, U. Molecular discrimination of toxic and non-toxic Alexandrium species (Dinophyta) in natural phytoplankton assemblages from the Scottish coast of the North Sea. Eur. J. Phycol. 2013, 48, 12–26. [Google Scholar] [CrossRef]
- Yang, I.; John, U.; Beszteri, S.; Glöckner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genom. 2010, 11, 248. [Google Scholar] [CrossRef]
- McLean, T.I. “Eco-omics”: A review of the application of genomics, transcriptomics, and proteomics for the study of the ecology of harmful algae. Microb. Ecol. 2013, 65, 901–915. [Google Scholar] [CrossRef]
- Zhang, S.; Sui, Z.; Chang, L.; Kang, K.; Ma, J.; Kong, F.; Zhou, W.; Wang, J.; Guo, L.; Geng, H.; et al. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 2014, 537, 285–293. [Google Scholar] [CrossRef]
- Feder, M.E.; Walser, J.C. The biological limitations of transcriptomics in elucidating stress and stress responses. J. Evol. Biol. 2005, 18, 901–910. [Google Scholar] [CrossRef]
- Chan, L.L.; Hodgkiss, I.J.; Lam, P.K.S.; Wan, J.M.F.; Chou, H.N.; Lum, J.H.K.; Lo, S.C.L. Use of two-dimensional gel electrophoresis to differentiate morphospecies of Alexandrium minutum, a paralytic shellfish poisoning toxin-producing dinoflagellate of harmful algal blooms. Proteomics 2005, 5, 1580–1593. [Google Scholar] [CrossRef]
- Wang, D.Z.; Li, C.; Xie, Z.X.; Dong, H.P.; Lin, L.; Hong, H.S. Homology-driven proteomics of dinoflagellates with unsequenced genomes using MALDI-TOF/TOF and automated de novo sequencing. Evid. Based Complement. Altern. Med. 2011. [Google Scholar] [CrossRef]
- Wang, D.Z.; Zhang, Y.J.; Zhang, S.F.; Lin, L.; Hong, H.S. Quantitative proteomic analysis of cell cycle of the dinoflagellate Prorocentrum donghaiense (Dinophyceae). PLoS ONE 2013, 8, e63659. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.Y.; Lü, S. Molecular ecological responses of dinoflagellate, Karenia mikimotoi to environmental nitrate stress. Mar. Pollut. Bull. 2011, 62, 2692–2699. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Klochkova, T.A.; Han, J.W.; Kim, G.H.; Yoo, Y.; Jeong, H.J. Comparative proteomics of the mixotrophic dinoflagellate Prorocentrum micans growing in different trophic modes. Algae 2011, 26, 87–96. [Google Scholar] [CrossRef][Green Version]
- Wang, D.Z.; Li, C.; Zhang, Y.J.; Wang, Y.Y.; He, Z.P.; Lin, L.; Hong, H. Quantitative proteomic analysis of differentially expressed proteins in the toxicity-lost mutant of Alexandrium catenella (Dinophyceae) in the exponential phase. J. Proteom. 2012, 75, 5564–5577. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.J.; Xie, Z.X.; He, Z.P.; Lin, L.; Wang, D.Z. Quantitative proteomic analysis reveals evolutionary divergence and species-specific peptides in the Alexandrium tamarense complex (Dinophyceae). J. Proteom. 2013, 86, 85–96. [Google Scholar] [CrossRef]
- Li, C.; Wang, D.Z.; Dong, H.P.; Xie, Z.X.; Hong, H.S. Proteomics of a toxic dinoflagellate Alexandrium catenella DH01: Detection and identification of cell surface proteins using fluorescent labeling. Chin. Sci. Bull. 2012, 57, 3320–3327. [Google Scholar] [CrossRef]
- Minge, M.A.; Shalchian-Tabrizi, K.; Tørresen, O.K.; Takishita, K.; Probert, I.; Inagaki, Y.; Klaveness, D.; Jakobsen, K.S. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 2010, 10, 191–202. [Google Scholar] [CrossRef]
- Peng, S.E.; Luo, Y.J.; Huang, H.J.; Lee, I.T.; Hou, L.S.; Chen, W.N.; Fang, L.S.; Chen, C.S. Isolation of tissue layers in hermatypic corals by N- acetylcysteine: Morphological and proteomic examinations. Coral Reefs 2008, 27, 133–142. [Google Scholar] [CrossRef]
- Chan, L.L.; Sit, W.H.; Lam, P.K.S.; Hsieh, D.P.H.; Hodgkiss, I.J.; Wan, J.M.F.; Dudgeon, D. Identification and characterization of a “biomarker of toxicity” from the proteome of the paralytic shellfish toxin-producing dinoflagellate Alexandrium tamarense (Dinophyceae). Proteomics 2006, 6, 654–666. [Google Scholar] [CrossRef]
- Gas, F.; Baus, B.; Pinto, L.; Compere, C.; Tanchou, V.; Quéméneur, E. One step immunochromatographic assay for the rapid detection of Alexandrium minutum. Biosens. Bioelectro. 2010, 25, 1235–1239. [Google Scholar] [CrossRef][Green Version]
- Carrera, M.; Garet, E.; Barreiro, A.; Garcés, E.; Pérez, D.; Guisande, C.; González-Fernández, Á. Generation of monoclonal antibodies for the specific immunodetection of the toxic dinoflagellate Alexandrium minutum Halim from Spanish waters. Harmful Algae 2010, 9, 272–280. [Google Scholar] [CrossRef]
- Cembella, A.D.; Quilliam, M.A.; Lewis, N.I.; Bauder, A.G.; Dell’Aversano, C.; Thomas, K.; Cusack, R.R. The toxigenic marine dinoflagellate Alexandrium tamarense as the probable cause of mortality of caged salmon in Nova Scotia. Harmful Algae 2002, 1, 313–325. [Google Scholar] [CrossRef]
- Escobar, M.T.L.; Sotto, L.P.A.; Jacinto, G.S.; Benico, G.A.; Azanza, R.V. Eutrophic conditions during the 2010 fish kill in Bolinao and Anda, Pangasinan, Philippines. J. Environ. Sci. Manag. 2013, 1, 29–35. [Google Scholar]
- Mardones, J.I.; Dorantes-Aranda, J.J.; Nichols, P.D.; Hallegraeff, G.M. Fish gill damage by the dinoflagellate Alexandrium catenella from Chilean fjords: Synergistic action of ROS and PUFA. Harmful Algae 2015, 49, 40–49. [Google Scholar] [CrossRef]
- Chou, H.N.; Chen, Y.M.; Chen, C.Y. Variety of PSP toxins in four culture strains of Alexandrium minutum collected from southern Taiwan. Toxicon 2004, 43, 337–340. [Google Scholar] [CrossRef]
- Zhang, S.F.; Zhang, Y.; Xie, Z.X.; Zhang, H.; Lin, L.; Wang, D.Z. iTRAQ-based quantitative proteomic analysis of a toxigenic dinoflagellate Alexandrium catenella and its non-toxic mutant. Proteomics 2015, 15, 4041–4050. [Google Scholar] [CrossRef]
- Figueroa, R.I.; Garces, E.; Bravo, I. Comparative study of the life cycles of Alexandrium tamutum and Alexandrium minutum (Gonyaulacales, Dinophyceae) in culture. J. Phycol. 2007, 43, 1039–1053. [Google Scholar] [CrossRef]
- Dagenais-Bellefeuille, S.; Morse, D. Putting the N in dinoflagellates. Front. Microbiol. 2013, 4, 369. [Google Scholar] [CrossRef]
- Moffatt, B.A.; Ashihara, H. Purine and pyrimidine nucleotide synthesis and metabolism. Arab. Book Am. Soc. Plant Biol. 2002, 1. [Google Scholar] [CrossRef]
- Menezes, M.; Branco, S.; Miotto, M.C.; Alves-de-Souza, C. The Genus Alexandrium (Dinophyceae, Dinophyta) in Brazilian coastal waters. Front. Mar. Sci. 2018, 5, 421. [Google Scholar] [CrossRef]
- Hazel, J.R. Thermal adaptation in biological membranes: Is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 1995, 57, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Appling, D.R. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991, 5, 2645–2651. [Google Scholar] [CrossRef] [PubMed]
- Tse, S.P.K.; Lee, F.W.F.; Mak, D.Y.L.; Kong, H.K.; Chan, K.K.Y.; Lo, P.Y.; Lo, S.C.L. Production of paralytic shellfish toxins (PSTs) in toxic Alexandrium catenella is intertwined with photosynthesis and energy production. Toxins 2020, 12, 477. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Z.; Gao, Y.; Lin, L.; Hong, H.S. Comparative proteomic analysis reveals proteins putatively involved in toxin biosynthesis in the marine dinoflagellate Alexandrium catenella. Mar. Drugs 2013, 11, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.; Salois, P.; Markovic, P.; Hastings, J.W. A nuclear-encoded form II RuBisCO in dinoflagellates. Science 1995, 268, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.W.; Wang, J.; Chan, L.L.; Lam, P.K.S.; Gu, J.D. Comparison of three protein extraction procedures from toxic and non-toxic dinoflagellates for proteomics analysis. Ecotoxicology 2015, 24, 1395–1406. [Google Scholar] [CrossRef]
- Wang, H.; Tomasch, J.; Michael, V.; Bhuju, S.; Jarek, M.; Petersen, J.; Wagner-Döbler, I. Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibae with the dinoflagellate Prorocentrum minimum. Front. Microbiol. 2015, 6, 1262. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Koch, F.; Gobler, C.J. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc. Natl. Acad. Sci. USA 2010, 107, 20756–20761. [Google Scholar] [CrossRef]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.J.; Smith, A.G. Algae acquire vitamin B 12 through a symbiotic relationship with bacteria. Nature 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Croft, M.T.; Warren, M.J.; Smith, A.G. Algae need their vitamins. Eukaryot. Cell 2006, 5, 1175–1183. [Google Scholar] [CrossRef]
- Cruz-López, R.; Maske, H. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front. Microbiol. 2016, 7, 560. [Google Scholar] [CrossRef] [PubMed]
- Vingiani, G.M.; Štālberga, D.; De Luca, P.; Ianora, A.; De Luca, D.; Lauritano, C. De novo tanscriptome of the non-saxitoxin producing Alexandrium tamutum reveals new insights on harmful dinoflagellates. Mar. Drugs 2020, 18, 386. [Google Scholar] [CrossRef] [PubMed]
- Kellmann, R.; Mihali, T.K.; Jeon, Y.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef] [PubMed]
- Sigaud-Kutner, T.C.S.; Aidar, E. The influence of osmotic shocks on the growth rate and chlorophyll-a content of planktonic algae species. Bol. Inst. Oceanográfico 1995, 43, 89–98. [Google Scholar] [CrossRef]
- Gendron, R.L.; Armstrong, E.; Paradis, H.; Haines, L.; Desjardins, M.; Short, C.E.; Driedzic, W.R. Osmotic pressure-adaptive responses in the eye tissues of rainbow smelt (Osmerus mordax). Mol. Vis. 2011, 17, 2596. [Google Scholar]
- Errera, R.M.; Campbell, L. Osmotic stress triggers toxin production by the dinoflagellate Karenia brevis. Proc. Natl. Acad. Sci. USA 2011, 108, 10597–10601. [Google Scholar] [CrossRef]
- Sunda, W.G.; Burleson, C.; Hardison, D.R.; Morey, J.S.; Wang, Z.; Wolny, J.; Van Dolah, F.M. Osmotic stress does not trigger brevetoxin production in the dinoflagellate Karenia brevis. Proc. Natl. Acad. Sci. USA 2013, 110, 10223–10228. [Google Scholar] [CrossRef]
- Ravanel, S.; Block, M.A.; Rippert, P.; Jabrin, S.; Curien, G.; Rébeillé, F.; Douce, R. Methionine metabolism in plants chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 2004, 279, 22548–22557. [Google Scholar] [CrossRef]
- Jackson, M.I.; Cao, J.; Zeng, H.; Uthus, E.; Combs, G.F. S-adenosylmethionine-dependent protein methylation is required for expression of selenoprotein P and gluconeogenic enzymes in HepG2 human hepatocytes. J. Biol. Chem. 2012, 287, 36455–36464. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, Y.G. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 2016, 11, 583–597. [Google Scholar] [CrossRef]
- Azanza-Corrales, R.; Hall, S. Isolation and culture of Pyrodinium bahamense var. compressum from the Philippines. In Toxic Phytoplankton Blooms in the Sea; Smayda, T.J., Shimizu, Y., Eds.; Springer-Verlag: Berlin, Heidelberg, 1993; pp. 725–730. [Google Scholar]
- Oshima, Y. Postcolumn derivatization liquid chromatographic method for paralytic shellfish toxins. J. AOAC Int. 1995, 78, 528–532. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Ménard, C. Determination of marine toxins by liquid chromatography. Fresen. J. Anal. Chem. 1991, 339, 494–498. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Niedzwiadek, B.; Menard, C. Quantitative determination of paralytic shellfish poisoning toxins in shellfish using prechromatographic oxidation and liquid chromatography with fluorescence detection: Collaborative study. J. AOAC Int. 2005, 88, 1714–1732. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.P.; Cembella, A.D. Effects of salinity, light and inorganic nitrogen on growth and toxigenicity of the marine dinoflagellate Alexandrium tamarense from northeastern Canada. J. Plankton Res. 1999, 21, 939–955. [Google Scholar] [CrossRef]
- Borkman, D.G.; Smayda, T.J.; Schwarz, E.N.; Flewelling, L.J.; Tomas, C.R. Recurrent vernal presence of the toxic Alexandrium tamarense/Alexandrium fundyense (Dinoflagellata) species complex in Narragansett Bay, USA. Harmful Algae 2014, 32, 73–80. [Google Scholar] [CrossRef]
- Lin, S.; Carpenter, E.J. An empirical protocol for whole-cell immunofluorescence of marine phytoplankton. J. Phycol. 1996, 32, 1083–1094. [Google Scholar] [CrossRef]
- Bankhead, P. Analyzing fluorescence microscopy images with ImageJ. ImageJ 2014, 1, 10–1109. [Google Scholar]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2008, 11, 2301. [Google Scholar] [CrossRef]
- Saltzman, A.B.; Leng, M.; Bhatt, B.; Singh, P.; Chan, D.W.; Dobrolecki, L.; Lewis, M.T. gpGrouper: A peptide grouping algorithm for gene- centric inference and quantitation of bottom-up proteomics data. Mol. Cell. Proteom. 2018, 17, 2270–2283. [Google Scholar] [CrossRef]
- Shirran, S.L.; Botting, C.H. A comparison of the accuracy of iTRAQ quantification by nLC-ESI MSMS and nLC-MALDI MSMS methods. J. Proteom. 2010, 73, 1391–1403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Martin, M.J. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Camon, E.; Magrane, M.; Barrell, D.; Lee, V.; Dimmer, E.; Maslen, J.; Apweiler, R. The gene ontology annotation (goa) database: Sharing knowledge in uniprot with gene ontology. Nucleic Acids Res. 2004, 32, D262–D266. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, J.S. Protein identification using MS/MS data. J. Proteom. 2011, 74, 1842–1851. [Google Scholar] [CrossRef] [PubMed]

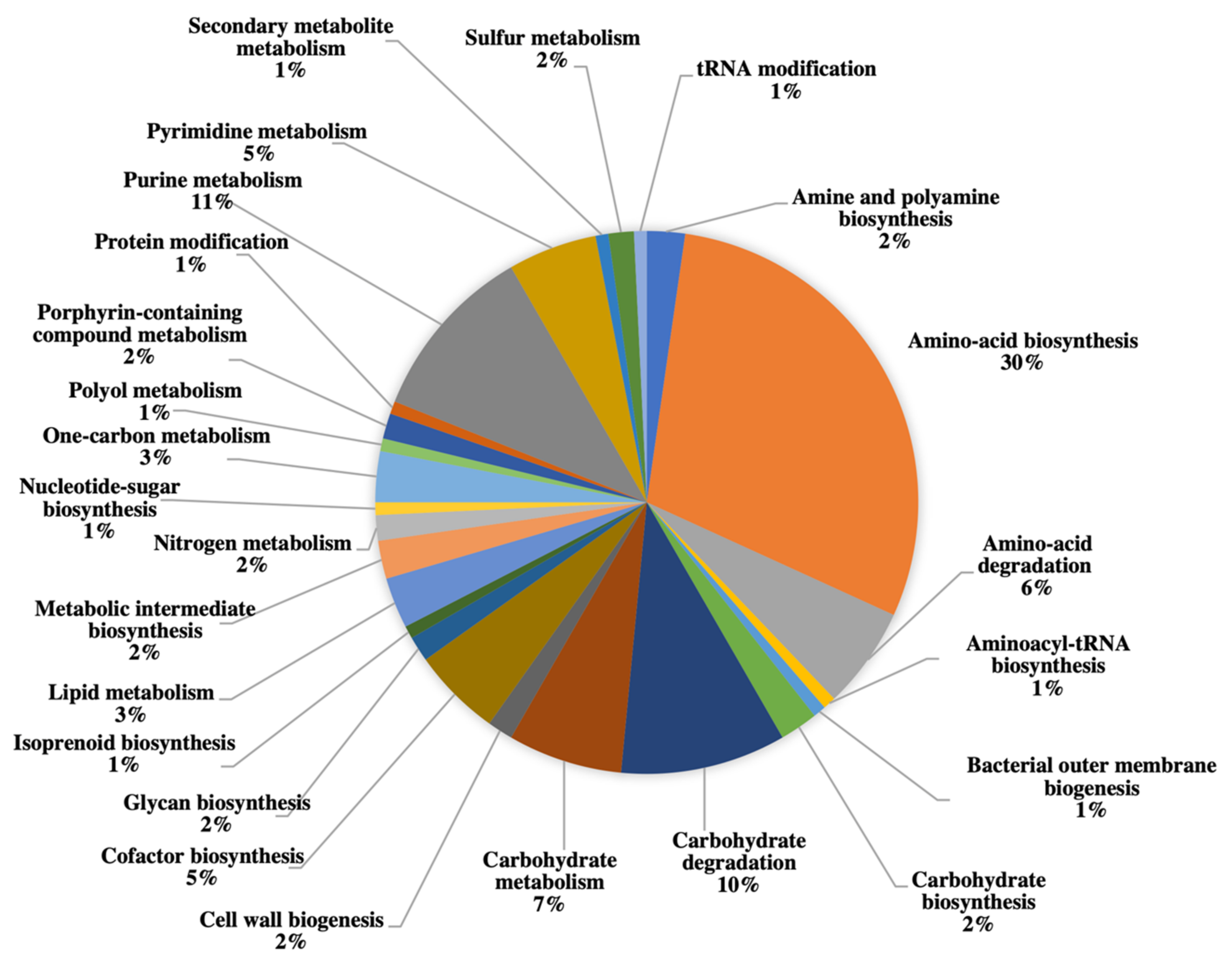
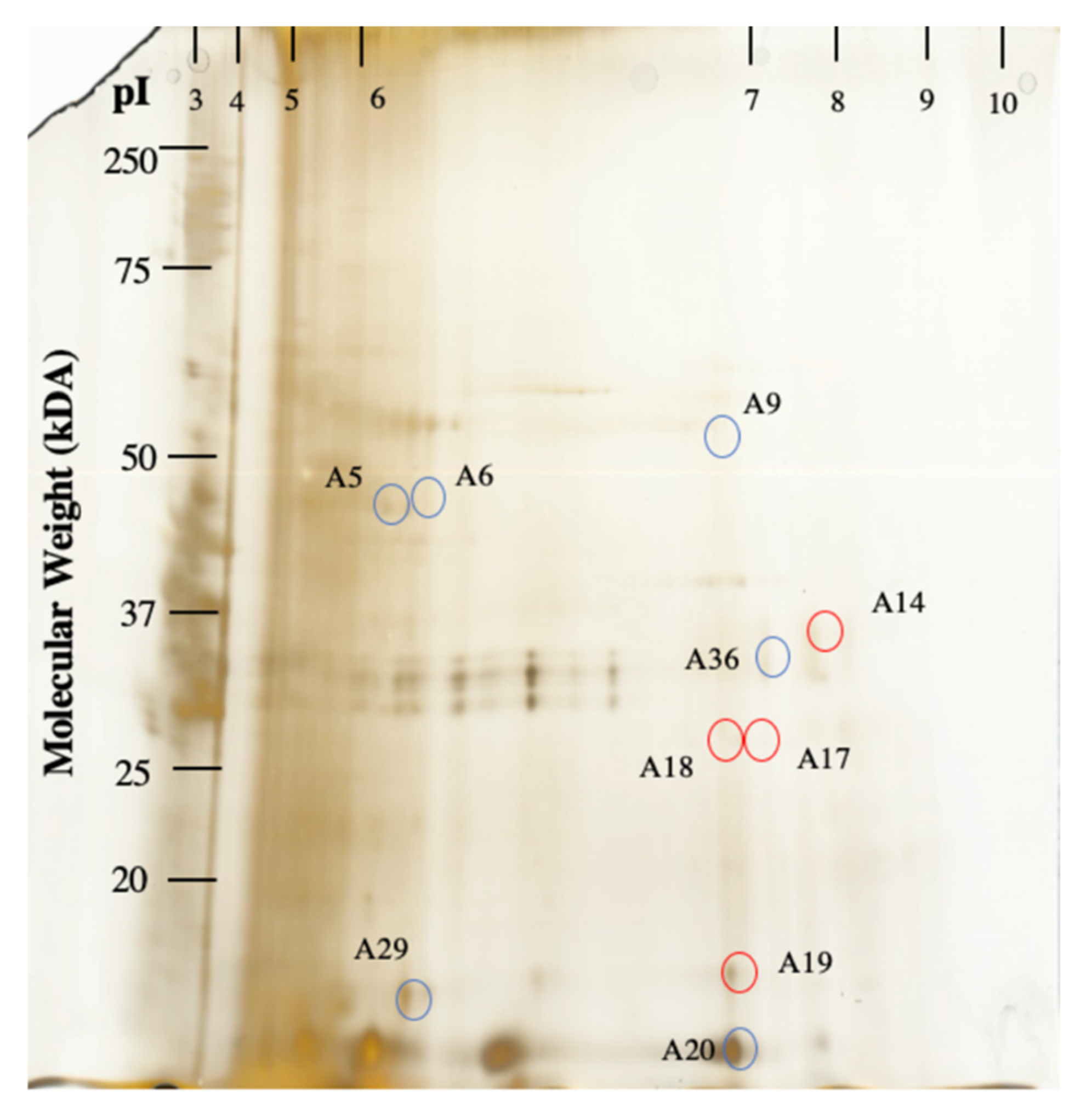
| Protein | Number of Significant Sequences | Sequence Coverage (%) | Representative Ratio (A.T/A.M.) a | Score | E-Value | Function |
|---|---|---|---|---|---|---|
| Osmotic Growth Proteins [Pbc transcript] | 6 | 27 | 0.196 | 392 | 3.2E −39 | succinate dehydrogenase activity |
| Type 1 Polyketide Synthase ** Sequence: R.SPASTGQSLGR.R [Pbc transcript] | 1 | 4 | 0.517 | 40 * | 0.00050 | polyketide synthesis |
| Serine/Arginine Splicing Factor 6 ** Sequence: R.GGLAQGSPRP [Pbc transcript] | 1 | 3 | 0.746 | 29 * | 0.0063 | mRNA splicing |
| Mitochondrial Ubiquinol Cytochrome C Oxidoreductase Core Beta Subunit-Like Protein 5 ** Sequence: R.GIPPAEMLLR.I [Pbc transcript] | 1 | 4 | 0.772 | 45 * | 0.00016 | redox reaction |
| Contig06626_2 ** Sequence: K.LAGVDAK.D [Pbc transcript] | 1 | 3 | 0.431 | 53 * | 2.5E −05 | No annotation |
| Contig00658_4 ** Sequence: R.SVVQLLR.R [Pbc transcript] | 1 | 2 | 0.514 | 87 | 10E −09 | No annotation |
| ATP Synthase Subunit [Pbc transcript] | 9 | 22 | 1.30 | 1173 | 2.5E −117 | ATP production |
| 3-Hydroxyacyl-CoA Dehydrogenase [Pbc transcript] | 3 | 18 | 1.36 | 117 | 10E −12 | redox reaction |
| Photosystem II protein D2 [Pbc transcript] | 5 | 14 | 1.38 | 87 | 10E −09 | photosynthesis |
| Light-Harvesting Protein [Pbc transcript] | 2 | 9 | 1.38 | 215 | 1.6E −21 | photosynthesis |
| Eukaryotic Translation Initiation Factor [Pbc transcript] | 3 | 8 | 1.56 | 154 | 2.0E −15 | translation |
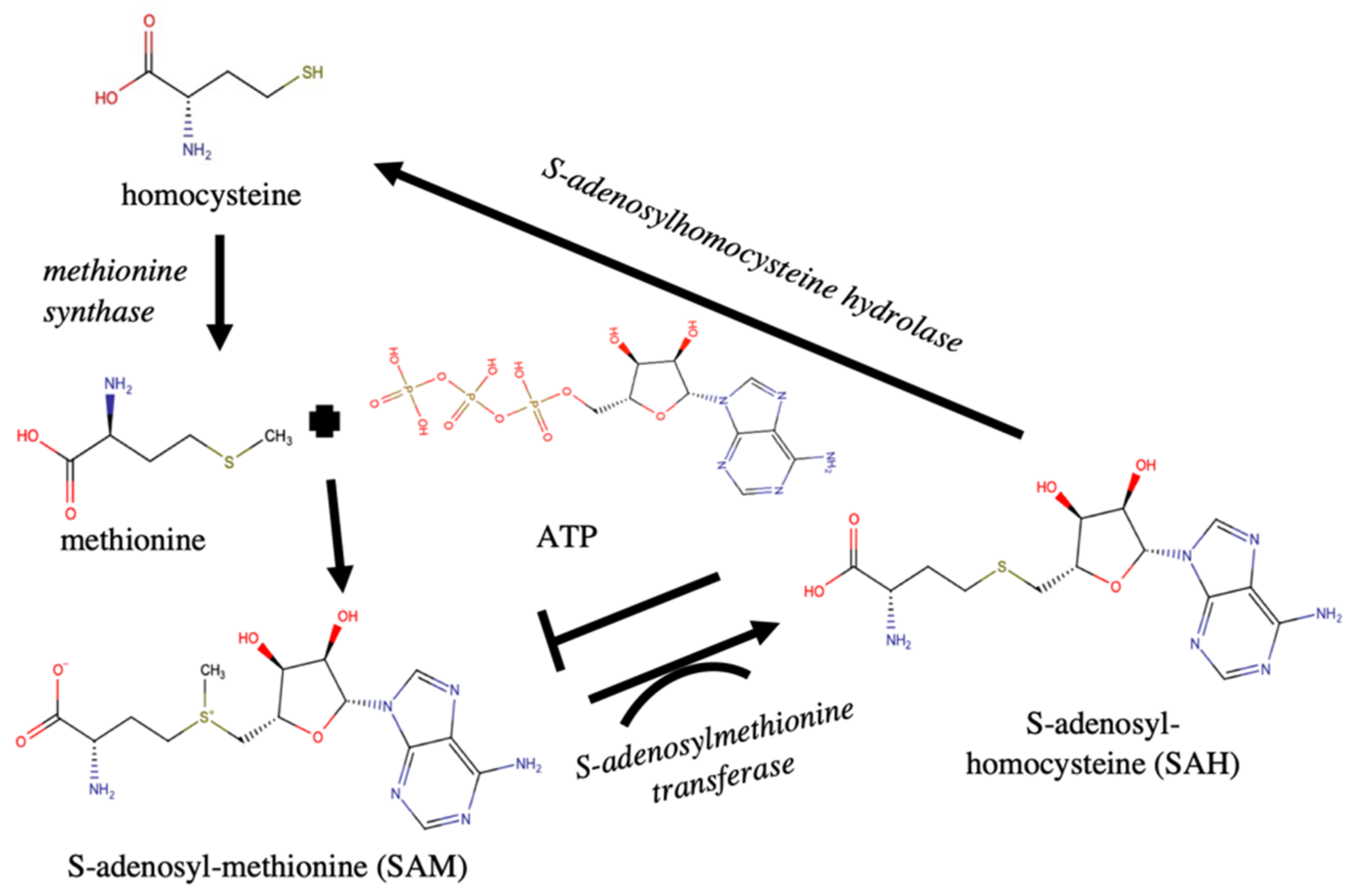
| Methionine Synthase ** Sequence: K.GLNMSIVNPGGLPR.Y [Pbc transcript] | 1 | 5 | 1.84 | 27 * | 0.010 | methionine production |
| Heat Shock Protein 90 [Pbc transcript] | 6 | 8 | 2.10 | 67 | 10E −07 | protein stabilization against heat stress |
| Plastid C1 Class II Fructose Bisphosphate Aldolase [Pbc transcript] | 3 | 8 | 2.30 | 206 | 1.3E −20 | glycolytic process |
| Malate Dehydrogenase ** Sequence: R.SVLAGLSGR.K [Pbc transcript] | 1 | 2 | 2.31 | 84 | 2.0E −08 | citric acid cycle |
| Zn-Dependent Alcohol Dehydrogenase ** Sequence: K.LNTGITPLEVAPMADAGITAYR.A [Marinovum Algicola] | 1 | 16 | 0.456 | 153 | 2.5E −15 | aldehyde production |
| Type-I Polyketide Synthase Sequence: R.SPASTGQSLGR.R [Pbc transcript] | 1 | 3 | 0.517 | 40 * | 0.0005 | polyketide synthesis |
| S-Adenosyl-Methionine Transferases [Pbc transcript] | 4 | 9 | 3.846 | 314 | 2.0E −31 | methylation |
| Protein | Number of Significant Sequences | Sequence Coverage (%) | Score | E-Value | Function |
|---|---|---|---|---|---|
| Argininosuccinate Synthase [Ruegeria atlantica] | 2 | 10 | 72 | 3.2E −07 | urea cycle |
| S-Adenosylhomocysteine Hydrolase ** Sequence: R.ATDVMIGGK.R [Alexandrium] | 1 | 6 | 109 | 6.3E −11 | adenosine and homocysteine production |
| S-Adenosylmethionine Synthase [Mameliella Alba] | 2 | 7 | 71 | 4.0E −07 | SAM synthesis |
| Histidine Kinase YycG ** Sequence: K.NPLASLRSAVGSLR.M [Mameliella Alba] | 1 | 1 | 52 * | 3.1E −05 | signal transduction |
| Aminomethyltransferase ** Sequence: K.AGLMDVSGLK.K [Ruegeria Atlantica] | 1 | 3 | 50 * | 0.00005 | methylenetetra-hydrofolate catabolism |
| Ferredoxin II ** Sequence: K.FSEQWPVIVTK.K [Ruegeria Atlantica] | 1 | 12 | 35 * | 0.0020 | electron transfer |
| RHS Repeat-Associated Core Domain Protein ** Sequence: R.ATWAPGAAGDWQSTGGMLTNAGATGPATLTAATADPAR.G [Mameliella Alba] | 1 | 3 | 34 * | 0.0020 | bacterial exotoxin |
| Adenylate Kinase ** Sequence: R.TLAQADALDALLAK.H [Marinovum Algicola] | 1 | 8 | 33 * | 0.0030 | cellular energy homeostasis |
| Transposase ** Sequence: R.DWIGAVGAK.T [Ruegeria Atlantica] | 1 | 23 | 25 * | 0.020 | transposon transporter |
| Methyltransferase** Sequence: R.ITTGVGK.G [Marinovum Algicola] | 1 | 4 | 23 * | 0.030 | methylation |
| ATP Sulfurylase/Adenylylsulfate Kinase (apsK) ** Sequence: K.VYLGGPVTGIQQPVHYDFR.G [Marinovum Algicola] | 1 | 3 | 20 * | 0.05 | activated sulfate synthesis |
| Proteins abundant in A. minutum | |||||||
|---|---|---|---|---|---|---|---|
| Protein Spot Code | Protein | Number of Peptides | Sequence Coverage (%) | Average Relative Intensity (A.M./A.T.) a | Score | E-Value | Function |
| A5 * | Ribulose Bisphosphate Carboxylase [Acidithiobacillus Caldus] | 4 | 13 | 2.05 | 93 | 2.5E −11 | carbon fixation |
| A6 * | Ribulose Bisphosphate Carboxylase [Acidithiobacillus Caldus] | 4 | 13 | 3.84 | 103 | 2.5E −12 | carbon fixation |
| A9 | Penicillin-Binding Protein [Parcubacteria Bacterium] | 3 | 2 | 3.7 | 66 | 1.3E −08 | peptidoglycan biosynthesis |
| A20 | Uncharacterized Protein ** Sequence: R.IAAELADGR.R [Streptosporangium Roseum] | 1 | 3 | 8.9 | 164 | 2.0E −18 | no annotation |
| A23 | Histidine Kinase [Geobacter Pickeringii] | 3 | 1 | 2.81 | 92 | 3.2E −11 | signal transduction |
| A36 | Uncharacterized Protein [Streptosporangium Roseum] | 3 | 3 | 2.71 | 68 | 7.9E −09 | no annotation |
| Proteins abundant in A. tamutum | |||||||
| Protein Spot Code | Protein | Number of Peptides | Sequence Coverage (%) | Average Relative Intensity (A.T./A.M.) a | Score | E-Value | Function |
| A14 | Thiamine-Monophosphate Kinase [Bacteroides Vulgatus] | 3 | 3 | 4.11 | 68 | 7.92E −09 | thiamine biosynthesis |
| A17 | tRNA Modification GTPase MnmE [Bosea sp. 117] | 2 | 3 | 3.27 | 80 | 5.0E −10 | tRNA methylation |
| A18 | Hypothetical Protein [Actinoplanes Globisporus] | 2 | 1 | 4.64 | 65 | 1.6E −08 | No annotation |
| A19 | Uncharacterized Protein [Lautropia Mirabilis ATCC 51599] | 2 | 2 | 2.37 | 68 | 8.0E −09 | No annotation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subong, B.J.J.; Lluisma, A.O.; Azanza, R.V.; Salvador-Reyes, L.A. Differentiating Two Closely Related Alexandrium Species Using Comparative Quantitative Proteomics. Toxins 2021, 13, 7. https://doi.org/10.3390/toxins13010007
Subong BJJ, Lluisma AO, Azanza RV, Salvador-Reyes LA. Differentiating Two Closely Related Alexandrium Species Using Comparative Quantitative Proteomics. Toxins. 2021; 13(1):7. https://doi.org/10.3390/toxins13010007
Chicago/Turabian StyleSubong, Bryan John J., Arturo O. Lluisma, Rhodora V. Azanza, and Lilibeth A. Salvador-Reyes. 2021. "Differentiating Two Closely Related Alexandrium Species Using Comparative Quantitative Proteomics" Toxins 13, no. 1: 7. https://doi.org/10.3390/toxins13010007
APA StyleSubong, B. J. J., Lluisma, A. O., Azanza, R. V., & Salvador-Reyes, L. A. (2021). Differentiating Two Closely Related Alexandrium Species Using Comparative Quantitative Proteomics. Toxins, 13(1), 7. https://doi.org/10.3390/toxins13010007