Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox
Abstract
1. Introduction
2. Conserved Mechanisms of Two-Component Systems Apply to Quorum-Mediated Virulence Regulation in S. aureus
3. Agr Signaling Mechanisms Regulate Toxin and Immunomodulatory Protein Production during S. aureus Osteomyelitis
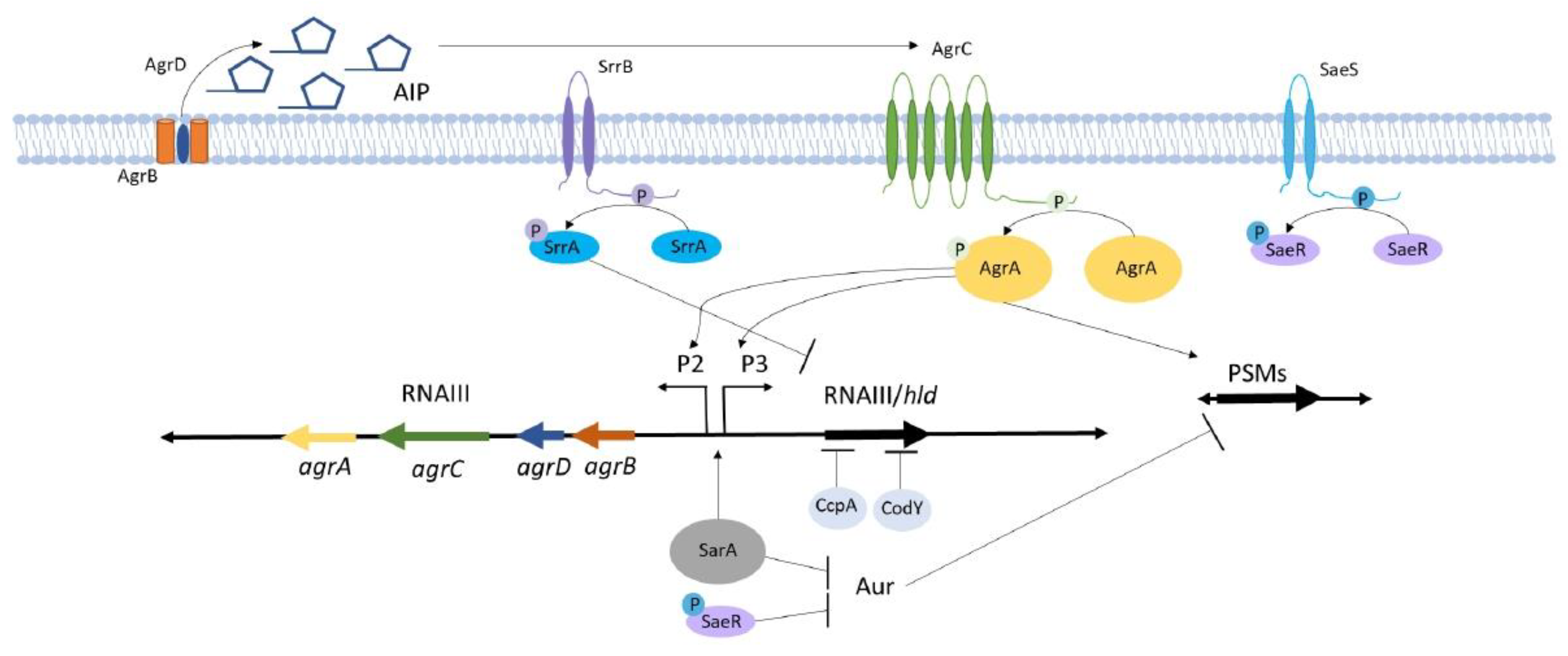
3.1. Agr Signaling Components Interact to Control Virulence Genes
3.2. S. aureus Cytolytic Toxins Regulated by Agr
3.2.1. Phenol-Soluble Modulins (PSMs)
3.2.2. Alpha-Hemolysin (Hla)
3.2.3. Panton–Valentine Leukocidin (PVL)
3.3. S. aureus Immunomodulatory Molecules Regulated by Agr
3.3.1. Protein A (SpA)
3.3.2. MHC Class II Analog Protein (Map)
3.4. S. aureus Coagulases Regulated by Agr
3.4.1. Staphylocoagulase (Coa)
3.4.2. Von Willebrand Factor-Binding Protein (vWbp)
4. Regulatory Control of AIP Is Strain and Species Specific
5. S. aureus Clinical Isolates with agr Mutations
5.1. Characteristic S. aureus agr Mutations Are Isolated from Patients with Chronic Infection and Have the Ability to Revert to Wildtype
5.2. Agr Expression Is Temporally Regulated to Promote S. aureus Adhesion and Toxin Production at Distinct Phases of Infection
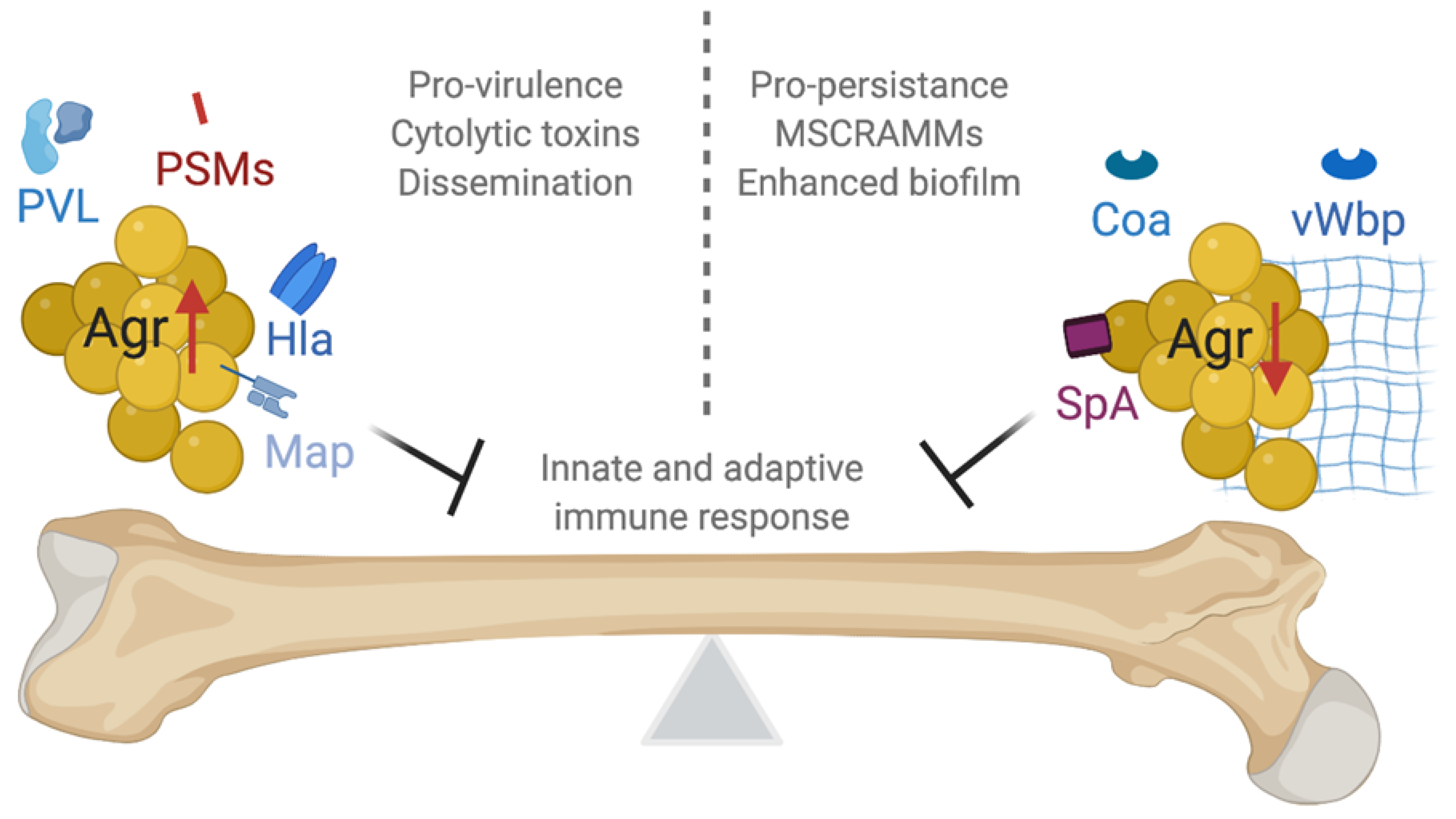
5.3. The Agr System Has Paradoxical Effects during Infection
6. Environmental Factors Influence S. aureus Agr Regulation Through Crosstalk with Global Regulators
6.1. Staphylococcal Respiratory Response AB (SrrAB) Senses and Responds to Changes in Cellular Redox
6.2. Staphylococcal Accessory Regulator A (SarA) Is a Critical Global Regulator that Crosstalks with the Agr System and Contributes to Osteomyelitis Pathogenesis
6.3. The S. aureus Exoprotein Expression (SaeRS) Two-Component System Regulates Proteases that Influence Osteomyelitis-Associated Bone Destruction
6.4. Metabolite-Responsive Transcription Factors Regulate S. aureus Metabolism and Virulence
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lew, D.P.; Waldvogel, F.A. Osteomyelitis. Lancet 2004, 364, 369–379. [Google Scholar] [CrossRef]
- Harik, N.S.; Smeltzer, M.S. Management of acute hematogenous osteomyelitis in children. Expert Rev. Anti Infect. Ther. 2010, 8, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hatzenbuehler, J.; Pulling, T.J. Diagnosis and management of osteomyelitis. Am. Fam. Physician 2011, 84, 1027–1033. [Google Scholar] [PubMed]
- Spagnolo, N.; Greco, F.; Rossi, A.; Ciolli, L.; Teti, A.; Posteraro, P. Chronic staphylococcal osteomyelitis: A new experimental rat model. Infect. Immun. 1993, 61, 5225–5230. [Google Scholar] [CrossRef] [PubMed]
- Power, M.E.; Olson, M.E.; Domingue, P.A.G.; Costerton, J.W. A rat model of Staphylococcus aureus chronic osteomyelitis that provides a suitable system for studying the human infection. J. Med. Microbiol. 1990, 33, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Alderson, M.; Speers, D.; Emslie, K.; Nade, S. Acute haematogenous osteomyelitis and septic arthritis—A single disease. A hypothesis based upon the presence of transphyseal blood vessels. J. Bone Jt. Surg. Br. 1986, 68, 268–274. [Google Scholar] [CrossRef]
- Cassat, J.E.; Hammer, N.D.; Campbell, J.P.; Benson, M.A.; Perrien, D.S.; Mrak, L.N.; Smeltzer, M.S.; Torres, V.J.; Skaar, E.P. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 2013, 13, 759–772. [Google Scholar] [CrossRef]
- Hannan, C.; Attinger, C. Special considerations in the management of osteomyelitis defects (diabetes, the ischemic or dysvascular bed, and irradiation). Semin. Plast. Surg. 2009, 23, 132–140. [Google Scholar] [CrossRef]
- De Mesy Bentley, K.L.; Trombetta, R.; Nishitani, K.; Bello-Irizarry, S.N.; Ninomiya, M.; Zhang, L.; Li, C.H.; McGrath, J.L.; Daiss, J.L.; Awad, H.A.; et al. Evidence of Staphylococcus aureus deformation, proliferation, and migration in canaliculi of live cortical bone in murine models of osteomyelitis. J. Bone Min. Res. 2017, 32, 985–990. [Google Scholar] [CrossRef]
- Lowy, F.D. Medical progress: Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Kong, C.; Neoh, H.M.; Nathan, S. Targeting staphylococcus aureus toxins: A potential form of anti-virulence therapy. Toxins 2016, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Brandis, G.; Cao, S.; Huseby, D.L.; Hughes, D. Having your cake and eating it-Staphylococcus aureus small colony variants can evolve faster growth rate without losing their antibiotic resistance. Microb. Cell 2017, 4, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Bagnoli, F. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation. Curr. Top. Microbiol. 2017, 409, 145–198. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Steiner, B.D.; Eberly, A.R.; Hurst, M.N.; Zhang, E.W.; Green, H.D.; Behr, S.; Jung, K.; Hadjifrangiskou, M. Evidence of cross-regulation in two closely related pyruvate-sensing systems in uropathogenic Escherichia coli. J. Membr. Biol. 2018, 251, 65–74. [Google Scholar] [CrossRef]
- Mike, L.A.; Choby, J.E.; Brinkman, P.R.; Olive, L.Q.; Dutter, B.F.; Ivan, S.J.; Gibbs, C.M.; Sulikowski, G.A.; Staugg, D.L.; Skaar, E.P. Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef]
- Sloan, T.J.; Murray, E.; Yokoyama, M.; Massey, R.C.; Chan, W.C.; Bonev, B.B.; Williams, P. Timing is everything: Impact of naturally occurring Staphylococcus aureus AgrC cytoplasmic domain adaptive mutations on autoinduction. J. Bacteriol. 2019, 201, 409–428. [Google Scholar] [CrossRef]
- Lebeau, C.; Vandenesch, F.; Greenland, T.; Novick, R.P.; Etienne, J. Coagulase expression in Staphylococcus aureus is positively and negatively modulated by an agr-dependent mechanism. J. Bacteriol. 1994, 176, 5534–5536. [Google Scholar] [CrossRef]
- Suligoy, C.M.; Lattar, S.M.; Noto Llana, M.; Gonzalez, C.D.; Alvarez, L.P.; Robinson, D.A.; Gomez, M.I.; Buzzola, F.R.; Sordelli, D.O. Mutation of agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front. Cell Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Voyich, J.M.; Vuong, C.; De Wald, M.; Nygaard, T.K.; Kocianova, S.; Griffith, S.; Jones, J.; Iverson, C.; Sturdevant, D.E.; Braughton, K.R.; et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 2009, 199, 1698–1706. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.M.; Anderson, K.L.; Beenken, K.E.; Smeltzer, M.S.; Dunman, P.M. The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front. Cell Infect. Microbiol. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Somerville, G.A.; Proctor, R.A. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 2009, 73, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Pragman, A.A.; Yarwood, J.M.; Tripp, T.J.; Schlievert, P.M. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2430–2438. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.D.; Snyder, D.J.; Putnam, N.E.; Valentino, M.D.; Hammer, N.D.; Lonergan, Z.R.; Hinger, S.A.; Aysanoa, E.E.; Blanchard, C.; Dunman, P.M.; et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015, 11, e1005341. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Todd, D.A.; Velázquez, J.V.; Cech, N.B.; Sonenshein, A.L. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol. 2014, 196, 1184–1196. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Sadykov, M.R.; Luong, T.T.; Lee, C.; Somerville, G.A.; Sonenshein, A.L. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 2008, 190, 2257–2265. [Google Scholar] [CrossRef]
- Ji, G.; Beavis, R.C.; Novick, R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 1995, 92, 12055–12059. [Google Scholar] [CrossRef]
- Novick, R.R.; Projan, S.J.; Kornblum, J.; Ross, H.F. The agr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 1995, 248, 446–458. [Google Scholar] [CrossRef]
- Rajasree, K.; Fasim, A.; Gopal, B. Conformational features of the Staphylococcus aureus AgrA-promoter interactions rationalize quorum-sensing triggered gene expression. Biochem. Biophys. Rep. 2016, 6, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Vandenesch, F.; Kornblum, J.; Novick, R.P. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 1991, 173, 6313–6320. [Google Scholar] [CrossRef] [PubMed]
- Geisinger, E.; Adhikari, R.P.; Jin, R.; Ross, H.F.; Novick, R.P. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006, 61, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. Embo. J. 1995, 14, 4569–4577. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.-H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the Agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef]
- Arvidson, S.; Tegmark, K. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 2001, 291, 159–170. [Google Scholar] [CrossRef]
- Phonimdaen, P.; O’Reilly, M.; O’Toole, P.W.; Foster, T.J. Molecular cloning and expression of the coagulase gene of Staphylococcus aureus 8325-4. J. Gen. Microbiol. 1988, 134, 75–83. [Google Scholar] [CrossRef]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. Embo. J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Patel, A.H.; Kornblum, J.; Kreiswirth, B.; Novick, R.; Foster, T.J. Regulation of the protein A-encoding gene in Staphylococcus aureus. Gene 1992, 114, 25–34. [Google Scholar] [CrossRef]
- Chabelskaya, S.; Rie Bordeau, V.; Felden, B. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic. Acid 2014, 42, 4847–4858. [Google Scholar] [CrossRef]
- Liu, Y.; Mu, C.; Ying, X.; Li, W.; Wu, N.; Dong, J.; Gao, Y.; Shao, N.; Fan, M.; Yang, G. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. Febs. Lett. 2011, 585, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Boisset, S.; Saveanu, C.; Benito, Y.; Geissmann, T.; Namane, A.; Lina, K.; Etienne, J.; Ehresmann, B.; Ehresmann, C.; et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005, 24, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Luong, T.T.; Lee, C.Y. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 14036–14041. [Google Scholar] [CrossRef]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef]
- Gillaspy, A.F.; Hickmon, S.G.; Skinner, R.A.; Thomas, J.R.; Nelson, C.L.; Smeltzer, M.S. Role of the accessory gene regulator (Agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 1995, 63, 3373–3380. [Google Scholar] [CrossRef]
- Blevins, J.S.; Elasri, M.O.; Allmendinger, S.D.; Beenken, K.E.; Skinner, R.A.; Thomas, R.J.; Smeltzer, M.S. Role of sarA in the Pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 2003, 71, 516–523. [Google Scholar] [CrossRef]
- Hendrix, A.S.; Spoonmore, T.J.; Wilde, A.D.; Putnam, N.E.; Hammer, N.D.; Snyder, D.J.; Guelcher, S.A.; Skaar, E.P.; Cassat, J.E. Repurposing the nonsteroidal anti-inflammatory drug diflunisal as an osteoprotective, antivirulence therapy for Staphylococcus aureus osteomyelitis. Antimicrob. Agents Chemother. 2016, 60, 5322–5330. [Google Scholar] [CrossRef]
- Bouras, D.; Doudoulakakis, A.; Tsolia, M.; Vaki, I.; Giormezis, N.; Petropoulou, N.; Lebessi, E.; Gennimata, V.; Tsakris, A.; Spiliopoulou, I.; et al. Staphylococcus aureus osteoarticular infections in children: An 8-year review of molecular microbiology, antibiotic resistance and clinical characteristics. J. Med. Microbiol. 2018, 67, 1753–1760. [Google Scholar] [CrossRef]
- Krishna Mannala, G.; Koettnitz, J.; Mohamed, W.; Sommer, U.; Lips, K.S.; Sproer, C.; Bunk, B.; Overmann, J.; Hain, T.; Heiss, C.; et al. Whole-genome comparison of high and low virulent Staphylococcus aureus isolates inducing implant-associated bone infections. Int. J. Med. Microbiol. 2018, 208, 505–513. [Google Scholar] [CrossRef]
- Masters, E.A.; Salminen, A.T.; Begolo, S.; Luke, E.N.; Barrett, S.C.; Overby, C.T.; Gill, A.L.; de Mesy Bentley, K.L.; Awad, H.A.; Gill, S.R.; et al. An in vitro platform for elucidating the molecular genetics of S. aureus invasion of the osteocyte lacuno-canalicular network during chronic osteomyelitis. Nanomedicine 2019, 21. [Google Scholar] [CrossRef] [PubMed]
- Kwiecinski, J.M.; Jacobsson, G.; Horswill, A.R.; Josefsson, E.; Jin, T. Biofilm formation by Staphylococcus aureus clinical isolates correlates with the infection type. Infect. Dis. 2019, 51, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Joo, H.S.; Duong, A.C.; Dieringer, T.D.; Tan, V.Y.; Song, T.; Fischer, E.R.; Cheung, G.Y.C.; Li, M.; Otto, M. Essential Staphylococcus aureus toxin export system. Nat. Med. 2013, 19, 364–367. [Google Scholar] [CrossRef]
- Rasigade, J.-P.; Trouillet-Assant, S.; Ferry, T.; Diep, B.A.; Sapin, A.; Lhoste, Y.; Ranfaing, J.; Badiou, C.; Bento, Y.; Bes, M.; et al. PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS ONE 2013, 8, e63176. [Google Scholar] [CrossRef]
- Kretschmer, D.; Gleske, A.-K.; Rautenberg, M.; Wang, R.; Koberle, M.; Bohn, E.; Schoneberg, T.; Rabiet, M.-J.; Boulay, F.; Klebanoff, S.J.; et al. Human formyl peptide receptor 2 (FPR2/ALX) senses highly pathogenic Staphylococcus aureus. Cell Host Microbe. 2010, 7, 463–473. [Google Scholar] [CrossRef]
- Surewaard, B.G.J.; De Haas, C.J.C.; Vervoort, F.; Rigby, K.M.; Deleo, F.R.; Otto, M.; Van Stijp, J.A.G.; Nijland, R. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol. 2013, 15, 1427–1437. [Google Scholar] [CrossRef]
- Brandt, S.L.; Putnam, N.E.; Cassat, J.E.; Serezani, H. Innate immunity to Staphylococcus aureus: Evolving paradigms in soft tissue and invasive infection. J. Immunol. 2019, 200, 3871–3880. [Google Scholar] [CrossRef]
- Füssle, R.; Bhakdi, S.; Sziegoleit, A.; Tranum-Jensen, J.; Kranz, T.; Wellensiek, H.-J. On the mechanism of membrane damage by Staphylococcus aureus a-Toxin. J. Cell Biol. 1981, 91, 83–94. [Google Scholar] [CrossRef]
- Ward, R.J.; Leonard, K. The Staphylococcus aureus α-toxin channel complex and the effect of Ca2+ ions on its interaction with lipid layers. J. Struct. Biol. 1992, 109, 129–141. [Google Scholar] [CrossRef]
- Tobkes, N.; Wallace, B.A.; Bayley, H. Secondary structure and assembly mechanism of an oligomeric channel protein. Biochemistry 1985, 24, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Wilke, G.A.; Wardenburg, J.B. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. USA 2010, 107, 13473–13478. [Google Scholar] [CrossRef] [PubMed]
- Berube, B.J.; Wardenburg, J.B. Staphylococcus aureus α-toxin: Nearly a century of intrigue. Toxins 2013, 5, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gulbins, E.; Edwards, M.J.; Caldwell, C.C.; Fraunholz, M.; Becker, K.A. Staphylococcus aureus α-toxin induces inflammatory cytokines via lysosomal acid sphingomyelinase and ceramides. Cell. Physiol. Biochem. 2017, 43, 2170–2184. [Google Scholar] [CrossRef] [PubMed]
- Craven, R.R.; Gao, X.; Allen, I.C.; Gris, D.; Bubeck Wardenburg, J.; McElvania-TeKippe, E.; Ting, J.P.; Duncan, J.A. Staphylococcus aureus α-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Nilsson, I.M.; Hartford, O.; Foster, T.; Tarkowski, A. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect. Immun. 1999, 67, 1045–1049. [Google Scholar] [CrossRef]
- Smith, I.D.M.; Milto, K.M.; Doherty, C.J.; Amyes, S.G.B.; Simpson, A.H.R.W.; Hall, A.C. A potential key role for alpha-haemolysin of Staphylococcus aureus in mediating chondrocyte death in septic arthritis. Bone Jt. Res. 2018, 7, 457–467. [Google Scholar] [CrossRef]
- Yoong, P.; Torres, V.J. The effects of Staphylococcus aureus leukotoxins on the host: Cell lysis and beyond. Curr. Opin. Microbiol. 2013, 16, 63–69. [Google Scholar] [CrossRef]
- Colin, D.A.; Mazurier, I.; Sire, S.; Finck-Barbancon, V. Interaction of the two components of leukocidin from Staphylococcus aureus with human polymorphonuclear leukocyte membranes: Sequential binding and subsequent activation. Infect. Immun. 1994, 62, 3184–3188. [Google Scholar] [CrossRef]
- Menestrina, G.; Dalla Serra, M.; Comai, M.; Coraiola, M.; Viero, G.; Werner, S.; Colin, D.A.; Monteil, H.; Prevost, G. Ion channels and bacterial infection: The case of β-barrel pore-forming protein toxins of Staphylococcus aureus. FEBS Lett. 2003, 552, 54–60. [Google Scholar] [CrossRef]
- Cré Mieux, A.-C.; Dumitrescu, O.; Lina, G.; Vallee, C.; Cote, J.-F.; Muffat-Joly, M.; Lilin, T.; Etienne, J.; Vandenesch, F.; Saleh-Mghir, A. Panton-Valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. PLoS ONE 2019, 4, e7204. [Google Scholar] [CrossRef]
- Holzinger, D.; Gieldon, L.; Mysore, V.; Nippe, N.; Taxman, D.J.; Duncan, J.; Broglie, P.M.; Marketon, K.; Austermann, J.; Vogl, T.; et al. Staphylococcus aureus Panton-Valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J. Leukoc. Biol. 2012, 92, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Wang, Y.; Feng, Z.; Xu, L.; Zhao, S.; Gong, Y.; Zhang, C.; Luo, X.; Li, S.; Rao, X.; et al. Panton-Valentine leucocidin (PVL) as a potential indicator for prevalence, duration, and severity of Staphylococcus aureus osteomyelitis. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bocchini, C.E.; Hulten, K.G.; Mason, E.O.; Gonzalez, B.E.; Hammerman, W.A.; Kaplan, S.L. Panton-Valentine leukocidin genes are associated with enhanced inflammatory response and local disease in acute hematogenous Staphylococcus aureus osteomyelitis in children. Pediatrics 2006, 117, 433–440. [Google Scholar] [CrossRef]
- Jin, T.; Zhu, Y.; Li, J.; Shi, J.; He, X.Q.; Ding, J.; Xu, Y.Q. Staphylococcal protein A, Panton-Valentine leukocidin and coagulase aggravate the bone loss and bone destruction in osteomyelitis. Cell Physiol. Biochem. 2013, 32, 322–333. [Google Scholar] [CrossRef]
- Dohin, B.; Gillet, Y.; Kohler, R.; Lina, G.; Vandenesch, F.; Vanhems, P.; Floret, D.; Etienne, J. Pediatric bone and joint infections caused by Panton-Valentine leukocidin-positive Staphylococcus aureus. Pediatr. Infect. Dis. J. 2007, 26, 1042–1048. [Google Scholar] [CrossRef]
- Wardenburg, J.B.; Palazzolo-Ballance, A.M.; Otto, M.; Schneewind, O.; DeLeo, F.R. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 2008, 198, 1166–1170. [Google Scholar] [CrossRef]
- Tromp, A.T.; Van Gent, M.; Abrial, P.; Martin, A.; Jansen, J.P.; De Haas, C.J.C.; Van Kessel, K.P.M.; Bardoel, B.W.; Kruse, E.; Bourdonnay, E.; et al. Human CD45 is an f-component-specific receptor for the staphylococcal toxin Panton-Valentine leukocidin. Nat. Microbiol. 2018, 3, 708–717. [Google Scholar] [CrossRef]
- Löffler, B.; Hussain, M.; Grundmeier, M.; Bruck, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peter, G. Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef]
- Dossett, J.H.; Kronvall, G.; Williams, R.C.; Quie, P.G. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. J. Immunol. 1969, 103, 1405–1410. [Google Scholar]
- Kobayashi, S.D.; DeLeo, F.R. Staphylococcus aureus protein A promotes immune suppression. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Goodyear, C.S.; Silverman, G.J. Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a staphylococcal toxin. J. Exp. Med. 2003, 197, 1125–3119. [Google Scholar] [CrossRef] [PubMed]
- Keener, A.B.; Thurlow, L.T.; Kang, S.; Spidale, N.A.; Clarke, S.H.; Cunnion, K.M.; Tisch, R.; Richardson, A.R.; Vilen, B.J. Staphylococcus aureus protein A disrupts immunity mediated by long-lived plasma cells. J. Immunol. 2017, 198, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Claro, T.; Widaa, A.; O’Seaghdha, M.; Miajlovic, H.; Foster, T.J.; O’Brien, F.J.; Kerrigan, S.W. Staphylococcus aureus Protein A binds to osteoblasts and triggers signals that weaken bone in osteomyelitis. PLoS ONE 2011, 6, e18748. [Google Scholar] [CrossRef] [PubMed]
- Claro, T.; Widaa, A.; McDonnell, C.; Foster, T.J.; O’Brien, F.J.; Kerrigan, S.W. Staphylococcus aureus protein A binding to osteoblast tumour necrosis factor receptor 1 results in activation of nuclear factor kappa B and release of interleukin-6 in bone infection. Microbiology 2013, 159, 147–154. [Google Scholar] [CrossRef]
- Widaa, A.; Claro, T.; Foster, T.J.; O’Brien, F.J.; Kerrigan, S.W. Staphylococcus aureus protein A plays a critical role in mediating bone destruction and bone loss in osteomyelitis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Mendoza Bertelli, A.; Delpino, M.V.; Lattar, S.; Giai, C.; Noto Llana, M.; Sanjuan, N.; Cassat, J.E.; Sordelli, D.; Gomez, M. Staphylococcus aureus protein A enhances osteoclastogenesis via TNFR1 and EGFR signaling. Biochim. Biophys. Acta 2016, 1862, 1975–1983. [Google Scholar] [CrossRef]
- Chavakis, T.; Hussain, M.; Kanse, S.M.; Peters, G.; Bretzel, R.G.; Flock, J.-I.; Herrmann, M.; Preissner, K.T. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 2002, 8, 687–693. [Google Scholar] [CrossRef]
- Haggar, A.; Ehrnfelt, C.; Holgersson, J.; Flock, J.-I. The extracellular adherence protein from Staphylococcus aureus inhibits neutrophil binding to endothelial cells. Infect. Immun. 2004, 72, 6164–6167. [Google Scholar] [CrossRef]
- Lee, L.Y.; Miyamoto, Y.J.; McIntyre, B.W.; Hook, M.; McCrea, K.W.; McDevill, D.; Brown, E.L. The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell-mediated responses. J. Clin. Investig. 2002, 110, 1461–1471. [Google Scholar] [CrossRef]
- Wolz, C.; McDevitt, D.; Foster, T.J.; Cheung, A.L. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 1996, 64, 3142–3147. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; You, Y.; Shang, F.; Sun, B. Rot and Agr system modulate fibrinogen-binding ability mainly by regulating clfB expression in Staphylococcus aureus NCTC8325. Med. Microbiol. Immunol. 2012, 201, 81–92. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, R.; Cockayne, A.; Humphreys, H. Clinical and molecular aspects of the pathogenesis of Staphylococcus aureus bone and joint infections. J. Med. Microbiol. 1996, 44, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Panizzi, P.; Friedrich, R.; Fuentes-Prior, P.; Richter, K.; Bock, P.E.; Bode, W. Fibrinogen substrate recognition by staphylocoagulase (pro)thrombin complexes. J. Biol. Chem. 2006, 281, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.G.; McAdow, M.; Kim, H.K.; Bae, T.; Missiakas, D.M.; Schneewind, O. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010, 6, e1001036. [Google Scholar] [CrossRef]
- Guggenberger, C.; Wolz, C.; Morrissey, J.A.; Heesemann, J. Two distinct coagulase-dependent barriers protect Staphylococcus aureus from neutrophils in a three dimensional in vitro infection model. PLoS Pathog. 2012, 8, e1002434. [Google Scholar] [CrossRef]
- Thomas, S.; Liu, W.; Arora, S.; Ganesh, V.; Ko, Y.-P.; Hook, M. The complex fibrinogen interactions of the Staphylococcus aureus coagulases. Front. Cell Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Dufour, P.; Jarraud, S.; Vandenesch, F.; Greenland, T.; Novick, R.P.; Bes, M.; Etienee, J.; Lina, G. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 2002, 184, 1180–1186. [Google Scholar] [CrossRef]
- Jarraud, S.; Lyon, G.J.; Figueiredo, A.M.; Lina, G.; Vandenesch, F.; Etienee, J.; Muir, T.W.; Novick, R.P. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 2000, 182, 6517–6522. [Google Scholar] [CrossRef]
- Guangyong, J.; Beavis, R.; Novick, R. Bacterial interference caused by autoinducing peptide variants. Science 1997, 27, 2027–2030. [Google Scholar]
- Zhang, L.; Ji, G. Identification of a staphylococcal AgrB segment(s) responsible for group-specific processing of AgrD by gene swapping. J. Bacteriol. 2004, 186, 6706–6713. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Biswas, R.; Götz, F.; Biswas, L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immune. 2014, 82, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Parlet, C.P.; Chung, N.; Todd, D.A.; Rodriguez, L.I.; Van Dyke, M.J.; Cech, N.B.; Horswill, A.R. Coagulase-negative Staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 2017, 22, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Souza Jorge, L.; Silva Fucuta, P.; Oliveira, M.G.L.; Arruda Nakazone, M.; Arruda de Matos, J.; Gomes Chueire, A.; Costa Salles, M.J. Outcomes and risk factors for polymicrobial posttraumatic osteomyelitis. J. Bone Joint. Infect. 2018, 3, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Matias, C.; Serrano, I.; Van-Harten, S.; Mottola, C.; Mendes, J.J.; Tavares, L.; Oliveira, M. Polymicrobial interactions influence the agr copy number in Staphylococcus aureus isolates from diabetic foot ulcers. Antonie Van Leeuwenhoek 2018, 111, 2225–2232. [Google Scholar] [CrossRef]
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus pathogenicity and polymicrobial interactions: Lessons beyond Koch’s postulates. J. Fungi. 2019, 5, 81. [Google Scholar] [CrossRef]
- Shopsin, B.; Drlica-Wagner, A.; Mathema, B.; Adhikari, R.P.; Kreiswirth, B.N.; Novick, R.P. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 2008, 198, 1171–1174. [Google Scholar] [CrossRef]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. Agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar] [CrossRef]
- Schweizer, M.L.; Furuno, J.P.; Sakoulas, G.; Johnson, J.K.; Harris, A.D.; Shardell, M.D.; McGregor, J.C.; Thom, K.A.; Perencevich, E.N. Increased mortality with accessory gene regulator (agr) dysfunction in Staphylococcus aureus among bacteremic patients. Antimicrob. Agents Chemother. 2011, 55, 1082–1087. [Google Scholar] [CrossRef]
- Altman, D.R.; Sullivan, M.J.; Chacko, K.I.; Balasubramanian, D.; Pak, T.R.; Sause, W.E.; Kumar, K.; Sebra, R.; Deikus, G.; Attie, O.; et al. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Gor, V.; Takemura, A.J.; Nishitani, M.; Higashide, M.; Medrano Romero, V.; Ohniwa, R.L.; Morikawa, K. Finding of Agr phase variants in Staphylococcus aureus. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Buckling, A.; Neilson, J.; Lindsay, J.; ffrench-Constant, R.; Enright, M.; Day, N.; Massey, R.C. Clonal distribution and phase-variable expression of a major histocompatibility complex analogue protein in Staphylococcus aureus. J. Bacteriol. 2005, 187, 2917–2919. [Google Scholar] [CrossRef] [PubMed]
- Traber, K.; Novick, R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ- and α-haemolysins. Mol. Microbiol. 2006, 59, 1519–1530. [Google Scholar] [CrossRef]
- Cheng, A.G.; DeDent, A.C.; Schneewind, O.; Missiakas, D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011, 19, 225–232. [Google Scholar] [CrossRef]
- Carnes, E.C.; Lopez, D.M.; Donegan, N.P.; Donegan, N.P.; Cheung, A.; Gresham, H.; Timmins, G.S.; Brinker, C.J. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 2010, 6, 41–45. [Google Scholar] [CrossRef]
- Kolar, S.L.; Antonio Ibarra, J.; Rivera, F.E.; Mootz, J.M.; Davenport, J.E.; Stevens, S.M.; Horswill, A.R.; Shaw, L.N. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2013, 2, 18–34. [Google Scholar] [CrossRef]
- Fowler, V.G.; Sakoulas, G.; Mcintyre, L.M.; Meka, V.G.; Arbeit, R.D.; Cabell, C.H.; Stryjewski, M.E.; Eliopoulos, G.M.; Reller, B.; Corey, R.; et al. Persistent bacteremia due to methicillin-resistant Staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. JID 2004, 190, 1140–1149. [Google Scholar] [CrossRef]
- Paulander, W.; Varming, A.N.; Bæk, K.T.; Haaber, J.; Frees, D.; Ingmer, H. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. Mbio 2012, 3. [Google Scholar] [CrossRef]
- He, L.; Le, K.Y.; Khan, B.A.; Nguyen, T.H.; Hung, R.L.; Bae, J.S.; Kabat, J.; Zheng, Y.; Cheung, G.Y.C.; Li, M.; et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat. Microbiol. 2019, 4, 1114–1119. [Google Scholar] [CrossRef]
- Nakamura, Y.; Takahashi, H.; Takaya, A.; Inoue, Y.; Katayama, Y.; Kusuya, Y.; Shoji, T.; Takada, S.; Nakagawa, S.; Oguma, R.; et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors 2013, 13, 5130–5166. [Google Scholar] [CrossRef]
- Khodaverdian, V.; Pesho, M.; Truitt, B.; Bollinger, L.; Patel, P.; Nithianantham, S.; Yu, G.; Delaney, E.; Jankowski, E.; Shoham, M. Discovery of antivirulence agents against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 3645–3652. [Google Scholar] [CrossRef] [PubMed]
- Spoonmore, T.J.; Ford, C.A.; Curry, J.M.; Guelcher, S.A.; Cassat, J.E. Concurrent local delivery of diflunisal limits bone destruction but fails to improve systemic vancomycin efficacy during Staphylococcus aureus osteomyelitis. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Nakayama, J.; Uemura, Y.; Nishiguchi, K.; Yoshimura, N.; Igarashi, Y.; Sonomoto, K. Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in gram-positive bacteria. Antimicrob. Agents Chemother. 2009, 53, 580–586. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench Agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365. [Google Scholar] [CrossRef]
- Brown, M.M.; Kwiecinski, J.M.; Cruz, L.M.; Shahbandi, A.; Todd, D.A.; Cech, N.B.; Horswill, A.R. Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Valour, F.; Rasigade, J.P.; Trouillet-Assant, S.; Gagnaire, J.; Bouaziz, A.; Karsenty, J.; Lacour, C.; Bes, M.; Lustig, S.; Benet, T.; et al. Delta-toxin production deficiency in Staphylococcus aureus: A diagnostic marker of bone and joint infection chronicity linked with osteoblast invasion and biofilm formation. Clin. Microbiol. Infect. 2015, 21, e1–e11. [Google Scholar] [CrossRef]
- Tiwari, N.; López-Redondo, M.; Miguel-Romero, L.; Kulhankova, K.; Cahill, M.P.; Tran, P.M.; Kinney, K.J.; Kilgore, S.H.; Al-Tameemi, H.; Herfst, C.A.; et al. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc. Natl. Acad. Sci. USA, 2020. [Google Scholar] [CrossRef]
- Mashruwala, A.A.; van de Guchte, A.; Boyd, J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 2017, 6. [Google Scholar] [CrossRef]
- Mashruwala, A.A.; Gries, C.M.; Scherr, T.D.; Kielian, T.; Boyd, J.M. SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed]
- Beenken, K.E.; Mrak, L.N.; Griffin, L.M.; Zielinska, A.K.; Shaw, L.N.; Rice, K.C.; Horswill, A.R.; Bayles, K.W.; Smeltzer, M.S. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Koomey, J.M.; Butler, C.A.; Projan, S.J.; Fischetti, V.A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 1992, 89, 6462–6466. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.L.; Projan, S.J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 1994, 176, 4168–4172. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.T.; Manna, A.C.; Cheung, A.L. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 1998, 30, 991–1001. [Google Scholar] [CrossRef]
- Reyes, D.; Andrey, D.O.; Monod, A.; Kelley, W.L.; Zhang, G.; Cheung, A.L. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J. Bacteriol. 2011, 193, 6020–6031. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Joo, H.-S.; Mrak, L.N.; Griffin, L.M.; Luong, T.T.; Lee, C.Y.; Otto, M.; Shaw, L.N.; Smeltzer, M.S. Defining the strain-dependent impact of the staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J. Bacteriol. 2011, 193, 2948–2958. [Google Scholar] [CrossRef]
- Lindsay, J.A.; Foster, S.J. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol. Gen. Genet. 1999, 262, 323–331. [Google Scholar] [CrossRef]
- Loughran, A.J.; Gaddy, D.; Beenken, K.E.; Meeker, D.G.; Morell, R.; Zhao, H.; Byrum, S.D.; Tackett, A.J.; Cassat, J.E.; Smeltzer, M.S. Impact of sarA and phenol-soluble modulins on the pathogenesis of osteomyelitis in diverse clinical isolates of Staphylococcus aureus. Infect. Immun. 2016, 84, 2586–2594. [Google Scholar] [CrossRef]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. SarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Li, C.; Jeong, D.; Sohn, C.; He, C.; Bae, T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR Binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 2010, 192, 2111–2127. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.W.; Cho, H.; Jones, M.B.; Shatzkes, K.; Sun, F.; Ji, Q.; Liu, Q.; Peterson, S.N.; He, C.; Bae, T. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 2012, 86, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Goerke, C.; Mainiero, M.; Kraus, D.; Wolz, C. The virulence regulator Sae of Staphylococcus aureus: Promoter activities and response to phagocytosis-related signals. J. Bacteriol. 2008, 190, 3419–3428. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Jeong, D.W.; Liu, Q.; Yeo, W.-S.; Vogl, T.; Skaar, E.P.; Chazen, W.J.; Bae, T. Calprotectin increases the activity of the SaeRS two component system and murine mortality during Staphylococcus aureus infections. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef]
- Weinrick, B.; Dunman, P.M.; McAleese, F.; Murphy, E.; Projan, S.J.; Fang, Y.; Novick, R.P. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 2004, 186, 8407–8423. [Google Scholar] [CrossRef]
- Geiger, T.; Francois, P.; Liebeke, M.; Fraunholz, M.; Goerke, C.; Krismer, B.; Schrenzel, J.; Lalk, M.; Wolz, C. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Path. 2012, 8, e1003016. [Google Scholar] [CrossRef]
- Pohl, K.; Francois, P.; Stenz, L.; Schlink, F.; Geiger, T.; Herbert, C.G.; Schrenzel, J.; Wolz, C. CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009, 191, 2953–2963. [Google Scholar] [CrossRef]
- Majerczyk, C.D.; Dunman, P.M.; Luong, T.T.; Lee, C.Y.; Sadykov, M.R.; Somerville, G.A.; Bodi, K.; Sonenshein, A.L. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010, 192, 2861–2877. [Google Scholar] [CrossRef]
- Seidl, K.; Stucki, M.; Rueg, M.; Goerke, C.; Wolz, C.; Harris, L.; Berger-Bachi, B.; Bischoff, M. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006, 50, 1183–1194. [Google Scholar] [CrossRef]
- Esen, E.; Long, F. Aerobic glycolysis in osteoblasts. Curr. Osteoporos. Rep. 2014, 12, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Jeong, D.; Kang, H.K.; Jung, S.Y.; Kang, S.S.; Min, B.-M. Osteoclast precursors display dynamic metabolic shifts toward accelerated glucose metabolism at an early stage of RANKL-stimulated osteoclast differentiation. Cell Physiol. Biochem. 2007, 20, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Potter, A.D.; Butrico, C.E.; Ford, C.A.; Curry, J.M.; Trenary, I.A.; Tummarakota, S.S.; Hendrix, A.S.; Young, J.D.; Cassat, J.E. Host nutrient milieu drives an essential role for aspartate biosynthesis during invasive Staphylococcus aureus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 12394–12401. [Google Scholar] [CrossRef]
- Bischoff, M.; Wonnenberg, B.; Nippe, N.; Nyffenegger-Jann, N.J.; Voss, M.; Beisswenger, C.; Sunderkotter, C.; Molle, V.; Dinh, Q.T.; Lammert, F.; et al. CcpA affects infectivity of Staphylococcus aureus in a hyperglycemic environment. Front. Cell Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.; Balasubramanian, D.; Ohneck, E.A.; Sause, W.E.; Chapman, J.; Mejia-Sosa, B.; Lhakhang, T.; Heguy, A.; Tsirigos, A.; Ueberheide, B.; et al. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. MBio 2018, 9. [Google Scholar] [CrossRef]
- Ky, L.; Otto, M. Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Mlynek, K.D.; Sause, W.E.; Moormeier, D.E.; Sadykov, M.R.; Hill, K.R.; Torres, V.J.; Bayles, K.W.; Brinsmade, S.R. Nutritional regulation of the Sae two-component system by CodY in Staphylococcus aureus. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Bronner, S.; Stoessel, P.; Gravet, A.; Monteil, H.; Prevost, G. Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl. Environ. Microbiol. 2000, 66, 3931–3938. [Google Scholar] [CrossRef]
| Publication | Year | Model | Critical Finding |
|---|---|---|---|
| Gillaspy, A.F. et al. [46] | 1995 | Rabbit | Agr increases osteomyelitis incidence and severity, but inactivation of agr does not inhibit S. aureus colonization of bone. |
| Blevins, J.S. et al. [47] | 2003 | Mouse | Agr mutation decreases osteomyelitis pathology scores, including number of abscesses, physis destruction, and histological inflammation. |
| Cassat, J.E. et al. [7] | 2013 | Mouse | Inactivation of agr decreases bone destruction, and Agr-regulated α-type PSM production contributes to bone destruction during S. aureus infection. |
| Hendrix, A.S. et al. [48] | 2016 | Mouse | Pharmacologic blockade of Agr signaling reduces osteomyelitis pathogenesis. |
| Wilde, A.D. et al. [26] | 2015 | Mouse | Agr-regulated PSM production is influenced by hypoxia and signaling through SrrAB during osteomyelitis. |
| Bouras, D. et al. [49] | 2018 | Human | Clinical pediatric methicillin resistant S. aureus (MRSA) isolates primarily contained agr-III systems. |
| Krishna Mannala, G. et al. [50] | 2018 | Human | A low-virulent clinical S. aureus bone isolate harbors frameshift mutations in the agrC gene. |
| Suligoy, C.M. et al. [21] | 2018 | Mouse | Lack of Agr-dependent factors decrease S. aureus virulence during chronic osteomyelitis while permitting adaptation for persistence. |
| Masters, E.A. et al. [51] | 2019 | Murine and in vitro | A S. aureus agr mutant retains its ability to invade the osteocyte lacuno-canalicular network of cortical bone. |
| Kwiecinski, J.M. et al. [52] | 2019 | Human | S. aureus agr type from clinical invasive infections correlates with biofilm formation. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butrico, C.E.; Cassat, J.E. Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox. Toxins 2020, 12, 516. https://doi.org/10.3390/toxins12080516
Butrico CE, Cassat JE. Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox. Toxins. 2020; 12(8):516. https://doi.org/10.3390/toxins12080516
Chicago/Turabian StyleButrico, Casey E., and James E. Cassat. 2020. "Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox" Toxins 12, no. 8: 516. https://doi.org/10.3390/toxins12080516
APA StyleButrico, C. E., & Cassat, J. E. (2020). Quorum Sensing and Toxin Production in Staphylococcus aureus Osteomyelitis: Pathogenesis and Paradox. Toxins, 12(8), 516. https://doi.org/10.3390/toxins12080516





