Purification and Characterization of the Pink-Floyd Drillipeptide, a Bioactive Venom Peptide from Clavus davidgilmouri (Gastropoda: Conoidea: Drilliidae)
Abstract
1. Introduction
2. Results
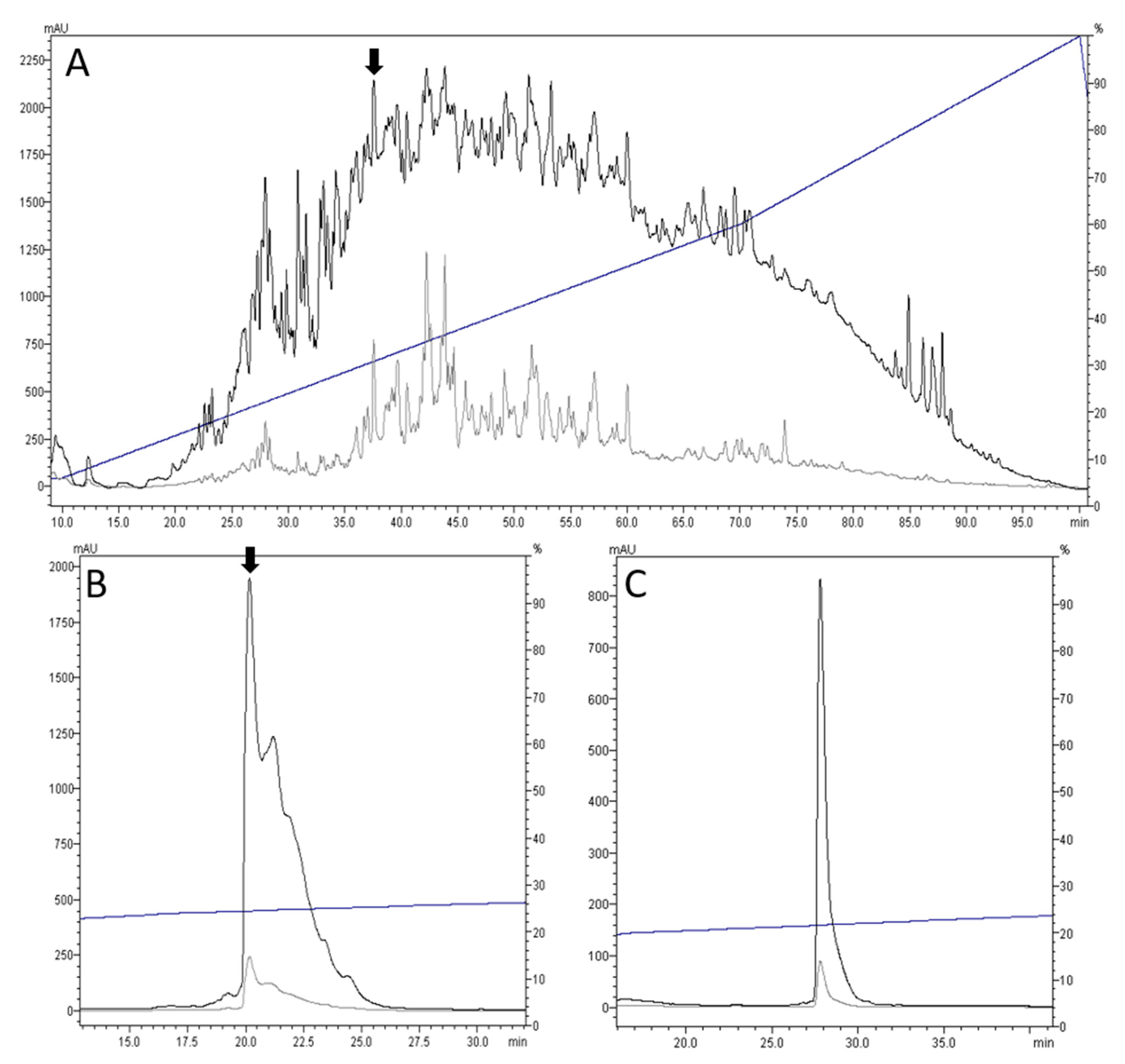
2.1. Identification and Initial Characterization of Drillipeptide cdg14a
2.2. Characterization of the Precursor of cdg14a
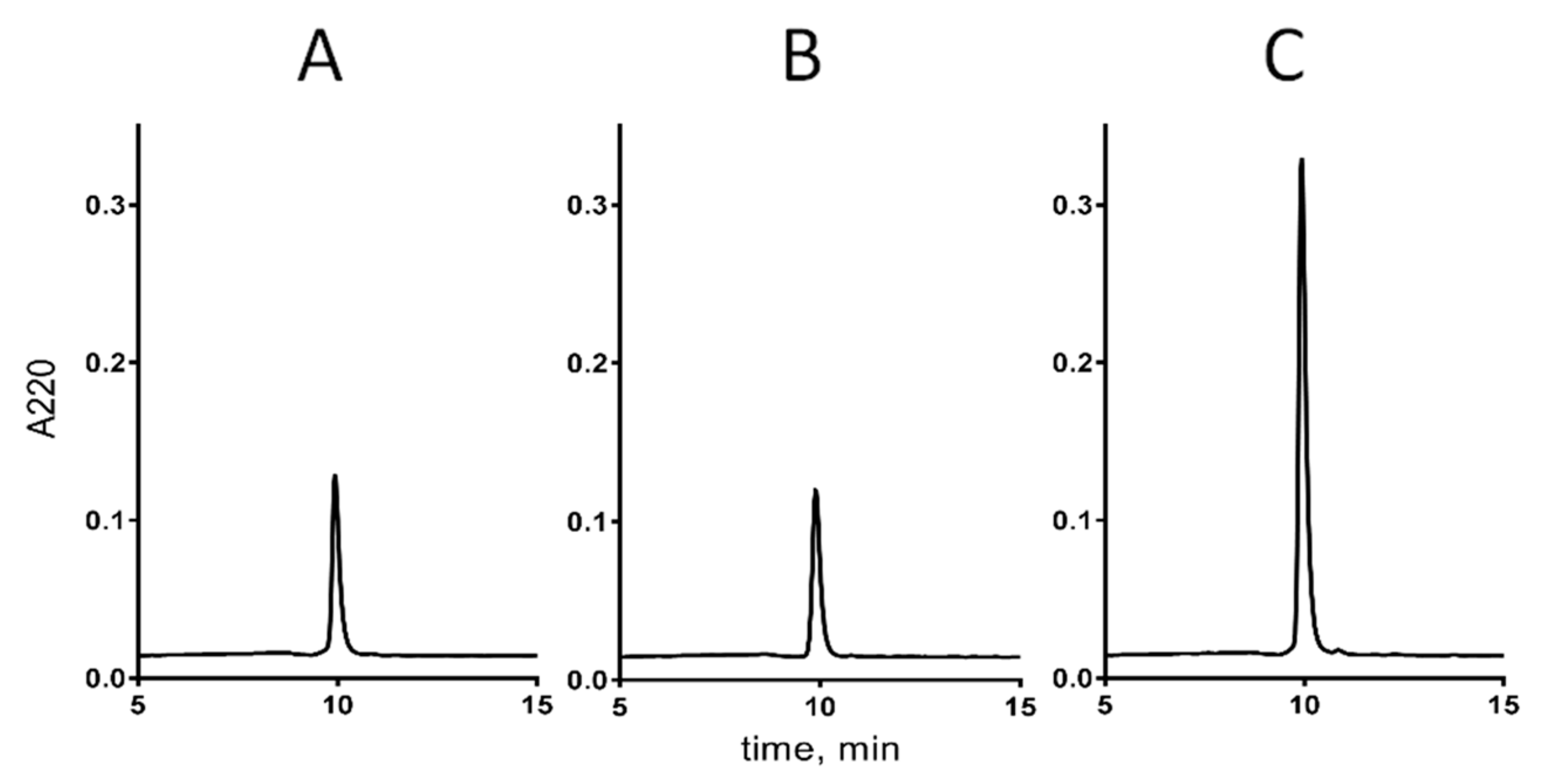
2.3. Chemical Synthesis of cdg14a
2.4. Behavioral Assay of cdg14a
2.5. Constellation Pharmacology
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Venom Extraction and Purification
4.3. Peptide Characterization
4.4. Intracranial Mouse Bioassay
4.5. RNA Sequencing and Transcriptome Library Assembly
4.6. Putative Toxin Identification
4.7. Peptide Synthesis
4.8. Oxidative Folding of cdg14a in the Presence of Oxidized and Reduced Glutathione
4.9. Co-Elution of the Native and Synthetic cdg14a
4.10. Constellation Pharmacology
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bouchet, P.; Lozouet, P.; Sysoev, A. An inordinate fondness for turrids. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2009, 56, 1724–1731. [Google Scholar] [CrossRef]
- Rosenberg, G. A New Critical Estimate of Named Species-Level Diversity of the Recent Mollusca. Am. Malacol. Bull. 2014, 32, 308–322. [Google Scholar] [CrossRef]
- Taylor, J.D.; Kantor, Y.I.; Sysoev, A.V. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda). Bull. Nat. Hist. Mus. Lond. Zool. 1993, 59, 125–170. [Google Scholar]
- Puillandre, N.; Samadi, S.; Boisselier, M.-C.; Sysoev, A.; Kantor, Y.I.; Cruaud, C.; Couloux, A.; Bouchet, P. Starting to unravel the toxoglossan knot: Molecular phylogeny of the “turrids” (Neogastropoda: Conoidea). Mol. Phylogenetics Evol. 2008, 47, 1122–1134. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Kantor, Y.I.; Sysoev, A.; Couloux, A.; Meyer, C.; Rawlings, T.; Todd, J.A.; Bouchet, P. The dragon tamed? A molecular phylogeny of the Conoidea (Gastropoda). J. Molluscan Stud. 2011, 77, 259–272. [Google Scholar] [CrossRef]
- Abdelkrim, J.; Aznar-Cormano, L.; Fedosov, A.E.; Kantor, Y.; Lozouet, P.; Phuong, M.A.; Zaharias, P.; Puillandre, N.; Fedosov, A.E.; Kantor, Y.; et al. Exon-Capture-Based Phylogeny and Diversification of the Venomous Gastropods (Neogastropoda, Conoidea). Mol. Biol. Evol. 2018, 35, 2355–2374. [Google Scholar] [CrossRef]
- Lu, A.; Watkins, M.; Li, Q.; Robinson, S.D.; Concepcion, G.P.; Yandell, M.; Weng, Z.; Olivera, B.M.; Safavi-Hemami, H.; Fedosov, A.E. Transcriptomic Profiling Reveals Extraordinary Diversity of Venom Peptides in Unexplored Predatory Gastropods of the Genus Clavus. Genome Biol. Evol. 2020, 12, 684–700. [Google Scholar] [CrossRef]
- Terlau, H.; Olivera, B.M. Conus Venoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef]
- Fedosov, A.E.; Puillandre, N. Integrative taxonomy of the Clavus canalicularis species complex (Drilliidae, Conoidea, Gastropoda) with description of four new species. Molluscan Res. 2020, 40, 251–266. [Google Scholar] [CrossRef]
- Kilburn, R.N.; Fedosov, A.; Kantor, Y. The shallow-water New Caledonia Drilliidae of genus Clavus Montfort, 1810 (Mollusca: Gastropoda: Conoidea). Zootaxa 2014, 3818, 1–69. [Google Scholar] [CrossRef]
- Bozzetti, L. Study of the collection of Mr. A. Guillot de Suduiraut with the descriptions of three new gastropod species (Fasciolariidae, Trochidae and Turridae). Bull. Inst. Malacol. Tokyo 1997, 3, 55–58. [Google Scholar]
- Imperial, J.S.; Bansal, P.S.; Alewood, P.F.; Daly, N.L.; Craik, D.J.; Sporning, A.; Terlau, H.; Vera, E.L.; Bandyopadhyay, P.K.; Olivera, B.M. A Novel Conotoxin Inhibitor of Kv1.6 Channel and nAChR Subtypes Defines a New Superfamily of Conotoxins. Biochemistry 2006, 45, 8331–8340. [Google Scholar] [CrossRef] [PubMed]
- Heralde, F.M.; Imperial, J.; Bandyopadhyay, P.K.; Olivera, B.M.; Concepcion, G.P.; Santos, A.D. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon 2008, 51, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Stauffer, D.A.; Karlin, A. Electrostatic potential of the acetylcholine binding sites in the nicotinic receptor probed by reactions of binding-site cysteines with charged methanethiosulfonates. Biochemistry 1994, 33, 6840–6849. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.L.B.; Imperial, J.S.; Morgenstern, D.; Ueberheide, B.; Gajewiak, J.; Antunes, A.; Robinson, S.D.; Espino, S.; Watkins, M.; Vasconcelos, V.; et al. Characterization of the First Conotoxin from Conus ateralbus, a Vermivorous Cone Snail from the Cabo Verde Archipelago. Mar. Drugs 2019, 17, 432. [Google Scholar] [CrossRef]
- Imperial, J.S.; Cabang, A.B.; Song, J.; Raghuraman, S.; Gajewiak, J.; Watkins, M.; Showers-Corneli, P.; Fedosov, A.; Concepcion, G.P.; Terlau, H.; et al. A family of excitatory peptide toxins from venomous crassispirine snails: Using Constellation Pharmacology to assess bioactivity. Toxicon 2014, 89, 45–54. [Google Scholar] [CrossRef]
- Imperial, J.S.; Chen, P.; Sporning, A.; Terlau, H.; Daly, N.; Craik, D.J.; Alewood, P.F.; Olivera, B.M. Tyrosine-rich Conopeptides Affect Voltage-gated K+ Channels. J. Biol. Chem. 2008, 283, 23026–23032. [Google Scholar] [CrossRef]
- Olivera, B.M.; Gray, W.R.; Zeikus, R.; McIntosh, J.M.; Varga, J.; Rivier, J.; De Santos, V.; Cruz, L.J. Peptide neurotoxins from fish-hunting cone snails. Science 1985, 230, 1338–1343. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Corpuz, G.O.; Layer, R.T.; Garrett, J.E.; Wagstaff, J.D.; Bulaj, G.; Vyazovkina, A.; Yoshikami, D.; Cruz, L.J.; Olivera, B.M. Isolation and Characterization of a NovelConusPeptide with Apparent Antinociceptive Activity. J. Biol. Chem. 2000, 275, 32391–32397. [Google Scholar] [CrossRef]
- Giacobassi, M.J.; Leavitt, L.S.; Raghuraman, S.; Alluri, R.K.; Chase, K.; Finol-Urdaneta, R.K.; Terlau, H.; Teichert, R.W.; Olivera, B.M. An integrative approach to the facile functional classification of dorsal root ganglion neuronal subclasses. Proc. Natl. Acad. Sci. USA 2020, 117, 5494–5501. [Google Scholar] [CrossRef]
- Teichert, R.W.; Memon, T.; Aman, J.W.; Olivera, B.M. Using constellation pharmacology to define comprehensively a somatosensory neuronal subclass. Proc. Natl. Acad. Sci. USA 2014, 111, 2319–2324. [Google Scholar] [CrossRef] [PubMed]
- Teichert, R.W.; Schmidt, E.W.; Olivera, B.M. Constellation pharmacology: A new paradigm for drug discovery. Annu. Rev. Pharm. Toxicol. 2015, 55, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Teichert, R.W.; Smith, N.J.; Raghuraman, S.; Yoshikami, J.; Light, A.R.; Olivera, B.M. Functional profiling of neurons through cellular neuropharmacology. Proc. Natl. Acad. Sci. USA 2012, 109, 1388–1395. [Google Scholar] [CrossRef] [PubMed]
- Ahorukomeye, P.; Disotuar, M.M.; Gajewiak, J.; Karanth, S.; Watkins, M.; Robinson, S.D.; Salcedo, P.F.; Smith, A.; Smith, B.J.; Schlegel, A.; et al. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. elife 2019, 8, 41574. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Kaas, Q.; Yu, R.; Jin, A.-H.; Dutertre, S.; Craik, D.J. ConoServer: Updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2011, 40, D325–D330. [Google Scholar] [CrossRef]
- Duckert, P.; Brunak, S.; Blom, N.S. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004, 17, 107–112. [Google Scholar] [CrossRef]
- Cordeiro, S.; Finol-Urdaneta, R.K.; Köpfer, D.; Markushina, A.; Song, J.; French, R.J.; Kopec, W.; De Groot, B.L.; Giacobassi, M.J.; Leavitt, L.S.; et al. Conotoxin κM-RIIIJ, a tool targeting asymmetric heteromeric Kv1 channels. Proc. Natl. Acad. Sci. USA 2018, 116, 1059–1064. [Google Scholar] [CrossRef]

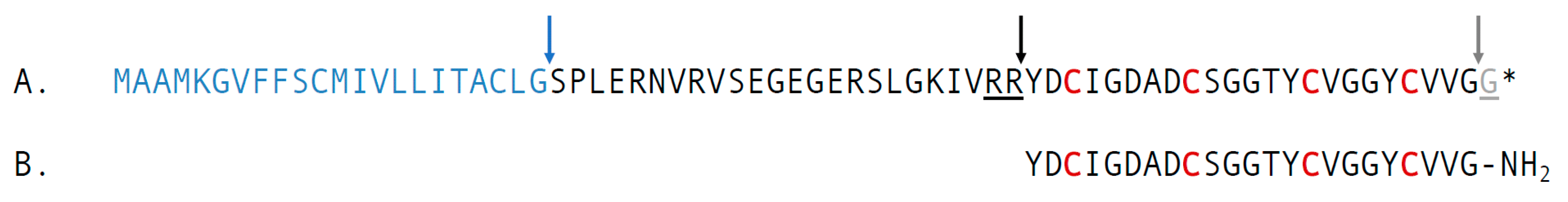



| Average Weight ± Deviation (g) | nmol of cdg14a in NSS | Observations (Intracranial Injection) |
|---|---|---|
| 12.1 ± 0.1 | 0 | No unusual behavior observed |
| 10.4 ± 0.3 | 1 | Mild shaking at 15 min; frequent scratching and grooming at 22 min lasting 2.5 h. |
| 10.6 ± 0.1 | 2.5 | Shaking at 7 min; frequent scratching and grooming at 11 min; licking of front paw and thumping of hind paw at 1 h lasting 2.5 h. One mouse had convulsions at 3.5 h but recovered. |
| 11.06 ± 0.26 | 5 | Shaking, frequent scratching, and grooming at 7 min; licking of front paw and thumping of hind paw at 18 min lasting 18 min; seizure at 40 min; lying on its side for the rest of the experiment (>3 h). |
| 11.6 ± 0.2 | 10 | Shaking, frequent scratching, and grooming at 5 min; licking of front paw and thumping of hind paw at 9 min lasting 18 min; seizure at 37 min; lying on its side for the rest of the experiment. Mice died at 2 h. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chua, V.M.; Gajewiak, J.; Watkins, M.; Espino, S.S.; Ramiro, I.B.L.; Omaga, C.A.; Imperial, J.S.; Carpio, L.P.D.; Fedosov, A.; Safavi-Hemami, H.; et al. Purification and Characterization of the Pink-Floyd Drillipeptide, a Bioactive Venom Peptide from Clavus davidgilmouri (Gastropoda: Conoidea: Drilliidae). Toxins 2020, 12, 508. https://doi.org/10.3390/toxins12080508
Chua VM, Gajewiak J, Watkins M, Espino SS, Ramiro IBL, Omaga CA, Imperial JS, Carpio LPD, Fedosov A, Safavi-Hemami H, et al. Purification and Characterization of the Pink-Floyd Drillipeptide, a Bioactive Venom Peptide from Clavus davidgilmouri (Gastropoda: Conoidea: Drilliidae). Toxins. 2020; 12(8):508. https://doi.org/10.3390/toxins12080508
Chicago/Turabian StyleChua, Victor M., Joanna Gajewiak, Maren Watkins, Samuel S. Espino, Iris Bea L. Ramiro, Carla A. Omaga, Julita S. Imperial, Louie Paolo D. Carpio, Alexander Fedosov, Helena Safavi-Hemami, and et al. 2020. "Purification and Characterization of the Pink-Floyd Drillipeptide, a Bioactive Venom Peptide from Clavus davidgilmouri (Gastropoda: Conoidea: Drilliidae)" Toxins 12, no. 8: 508. https://doi.org/10.3390/toxins12080508
APA StyleChua, V. M., Gajewiak, J., Watkins, M., Espino, S. S., Ramiro, I. B. L., Omaga, C. A., Imperial, J. S., Carpio, L. P. D., Fedosov, A., Safavi-Hemami, H., Salvador-Reyes, L. A., Olivera, B. M., & Concepcion, G. P. (2020). Purification and Characterization of the Pink-Floyd Drillipeptide, a Bioactive Venom Peptide from Clavus davidgilmouri (Gastropoda: Conoidea: Drilliidae). Toxins, 12(8), 508. https://doi.org/10.3390/toxins12080508





