Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates
Abstract
1. Introduction
2. Results
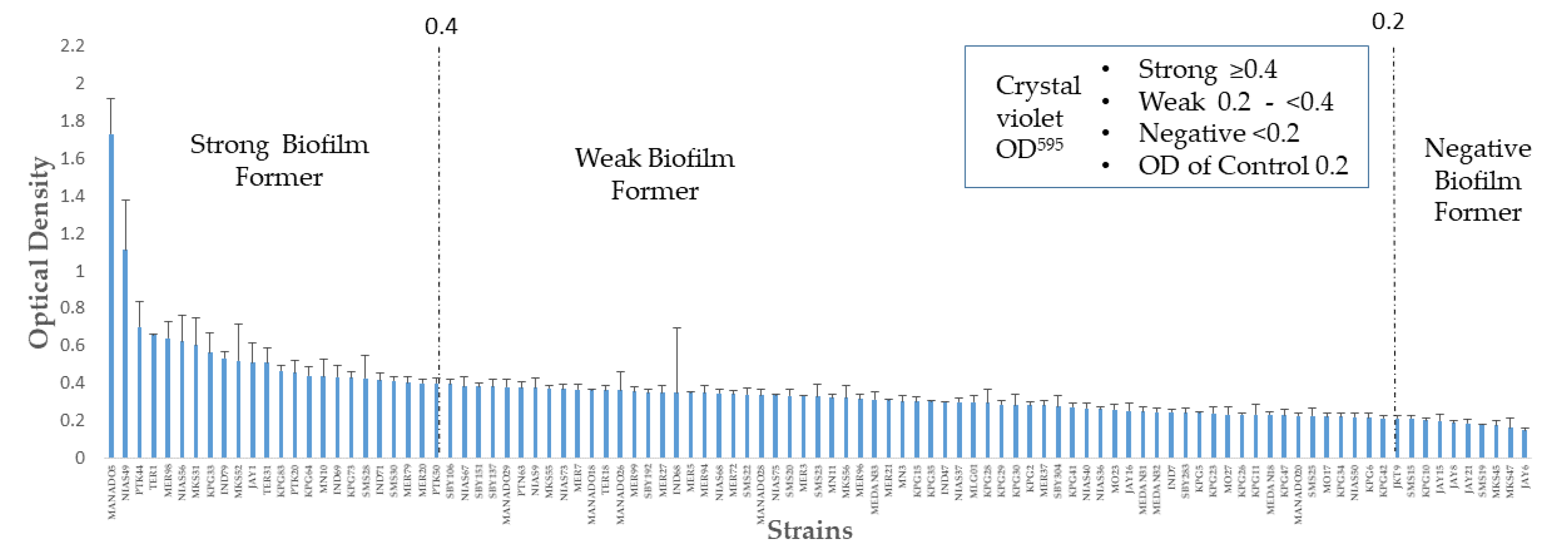
2.1. Distribution of Biofilm Formation
2.2. Biofilm Formation Level among Planktonic Sensitive and Resistant Isolates
2.3. Biofilm Formation and Planktonic Multidrug Resistance
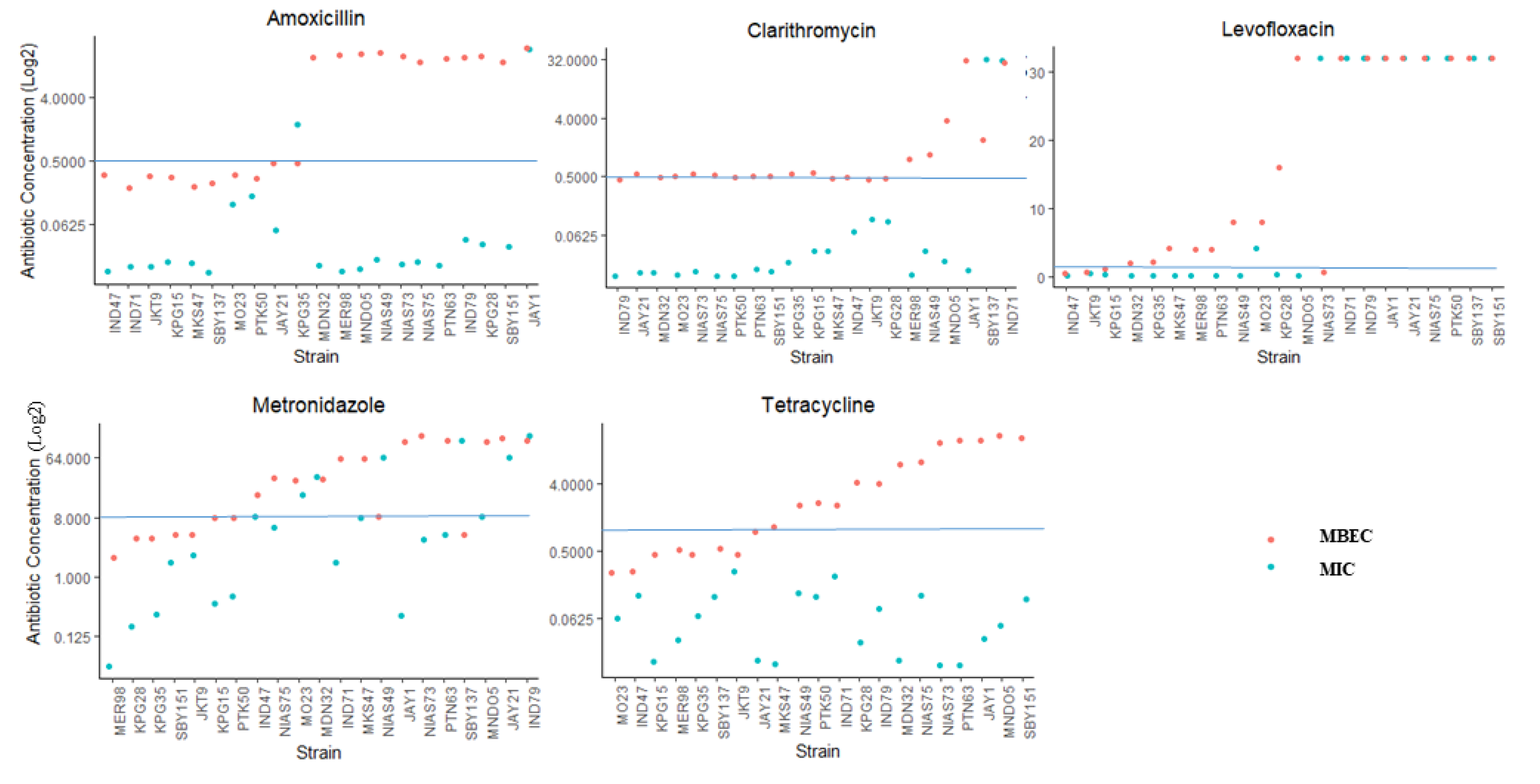
2.4. Antibiotic Resistance in the Biofilm Form and Planktonic Form
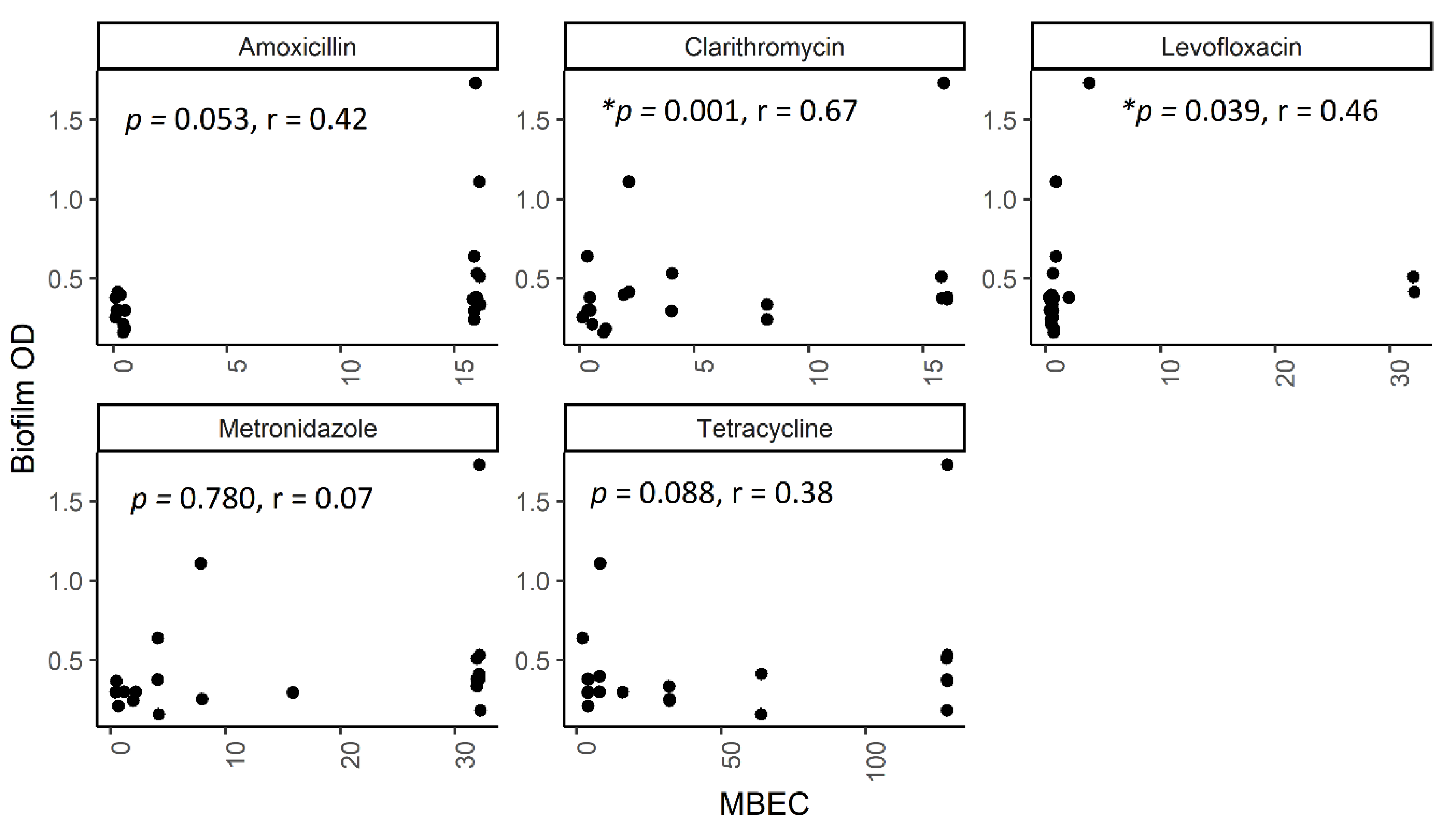
2.5. The Correlation between Biofilm Amount and Antibiotic Resistance in Biofilm-Forming Groups
2.6. The Effect of Antibiotic Exposure on Biofilm Formation
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Patient Sampling and H. pylori Isolates
5.2. Antibiotic Susceptibility Test
5.3. Biofilm Quantification
5.4. Biofilm Formation and MBEC
5.5. Scanning Electron Micrograph Analysis
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Savoldi, A.; Carrara, E.; Graham, P.D.Y.; Conti, M.; Tacconelli, E. Prevalence of antibiotic resistance in Helicobacter pylori: A systematic review and meta-analysis in World Health Organization regions. Gastroenterology 2018. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed]
- López-Góngora, S.; Puig, I.; Calvet, X.; Villoria, A.; Baylina, M.; Muñoz, N.; Sanchez-Delgado, J.; Suarez, D.; García-Hernando, V.; Gisbert, J.P. Systematic review and meta-analysis: Susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J. Antimicrob. Chemother. 2015, 70, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Shi, Z.; Lin, D.; Yang, N.; Meng, F.; Lin, L.; Jin, Z.; Zhou, Q.; Wu, J.; Zhang, J.; et al. Is tailored therapy based on antibiotic susceptibility effective? A multicenter, open-label, randomized trial. Front. Med. 2020, 14, 43–50. [Google Scholar] [CrossRef]
- Cammarota, G.; Sanguinetti, M.; Gallo, A.; Posteraro, B. biofilm formation by H elicobacter pylori as a target for eradication of resistant infection. Aliment. Pharmacol. Ther. 2012, 36, 222–230. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kamiya, S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. Biomed Res. Int. 2015, 2015, 914791. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Cole, S.P.; Harwood, J.; Lee, R.; She, R.; Guiney, D.G. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 2004, 186, 3124–3132. [Google Scholar] [CrossRef] [PubMed]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori biofilms in human gastric mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hanawa, T.; Kurata, S.; Ochiai, K.; Kamiya, S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS ONE 2013, 8, e73301. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.H.; Ng, C.G.; Chua, E.G.; Tay, A.C.; Peters, F.; Marshall, B.J.; Ho, B.; Goh, K.L.; Vadivelu, J.; Loke, M.F. Comparative Genomics Revealed Multiple Helicobacter pylori Genes Associated with Biofilm Formation In Vitro. PLoS ONE 2016, 11, e0166835. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320. [Google Scholar] [CrossRef]
- Sandoe, J.A.; Wysome, J.; West, A.P.; Heritage, J.; Wilcox, M.H. Measurement of ampicillin, vancomycin, linezolid and gentamicin activity against enterococcal biofilms. J. Antimicrob. Chemother. 2006, 57, 767–770. [Google Scholar] [CrossRef]
- Brady, A.J.; Laverty, G.; Gilpin, D.F.; Kearney, P.; Tunney, M. Antibiotic susceptibility of planktonic-and biofilm-grown staphylococci isolated from implant-associated infections: Should MBEC and nature of biofilm formation replace MIC? J. Med. Microbiol. 2017, 66, 461–469. [Google Scholar] [CrossRef]
- Macia, M.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef]
- Girard, L.P.; Ceri, H.; Gibb, A.P.; Olson, M.; Sepandj, F. MIC versus MBEC to determine the antibiotic sensitivity of Staphylococcus aureus in peritoneal dialysis peritonitis. Perit. Dial. Int. 2010, 30, 652–656. [Google Scholar] [CrossRef]
- Qi, L.; Li, H.; Zhang, C.; Liang, B.; Li, J.; Wang, L.; Du, X.; Liu, X.; Qiu, S.; Song, H. Relationship between Antibiotic Resistance, Biofilm Formation, and Biofilm-Specific Resistance in Acinetobacter baumannii. Front. Microbiol. 2016, 7, 483. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Syam, A.F.; Nusi, I.A.; Makmun, D.; Waskito, L.A.; Zein, L.H.; Akil, F.; Uwan, W.B.; Simanjuntak, D.; Wibawa, I.D.; et al. Surveillance of Helicobacter pylori Antibiotic Susceptibility in Indonesia: Different Resistance Types among Regions and with Novel Genetic Mutations. PLoS ONE 2016, 11, e0166199. [Google Scholar] [CrossRef]
- Herardi, R.; Syam, A.F.; Simadibrata, M.; Setiati, S.; Darnindro, N.; Abdullah, M.; Makmun, D. Comparison of 10-Day Course of Triple Therapy Versus 14-Day Course for Eradication of Helicobacter pylori Infection in an Indonesian Population: Double-Blinded Randomized Clinical Trial. Asian Pac. J. Cancer Prev. 2020, 21, 19–24. [Google Scholar] [CrossRef][Green Version]
- Heo, J.; Jeon, S.W. Optimal treatment strategy for Helicobacter pylori: Era of antibiotic resistance. World J. Gastroenterol. 2014, 20, 5654–5659. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.Y.; Fischbach, L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut 2010, 59, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-J.; Wu, M.-S.; Chen, C.-C.; Chen, C.-C.; Fang, Y.-J.; Bair, M.-J.; Chang, C.-Y.; Lee, J.-Y.; Hsu, W.-F.; Luo, J.-C.; et al. Impact of amoxicillin resistance on the efficacy of amoxicillin-containing regimens for Helicobacter pylori eradication: Analysis of five randomized trials. J. Antimicrob. Chemother. 2017, 72, 3481–3489. [Google Scholar] [CrossRef] [PubMed]
- Qaria, M.A.; Kumar, N.; Hussain, A.; Qumar, S.; Doddam, S.N.; Sepe, L.P.; Ahmed, N. Roles of Cholesteryl-α-Glucoside Transferase and Cholesteryl Glucosides in Maintenance of Helicobacter pylori Morphology, Cell Wall Integrity, and Resistance to Antibiotics. mBio 2018, 9, e01518–e01523. [Google Scholar] [CrossRef]
- Ge, X.; Cai, Y.; Chen, Z. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Hojo, F.; Kamiya, S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb. Pathog. 2019, 132, 100–108. [Google Scholar] [CrossRef]
- Attaran, B.; Falsafi, T.; Ghorbanmehr, N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 2017, 23, 1163–1170. [Google Scholar] [CrossRef]
- Reiter, K.C.; Villa, B.; da Silva Paim, T.G.; de Oliveira, C.F.; d’Azevedo, P.A. Inhibition of biofilm maturation by linezolid in meticillin-resistant Staphylococcus epidermidis clinical isolates: Comparison with other drugs. J. Med. Microbiol. 2013, 62, 394–399. [Google Scholar] [CrossRef]
- Wang, G.; Wilson, T.J.M.; Jiang, Q.; Taylor, D.E. Spontaneous Mutations That Confer Antibiotic Resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 2001, 45, 727–733. [Google Scholar] [CrossRef]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef]
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef]
- Brown, M.R.; Allison, D.G.; Gilbert, P. Resistance of bacterial biofilms to antibiotics: A growth-rate related effect? J. Antimicrob. Chemother. 1988, 22, 777–780. [Google Scholar] [CrossRef]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Wong, E.H.J.; Ng, C.G.; Goh, K.L.; Vadivelu, J.; Ho, B.; Loke, M.F. Metabolomic analysis of low and high biofilm-forming Helicobacter pylori strains. Sci. Rep. 2018, 8, 1409. [Google Scholar] [CrossRef]
- Awan, A.B.; Schiebel, J.; Böhm, A.; Nitschke, J.; Sarwar, Y.; Schierack, P.; Ali, A. Association of biofilm formation and cytotoxic potential with multidrug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI J. 2019, 18, 79. [Google Scholar]
- Pompilio, A.; Savini, V.; Fiscarelli, E.; Gherardi, G.; Di Bonaventura, G. Clonal Diversity, Biofilm Formation, and Antimicrobial Resistance among Stenotrophomonas maltophilia Strains from Cystic Fibrosis and Non-Cystic Fibrosis Patients. Antibiotics 2020, 9, 15. [Google Scholar] [CrossRef]
- Ramos-Vivas, J.; Chapartegui-González, I.; Fernández-Martínez, M.; González-Rico, C.; Fortún, J.; Escudero, R.; Marco, F.; Linares, L.; Montejo, M.; Aranzamendi, M. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Keays, T.; Ferris, W.; Vandemheen, K.L.; Chan, F.; Yeung, S.-W.; Mah, T.-F.; Ramotar, K.; Saginur, R.; Aaron, S.D. A retrospective analysis of biofilm antibiotic susceptibility testing: A better predictor of clinical response in cystic fibrosis exacerbations. J. Cyst. Fibros. 2009, 8, 122–127. [Google Scholar] [CrossRef]
- Wang, A.; Wang, Q.; Kudinha, T.; Xiao, S.; Zhuo, C. Effects of Fluoroquinolones and Azithromycin on Biofilm Formation of Stenotrophomonas maltophilia. Sci. Rep. 2016, 6, 29701. [Google Scholar] [CrossRef] [PubMed]
- Minardi, D.; Montanari, M.; Tili, E.; Cochetti, I.; Mingoia, M.; Varaldo, P.; Muzzonigro, G. Effects of fluoroquinolones on bacterial adhesion and on preformed biofilm of strains isolated from urinary double J stents. J. Chemother. 2008, 20, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.; Young, L.S.; Bermudez, L.E. A subinhibitory concentration of clarithromycin inhibits Mycobacterium avium biofilm formation. Antimicrob. Agents Chemother. 2004, 48, 4907–4910. [Google Scholar] [CrossRef]
- Dicicco, M.; Neethirajan, S.; Singh, A.; Weese, J.S. Efficacy of clarithromycin on biofilm formation of methicillin-resistant Staphylococcus pseudintermedius. BMC Vet. Res. 2012, 8, 225. [Google Scholar] [CrossRef]
- Singh, R.; Sahore, S.; Kaur, P.; Rani, A.; Ray, P. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef]
- Makipour, K.; Friedenberg, F.K. The potential role of N-acetylcysteine for the treatment of Helicobacter pylori. J. Clin. Gastroenterol. 2011, 45, 841–843. [Google Scholar] [CrossRef]
- Cai, J.; Huang, H.; Song, W.; Hu, H.; Chen, J.; Zhang, L.; Li, P.; Wu, R.; Wu, C. Preparation and evaluation of lipid polymer nanoparticles for eradicating H. pylori biofilm and impairing antibacterial resistance in vitro. Int. J. Pharm. 2015, 495, 728–737. [Google Scholar] [CrossRef]
- Fontes, L.E.S.; Martimbianco, A.L.C.; Zanin, C.; Riera, R. N-acetylcysteine as an adjuvant therapy for Helicobacter pylori eradication. Cochrane Database Syst. Rev. 2019, 2, Cd012357. [Google Scholar] [CrossRef]
- Chen, X.; Li, P.; Shen, Y.; Zou, Y.; Yuan, G.; Hu, H. Rhamnolipid-involved antibiotics combinations improve the eradication of Helicobacter pylori biofilm in vitro: A comparison with conventional triple therapy. Microb. Pathog. 2019, 131, 112–119. [Google Scholar] [CrossRef]
- Waskito, L.A.; Miftahussurur, M.; Lusida, M.I.; Syam, A.F.; Suzuki, R.; Subsomwong, P.; Uchida, T.; Hamdan, M.; Yamaoka, Y. Distribution and clinical associations of integrating conjugative elements and cag pathogenicity islands of Helicobacter pylori in Indonesia. Sci. Rep. 2018, 8, 6073. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Waskito, L.A.; Syam, A.F.; Nusi, I.A.; Wibawa, I.D.N.; Rezkitha, Y.A.A.; Siregar, G.; Yulizal, O.K.; Akil, F.; Uwan, W.B.; et al. Analysis of risks of gastric cancer by gastric mucosa among Indonesian ethnic groups. PLoS ONE 2019, 14, e0216670. [Google Scholar] [CrossRef] [PubMed]
- Doohan, D.; Miftahussurur, M.; Matsuo, Y.; Kido, Y.; Akada, J.; Matsuhisa, T.; Yee, T.T.; Htet, K.; Aftab, H.; Vilaichone, R.K.; et al. Characterization of a novel Helicobacter pylori East Asian-type CagA ELISA for detecting patients infected with various cagA genotypes. Med. Microbiol. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Syam, A.F.; Miftahussurur, M.; Makmun, D.; Nusi, I.A.; Zain, L.H.; Zulkhairi, A.F.; Uswan, W.B.; Simanjuntak, D.; Uchida, T.; Adi, P.; et al. Risk Factors and Prevalence of Helicobacter pylori in Five Largest Islands of Indonesia. A Preliminary Study. In PLoS ONE; 2015; Volume 10. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Dieases (ESCMID). Document EDEF. 3.1, June 2000: Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin. Microbiol. Infect. 2000, 6, 509–515. [Google Scholar]
- EUCAST. EUCAST clinical breakpoints for Helicobacter pylori. Eur. Soc. Clin. Microbiol. Infect. Dis. 2011. [Google Scholar]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Zaman, C.; Hanawa, T.; Kamiya, S. Assessment of in vitro biofilm formation by Helicobacter pylori. J. Gastroenterol. Hepatol. 2010, 25 (Suppl. 1), S90–S94. [Google Scholar] [CrossRef]
- Odeyemi, O.A. Microtiter plate assay methods of classification of bacterial biofilm formation. Food Control 2017, 245–246. [Google Scholar] [CrossRef]
- Niu, C.; Gilbert, E.S. Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 2004, 70, 6951–6956. [Google Scholar] [CrossRef] [PubMed]
- Kadurugamuwa, J.L.; Sin, L.V.; Yu, J.; Francis, K.P.; Kimura, R.; Purchio, T.; Contag, P.R. Rapid direct method for monitoring antibiotics in a mouse model of bacterial biofilm infection. Antimicrob. Agents Chemother. 2003, 47, 3130–3137. [Google Scholar] [CrossRef]

| Drug-Resistance Pattern | Median Biofilm Formed | n (%) | Biofilm Formation n (%) | ||
|---|---|---|---|---|---|
| Strong | Weak | Negative | |||
| All Sensitive | 0.314 | 33 (32.7) | 6 (26.1) | 23 (32.4) | 4 (57.1) |
| Single-Resistant | 0.333 | 42 (41.6) | 9 (39.1) | 31 (43.7) | 2 (28.5) |
| Double-Resistant | 0.344 | 21 (20.8) | 8 (34.8) | 12(16.9) | 1 (14.3) |
| Multidrug-Resistant | 0.276 | 5 (4.9) | 0 (0) | 5 (7.0) | 0 (0) |
| Total | 101 | 23 | 71 | 7 | |
| Resistance Proportion (%) | |||||
|---|---|---|---|---|---|
| Amoxicillin | Clarithromycin | Levofloxacin | Metronidazole | Tetracycline | |
| All MIC | 2/21 (9.5) | 2/21 (9.5) | 10/21 (47.6) | 6/21 (28.6) | 0/21 (0.0) |
| All MBEC | 11/21 (52.4) | 6/21 (28.6) | 17/21 (80.9) | 12/21 (57.1) | 12/21 (57.1) |
| p * | 0.006 | 0.24 | 0.051 | 0.12 | <0.001 |
| MIC | |||||
| Weak | 1/15 (6.7) | 1/15 (6.7) | 7/15 (46.7) | 4/15 (26.7) | 0/15 (0.0) |
| Strong | 1/6 (16.7) | 1/6 (16.7) | 3/6 (50) | 2/6 (33.3) | 0/6 (0.0) |
| p ** | 0.5 | 0.5 | 0.99 | 0.99 | - |
| MBEC | |||||
| Weak | 6/15 (40.0) | 1/15 (6.7) | 11/15 (73.3) | 8/15 (53.3) | 7/15 (46.7) |
| Strong | 5/6 (80.0) | 5/6 (83.3) | 6/6 (100) | 4/6 (66.7) | 5/6 (83.3) |
| p *** | 0.15 | 0.002 | 0.28 | 0.66 | 0.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473. https://doi.org/10.3390/toxins12080473
Fauzia KA, Miftahussurur M, Syam AF, Waskito LA, Doohan D, Rezkitha YAA, Matsumoto T, Tuan VP, Akada J, Yonezawa H, et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins. 2020; 12(8):473. https://doi.org/10.3390/toxins12080473
Chicago/Turabian StyleFauzia, Kartika Afrida, Muhammad Miftahussurur, Ari Fahrial Syam, Langgeng Agung Waskito, Dalla Doohan, Yudith Annisa Ayu Rezkitha, Takashi Matsumoto, Vo Phuoc Tuan, Junko Akada, Hideo Yonezawa, and et al. 2020. "Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates" Toxins 12, no. 8: 473. https://doi.org/10.3390/toxins12080473
APA StyleFauzia, K. A., Miftahussurur, M., Syam, A. F., Waskito, L. A., Doohan, D., Rezkitha, Y. A. A., Matsumoto, T., Tuan, V. P., Akada, J., Yonezawa, H., Kamiya, S., & Yamaoka, Y. (2020). Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins, 12(8), 473. https://doi.org/10.3390/toxins12080473







