AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken
Abstract
1. Introduction
2. Results
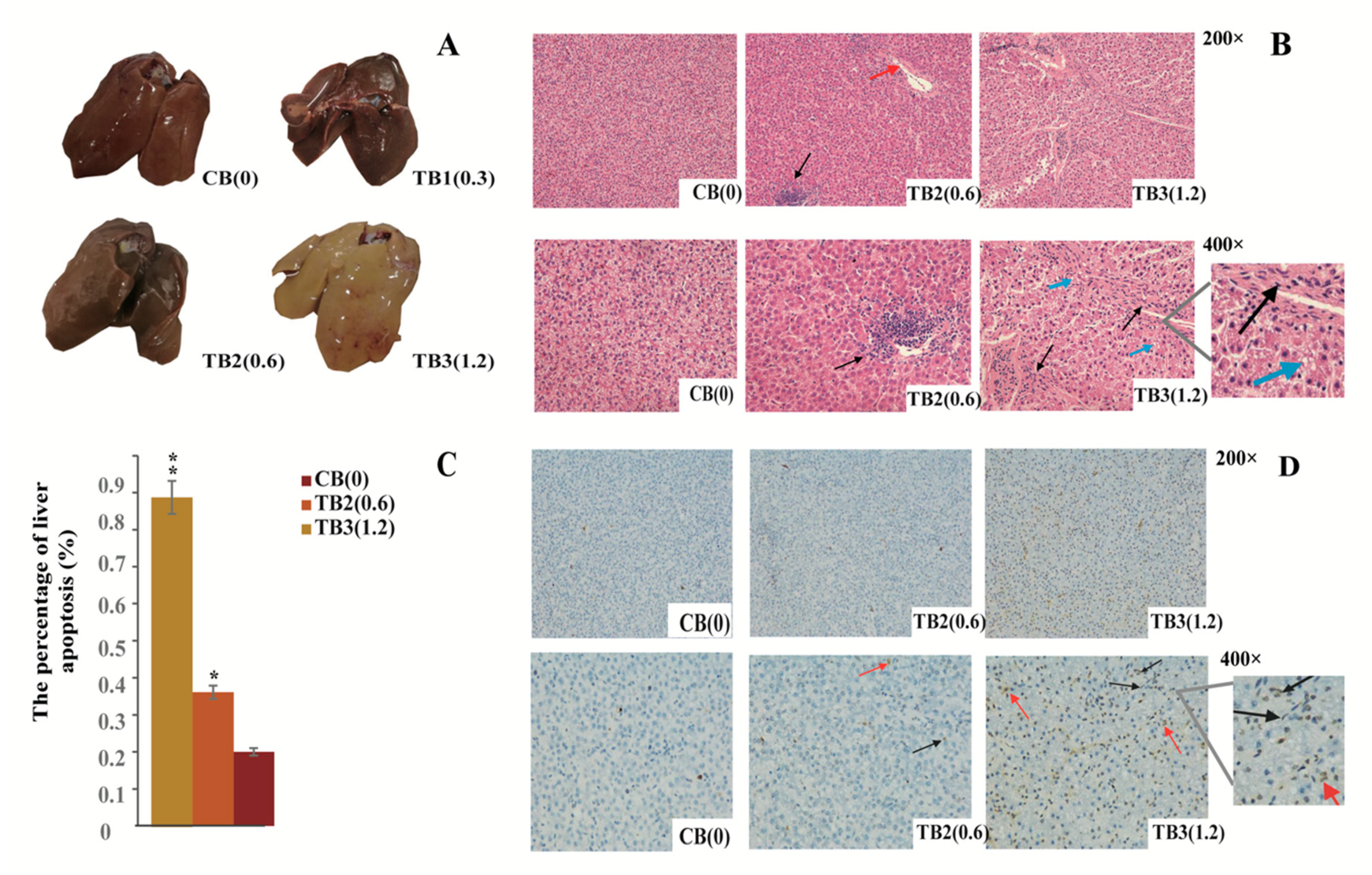
2.1. Histopathological Observations of Liver
2.2. AFB1 Triggered Apoptosis of Liver
2.3. Sequencing Data Statistics
2.4. Sample Correlation Analysis
2.5. Analysis of Differentially Expressed mRNAs lncRNAs and miRNAs
2.6. Genomic Characterization of lncRNAs miRNAs and mRNAs
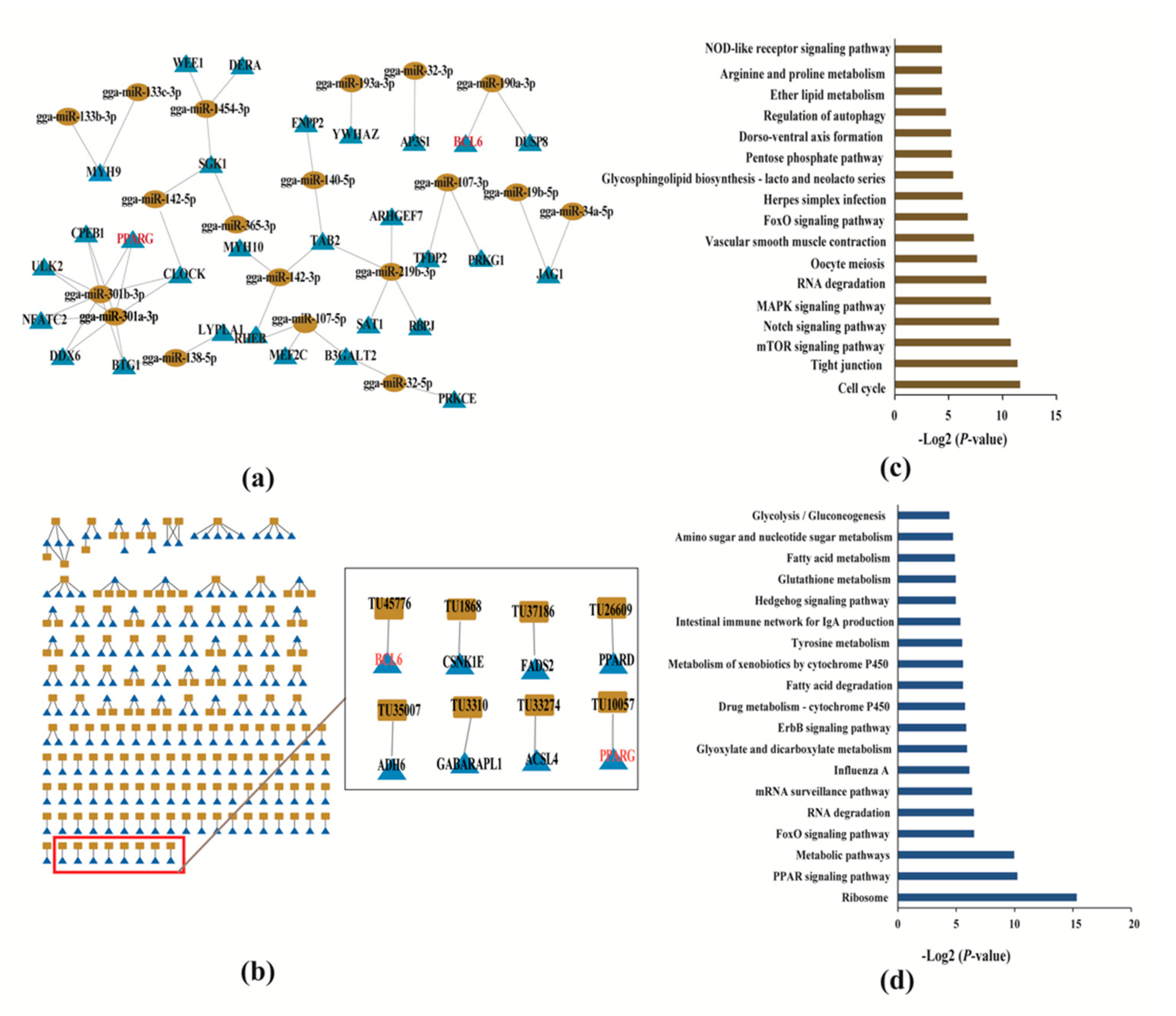
2.7. miRNA-mRNA targeted Association Analysis
2.8. Co-expression Network of mRNAs and lncRNAs
2.9. Functional Enrichment Analysis of Target Genes of miRNA and lncRNA
2.10. Confirmation of RNA-seq results by qRT-PCR
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Experimental Birds and Diets
5.2. Liver Histopathology
5.3. TUNEL Assay
5.4. RNA Isolation
5.5. Illumina Deep Sequencing
5.6. Transcriptomic Data Analysis
5.7. Classification of lncRNAs
5.8. Expression Analysis
5.9. Functional Enrichment Analyses for miRNA Target Genes and Protein Encoding Genes
5.10. miRNA-mRNA Targeted Association Analysis
5.11. Construction of the lncRNA mRNA Co-expression Network
5.12. Quantitative Real-time Polymerase Chain Reaction (qRT-PCR) Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sakamoto, M.I.; Murakami, A.E.; Fernandes, A.M.; Ospina-Rojas, I.C.; Hirata, A.K. Performance and serum biochemical profile of Japanese quail supplemented with silymarin and contaminated with aflatoxin B1. Poult. Sci. 2017, 97, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Coulombe, R.A.; Reed, K.M. Aflatoxicosis: Lessons from toxicity and responses to aflatoxin B1 in poultry. Agriculture 2015, 5, 742–777. [Google Scholar] [CrossRef]
- Zhao, J.; Shirley, R.B.; Dibner, J.D.; Uraizee, F.; Officer, M.; Kitchell, M.; Vazquez-Anon, M.; Knight, C.D. Comparison of hydrated sodium calcium aluminosilicate and yeast cell wall on counteracting aflatoxicosis in broiler chicks. Poult. Sci. 2010, 89, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.J.; Cortes, A.; Botero, L. Evaluation of the ability of a feed additive to ameliorate the adverse effects of aflatoxins in turkey poults. Br. Poult. Sci. 2009, 50, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Ankrah, N.A.; Addo, P.G.; Abrahams, C.A.; Ekuban, F.A.; Addae, M.M. Comparative effects of aflatoxins G1 and B1 at levels within human exposure limits on mouse liver and kidney. West Afr. J. Med. 1993, 12, 105–109. [Google Scholar] [PubMed]
- Sepahdari, A.; Ebrahimzadeh, M.H.; Sharifpour, I.; Khosravi, A.; Motalebi, A.; Mohseni, M.; Kakoolaki, S.; Pourali, H.; Halajian, A. Effects of different dietary levels of AFB1 on survival rate and growth factors of Beluga (Huso huso). Iran. J. Fish. Sci. 2010, 9, 141–150. [Google Scholar]
- Carlson, D.B.; Williams, D.E.; Spitsbergen, J.M.; Ross, P.F.; Bacon, C.W.; Meredith, F.I.; Riley, R.T. Fumonisin B1 promotes aflatoxin B1 and N-methyl-N′-nitro-nitrosoguanidine-initiated liver tumors in rainbow trout. Toxicol. Appl. Pharmacol. 2001, 172, 29–36. [Google Scholar] [CrossRef]
- Chen, J.; Chen, K.; Yuan, S.; Peng, X.; Fang, J.; Wang, F.; Cui, H.; Chen, Z.; Yuan, J.; Geng, Y. Effects of aflatoxin B1 on oxidative stress markers and apoptosis of spleens in broilers. Toxicol. Ind. Health 2016, 32, 278–284. [Google Scholar] [CrossRef]
- Sirajudeen, M.; Gopi, K.; Tyagi, J.S.; Moudgal, R.P.; Mohan, J.; Singh, R. Protective effects of melatonin in reduction of oxidative damage and immunosuppression induced by aflatoxin B1-contaminated diets in young chicks. Environ. Toxicol. 2011, 26, 153–160. [Google Scholar] [CrossRef]
- Yarru, L.P. Effects of aflatoxin on hepatic gene expression in a poultry model. Ph.D. Thesis, University of Missouri, Columbia, MI, USA, 2008. [Google Scholar]
- Yarru, L.; Settivari, R.; Antoniou, E.; Ledoux, D.; Rottinghaus, G. Toxicological and gene expression analysis of the impact of aflatoxin B1 on hepatic function of male broiler chicks. Poult. Sci. 2009, 88, 360–371. [Google Scholar] [CrossRef]
- Murarolli, R.A. Effects of aflatoxin B1 (AFB1) on hepatic gene expression in pigs and turkeys. Ph.D. Thesis, University of Missouri, Columbia, MI, USA, 2013. [Google Scholar]
- Monson, M.S.; Settlage, R.E.; Mendoza, K.M.; Rawal, S.; El-Nezami, H.S.; Coulombe, R.A.; Reed, K.M. Modulation of the spleen transcriptome in domestic turkey (Meleagris gallopavo) in response to aflatoxin B1 and probiotics. Immunogenetics 2015, 67, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Settlage, R.E.; McMahon, K.W.; Mendoza, K.M.; Rawal, S.; El-Nezami, H.S.; Coulombe, R.A.; Reed, K.M. Response of the hepatic transcriptome to aflatoxin B1 in domestic turkey (Meleagris gallopavo). PLoS ONE 2014, 9, e100930. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef]
- Legnini, I.; Morlando, M.; Mangiavacchi, A.; Fatica, A.; Bozzoni, I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 2014, 53, 506–514. [Google Scholar] [CrossRef]
- Muret, K.; Klopp, C.; Wucher, V.; Esquerré, D.; Legeai, F.; Lecerf, F.; Désert, C.; Boutin, M.; Jehl, F.; Acloque, H. Long noncoding RNA repertoire in chicken liver and adipose tissue. Genet. Sel. Evol. 2017, 49, 6. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Li, Y.; Bai, H.; Xue, F.; Xu, S.; Xu, H.; Shi, L.; Yang, N.; Chen, J. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; He, J.; Lin, J.; Sun, X.; Sun, F.; Ou, C.; Jiang, C. Distinct response of the hepatic transcriptome to Aflatoxin B 1 induced hepatocellular carcinogenesis and resistance in rats. Sci. Rep. 2016, 6, 31898. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.-T.; Hu, J.-W.; Yin, R.; Xu, L. Long noncoding RNA: An emerging paradigm of cancer research. Tumor Biol. 2013, 34, 613–620. [Google Scholar] [CrossRef]
- Omer, A.D.; Janas, M.M.; Novina, C.D. The chicken or the egg: MicroRNA-mediated regulation of mRNA translation or mRNA stability. Mol. Cell 2009, 35, 739–740. [Google Scholar] [CrossRef]
- Yang, W.; Lian, J.; Feng, Y.; Srinivas, S.; Guo, Z.; Zhong, H.; Zhuang, Z.; Wang, S. Genome-wide miRNA-profiling of aflatoxin B1-induced hepatic injury using deep sequencing. Toxicol. Lett. 2014, 226, 140–149. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Long, X.-D.; Xi, Z.-F.; Ma, Y.; Huang, X.-Y.; Yao, J.-G.; Wang, C.; Xing, T.-Y.; Xia, Q. MicroRNA-24 modulates aflatoxin B1-related hepatocellular carcinoma prognosis and tumorigenesis. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Ortatatli, M.; Oğuz, H. Ameliorative effects of dietary clinoptilolite on pathological changes in broiler chickens during aflatoxicosis. Res. Vet. Sci. 2001, 71, 59–66. [Google Scholar] [CrossRef]
- Brockmöller, J.; Ivar, R. Assessment of liver metabolic function. Clin. Pharmacokinet. 1994, 27, 216–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009, 8, 651–661. [Google Scholar] [CrossRef]
- Calne, R. Immunological tolerance–the liver effect. Immunol. Rev. 2000, 174, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Grijalva, J.; Vakili, K. Neonatal liver physiology. Semin. Pediatric Surg. 2013, 22, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.M.; Mendoza, K.M.; Coulombe, R.A. Differential transcriptome responses to aflatoxin B1 in the cecal tonsil of susceptible and resistant turkeys. Toxins 2019, 11, 55. [Google Scholar] [CrossRef]
- Marchese, S.; Polo, A.; Ariano, A.; Velotto, S.; Costantini, S.; Severino, L. Aflatoxin B1 and M1: Biological properties and their involvement in cancer development. Toxins 2018, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.-O.; Corcoran, M.; Grandér, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wei, M.; Lin, P.; Chen, Z.; Gong, P.; Quan, Z.; Tang, Z. Integrated mRNA and lncRNA expression profiling for exploring metastatic biomarkers of human intrahepatic cholangiocarcinoma. Am. J. Cancer Res. 2017, 7, 688. [Google Scholar]
- Diederichs, S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef]
- Shang, C.; Zhu, W.; Liu, T.; Wang, W.; Huang, G.; Huang, J.; Zhao, P.; Zhao, Y.; Yao, S. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol. Rep. 2016, 35, 3185–3197. [Google Scholar] [CrossRef]
- Chavez-Sanchez, M.C.; Palacios, C.M.; Moreno, I.O. Pathological effects of feeding young Oreochromis niloticus diets supplemented with different levels of aflatoxin B1. Aquaculture 1994, 127, 49–60. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W. Aflatoxin B1 impairs mitochondrial functions, activates ROS generation, induces apoptosis and involves Nrf2 signal pathway in primary broiler hepatocytes. Anim. Sci. J. 2016, 87, 1490–1500. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.P.; DeBaun, M.R.; Mitsuya, K.; Galonek, H.L.; Brandenburg, S.; Oshimura, M.; Feinberg, A.P. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith–Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. USA 1999, 96, 5203–5208. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Ponjavic, J.; Oliver, P.L.; Lunter, G.; Ponting, C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009, 5, e1000617. [Google Scholar] [CrossRef]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. BloodJ. Am. Soc. Hematol. 2009, 113, 2526–2534. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A. Long noncoding RNA programs active chromatin domain to coordinate homeotic gene activation. Nature 2011, 472, 120. [Google Scholar] [CrossRef]
- Kaoud, H.A. Innovative methods for the amelioration of aflatoxin (Afb1) effect in broiler chicks. Sci. J. Appl. Res. 2013, 1, 15–19. [Google Scholar]
- Hussain, Z. Aflatoxicosis in chicken: An inter-relationship between pathology, dietary and tissue aflatoxin levels. Ph.D. Thesis, University of Agriculture Faisalabad, Punjab, Pakistan, 2008. [Google Scholar]
- Rawal, S.; Mendoza, K.M.; Reed, K.M.; Coulombe, R.A. Structure, Genetic Mapping, and Function of the Cytochrome P450 3A37 Gene in the Turkey (Meleagris gallopavo). Cytogenet. Genome Res. 2009, 125, 67–73. [Google Scholar] [CrossRef]
- Diaz, G.; Murcia, H.; Cepeda, S. Cytochrome P450 enzymes involved in the metabolism of aflatoxin B1 in chickens and quail. Poult. Sci. 2010, 89, 2461–2469. [Google Scholar] [CrossRef]
- Wei, W.; Li, R.; He, L.; Wang, X.; Ji, X.; Zhou, Y. Evaluation of toxicological impacts on Sprague–Dawley rat by successively low dose of aflatoxin B1. J. Sci. Food Agric. 2014, 94, 3017–3022. [Google Scholar] [CrossRef]
- Kroemer, G.; Castedo, M.; Ferri, K.F. Mammalian Target of Rapamycin (mTOR): Pro- and Anti-Apoptotic. Cell Death Differ. 2002, 9, 99–100. [Google Scholar]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 2011, 1813, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Fajas, L.; Auboeuf, D.; Raspé, E.; Schoonjans, K.; Lefebvre, A.-M.; Saladin, R.; Najib, J.; Laville, M.; Fruchart, J.-C.; Deeb, S. The organization, promoter analysis, and expression of the human PPARγ gene. J. Biol. Chem. 1997, 272, 18779–18789. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.R.; Barrick, C.; Kim, K.-A.; Lindner, J.; Blondeau, B.; Fujimoto, Y.; Shiota, M.; Kesterson, R.A.; Kahn, B.B.; Magnuson, M.A. Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Cermak, L.; Pagan, J.K.; Rossi, M.; Martinengo, C.; di Celle, P.F.; Chapuy, B.; Shipp, M.; Chiarle, R.; Pagano, M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012, 481, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.X.; Ou, J.S.; Li, Y.; Su, J.J.; Ou, C.; Yang, C.; Yue, H.F.; Ban, K.C. Dynamic expression of apoptosis-related genes during development of laboratory hepatocellular carcinoma and its relation to apoptosis. World J. Gastroenterol. 2005, 11, 4740–4744. [Google Scholar] [CrossRef]
- Tayman, C.; Tonbul, A.; Kosus, A.; Hirfanoglu, I.M.; Haltas, H.; Uysal, S.; Tatli, M.M.; Andiran, F. Protective effects of caffeic acid phenethyl ester (CAPE) on intestinal damage in necrotizing enterocolitis. Pediatric Surg. Int. 2011, 27, 1179–1189. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Na, L.; Wei, C.; Nikolaus, R. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. In Genome Informatics 2009: Genome Informatics Series Vol. 23; World Scientific: Singapore, 2009; pp. 205–211. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hedan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Category | Sample Name | Raw Reads (Mb) | Clean Reads (Mb) | Effective Rate (%) | Raw Base (Gb) | Clean Base (Gb) | Mapping Rate (%) |
|---|---|---|---|---|---|---|---|
| lncRNA | CB-1 | 99.97 | 97.62 | 97.64 | 15.00 | 14.64 | 96.20 |
| CB-2 | 116.55 | 114.09 | 97.89 | 17.49 | 17.11 | 95.80 | |
| CB-3 | 89.33 | 87.35 | 97.78 | 13.39 | 13.10 | 95.70 | |
| TB3-1 | 52.69 | 52.23 | 99.13 | 15.81 | 15.67 | 92.10 | |
| TB3-2 | 57.12 | 56.64 | 99.17 | 17.14 | 16.99 | 92.70 | |
| TB3-3 | 53.94 | 53.53 | 99.25 | 16.18 | 16.06 | 92.80 | |
| miRNA | CB-1 | 12.58 | 10.96 | 87.15 | 0.63 | 0.24 | 76.00 |
| CB-2 | 12.18 | 10.75 | 88.28 | 0.61 | 0.24 | 74.20 | |
| CB-3 | 11.87 | 11.87 | 87.56 | 0.59 | 0.23 | 76.70 | |
| TB3-1 | 11.09 | 9.53 | 85.93 | 0.53 | 0.48 | 86.60 | |
| TB3-2 | 10.08 | 8.24 | 81.75 | 0.49 | 0.54 | 81.60 | |
| TB3-3 | 12.69 | 10.71 | 84.39 | 0.61 | 0.41 | 82.20 |
| Gene Symbol | Log (FC) | miRNA | Log (FC) | Gene Symbol | Log (FC) | miRNA | Log (FC) |
|---|---|---|---|---|---|---|---|
| ARHGEF7 | −1.10 | gga-miR-219b-3p | 6.04 | ENPP2 | −1.28 | gga-miR-140-5p | −3.69 |
| BCL6 | −2.40 | gga-miR-190a-3p | 5.44 | JAG1 | −1.62 | gga-miR-19b-5p | −5.22 |
| CLOCK | −1.22 | gga-miR-142-5p | 6.54 | JAG1 | −1.62 | gga-miR-34a-5p | −5.95 |
| CLOCK | −1.22 | gga-miR-301b-3p | 3.87 | JAG1 | −1.62 | gga-miR-34a-5p | −5.95 |
| CLOCK | −1.22 | gga-miR-301a-3p | 2.37 | MEF2C | −1.36 | gga-miR-107-5p | −4.00 |
| DDX6 | −1.23 | gga-miR-301b-3p | 3.87 | PRKCE | −1.07 | gga-miR-32-5p | −6.13 |
| DDX6 | −1.23 | gga-miR-301a-3p | 2.37 | SGK1 | −1.14 | gga-miR-365-3p | −4.59 |
| DUSP8 | −1.45 | gga-miR-190a-3p | 5.44 | TAB2 | −1.17 | gga-miR-140-5p | −3.69 |
| MYH10 | −1.45 | gga-miR-142-3p | 5.91 | BTG1 | 4.33 | gga-miR-301b-3p | 3.87 |
| MYH9 | −1.26 | gga-miR-133b-3p | 4.65 | BTG1 | 4.33 | gga-miR-138-5p | 3.30 |
| MYH9 | −1.26 | gga-miR-133c-3p | 2.60 | BTG1 | 4.33 | gga-miR-301a-3p | 2.37 |
| NFATC2 | −1.26 | gga-miR-301b-3p | 3.87 | CPEB1 | 2.97 | gga-miR-301b-3p | 3.87 |
| NFATC2 | −1.26 | gga-miR-301a-3p | 2.37 | CPEB1 | 2.97 | gga-miR-301a-3p | 2.37 |
| SGK1 | −1.14 | gga-miR-142-5p | 6.54 | DERA | 1.39 | gga-miR-1454-3p | 5.58 |
| SGK1 | −1.14 | gga-miR-1454-3p | 5.58 | LYPLA1 | 1.04 | gga-miR-138-5p | 3.30 |
| TAB2 | −1.17 | gga-miR-219b-3p | 6.04 | PPARG | 1.15 | gga-miR-301b-3p | 3.87 |
| TAB2 | −1.17 | gga-miR-142-3p | 5.91 | PPARG | 1.15 | gga-miR-301a-3p | 2.37 |
| TFDP2 | −1.01 | gga-miR-107-3p | 6.31 | PRKG1 | 3.44 | gga-miR-107-3p | 6.31 |
| ULK2 | −1.63 | gga-miR-301b-3p | 3.87 | RBPJ | 1.86 | gga-miR-219b-3p | 6.04 |
| ULK2 | −1.63 | gga-miR-301a-3p | 2.37 | RHEB | 2.12 | gga-miR-142-3p | 5.91 |
| AP3S1 | 1.44 | gga-miR-32-3p | −4.69 | SAT1 | 1.57 | gga-miR-219b-3p | 6.04 |
| RHEB | 2.12 | gga-miR-107-5p | −4.00 | WEE1 | 1.46 | gga-miR-1454-3p | 5.58 |
| B3GALT2 | −1.32 | gga-miR-107-5p | −4.00 | YWHAZ | 1.29 | gga-miR-193a-3p | 2.95 |
| B3GALT2 | −1.32 | gga-miR-32-5p | −6.13 |
| Lnc_id | Chr | Start | End | Gene Transcription ID | Gene Name | Start | End | PCC | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| TU10057 | 12 | 4774308 | 4829222 | ENSGALT00000007976 | PPARG | 4859854 | 4880521 | 0.87 | 0.025 |
| TU37186 | 5 | 16757648 | 16759893 | ENSGALT00000011621 | FADS2 | 16777263 | 16794363 | 0.88 | 0.019 |
| TU33274 | 4 | 13925955 | 13927633 | ENSGALT00000013126 | ACSL4 | 13835063 | 13847676 | −0.81 | 0.049 |
| TU35007 | 4 | 60355237 | 60362380 | ENSGALT00000077492 | ADH6 | 60350736 | 60361473 | 0.96 | 0.002 |
| TU35007 | 4 | 60355237 | 60362380 | ENSGALT00000052318 | ADH6 | 60363449 | 60368454 | 0.96 | 0.002 |
| TU35007 | 4 | 60355237 | 60362380 | ENSGALT00000037633 | ADH6 | 60350928 | 60355023 | 0.96 | 0.002 |
| TU35007 | 4 | 60355237 | 60362380 | ENSGALT00000032123 | ADH6 | 60350738 | 60361446 | 0.96 | 0.002 |
| TU35007 | 4 | 60355237 | 60362380 | ENSGALT00000032121 | ADH6 | 60356945 | 60368587 | 0.96 | 0.002 |
| TU26609 | 26 | 4070789 | 4074525 | ENSGALT00000004076 | PPARD | 4089638 | 4106262 | −0.87 | 0.023 |
| TU3310 | 1 | 78318443 | 78322786 | ENSGALT00000023745 | ATG8 | 78391093 | 78400942 | 0.89 | 0.019 |
| TU45776 | 9 | 14829770 | 14830278 | ENSGALT00000011898 | BCL6 | 14830574 | 14846807 | −0.83 | 0.043 |
| TU1868 | 1 | 50879489 | 50880054 | ENSGALT00000084251 | CSNK1E | 50966184 | 50982191 | −0.85 | 0.032 |
| TU1868 | 1 | 50879489 | 50880054 | ENSGALT00000053457 | CSNK1E | 50962355 | 50982806 | −0.85 | 0.032 |
| Transcript Name | Forward | Reverse |
|---|---|---|
| DTU45776 | CCTAAGACCGAAGCCAGCATC | AAGCGAATCCCACGAGCAG |
| DTU10057 | GGCAGCAAGGCATCAGAAGG | TGCAGAGTCGAAACAACCCATG |
| gga-miR-301b-3p | GCAGTGCAATAGTATTGTCAAAGCA | TGCTTTGACAATACTATTGCACTGC |
| gga-miR-190a-3p | CCGCCCTATATATCAAACATATTCC | GGAATATGTTTGATATATAGGGCGG |
| β- actin | GTCCACCGCAAATGCTTCTA | AGCCATGCCAACTCCGTCTT |
| U6 | GGAACGATACAGAGAAGATTAGC | TGGAACGCTTCACGAATTTGCG |
| SGK1 | AATGGCACAACCTCCACCTTC | AAGACTGCTCCAAGGCACCAC |
| PPARG | GACCAAAGCCAAGGCAAGG | TTGATCTGATCTTCTCCCATCCTTA |
| RHEB | ACGCACTCAGCACAAAGGACC | TTCCACCACCTTGCCCTGA |
| ATG8 | CCTGGCTCGGCTCCCTTCTC | ATCTTTGCCGACTCGACACATCTG |
| BCL-6 | CGAGGAGCCCCCGAGCAAATCAGG | CCGCGGTGCTCAGGAGAAGGGAACT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Kumar Mishra, S.; Wang, T.; Xu, Z.; Zhao, X.; Wang, Y.; Yin, H.; Fan, X.; Zeng, B.; Yang, M.; et al. AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken. Toxins 2020, 12, 290. https://doi.org/10.3390/toxins12050290
Liu X, Kumar Mishra S, Wang T, Xu Z, Zhao X, Wang Y, Yin H, Fan X, Zeng B, Yang M, et al. AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken. Toxins. 2020; 12(5):290. https://doi.org/10.3390/toxins12050290
Chicago/Turabian StyleLiu, Xueqin, Shailendra Kumar Mishra, Tao Wang, Zhongxian Xu, Xiaoling Zhao, Yan Wang, Huadong Yin, Xiaolan Fan, Bo Zeng, Mingyao Yang, and et al. 2020. "AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken" Toxins 12, no. 5: 290. https://doi.org/10.3390/toxins12050290
APA StyleLiu, X., Kumar Mishra, S., Wang, T., Xu, Z., Zhao, X., Wang, Y., Yin, H., Fan, X., Zeng, B., Yang, M., Yang, D., Ni, Q., Li, Y., Zhang, M., Zhu, Q., Chen, F., & Li, D. (2020). AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken. Toxins, 12(5), 290. https://doi.org/10.3390/toxins12050290





