A Bacillus thuringiensis Chitin-Binding Protein is Involved in Insect Peritrophic Matrix Adhesion and Takes Part in the Infection Process
Abstract
1. Introduction
2. Results
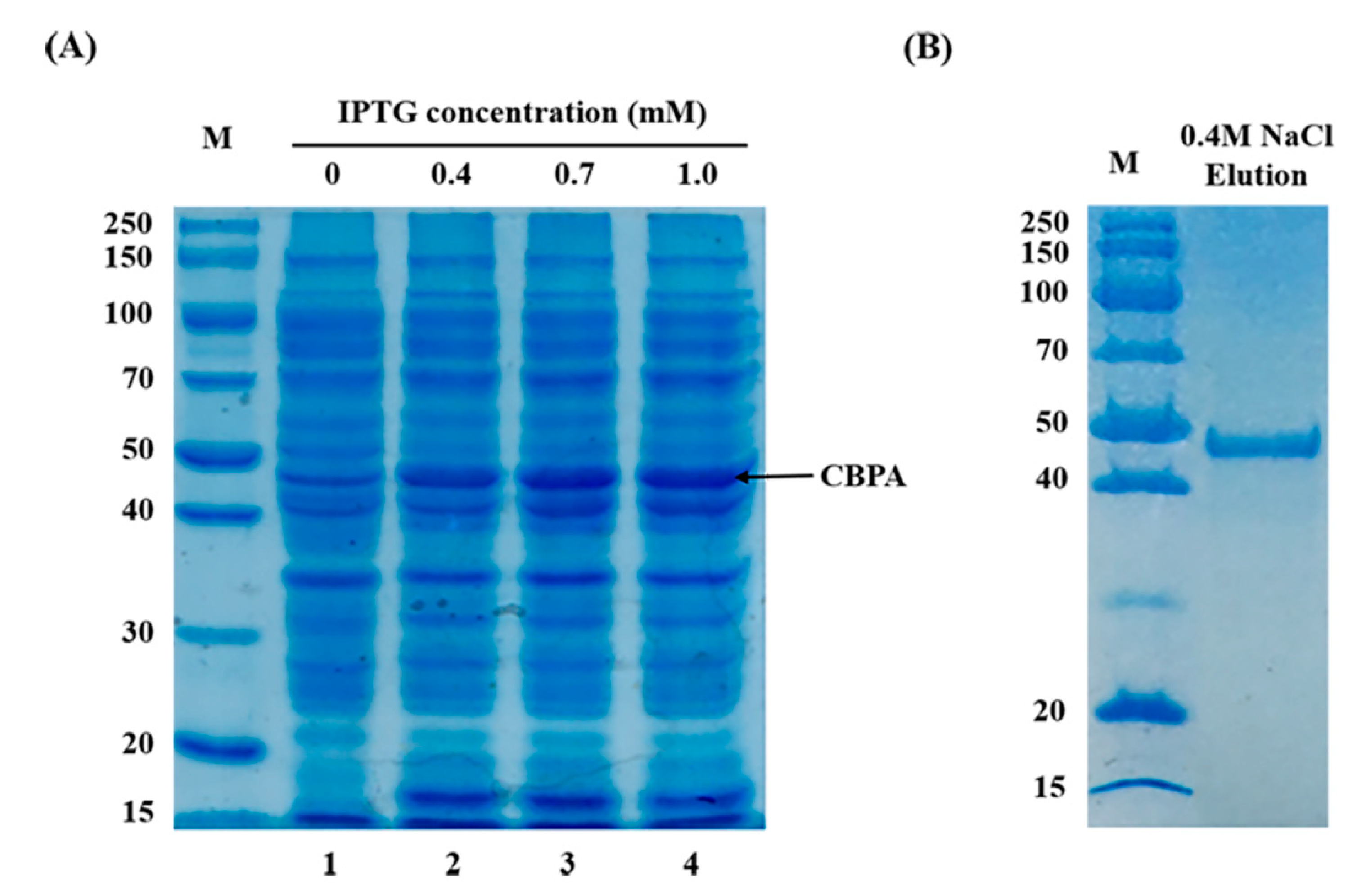
2.1. ∆cbpA Mutant, ∆cbpA::cbpA-Complemented Strain Construction
2.2. Role of CBPA in Ostrinia Furnacalis and Galleria Mellonella Mortality
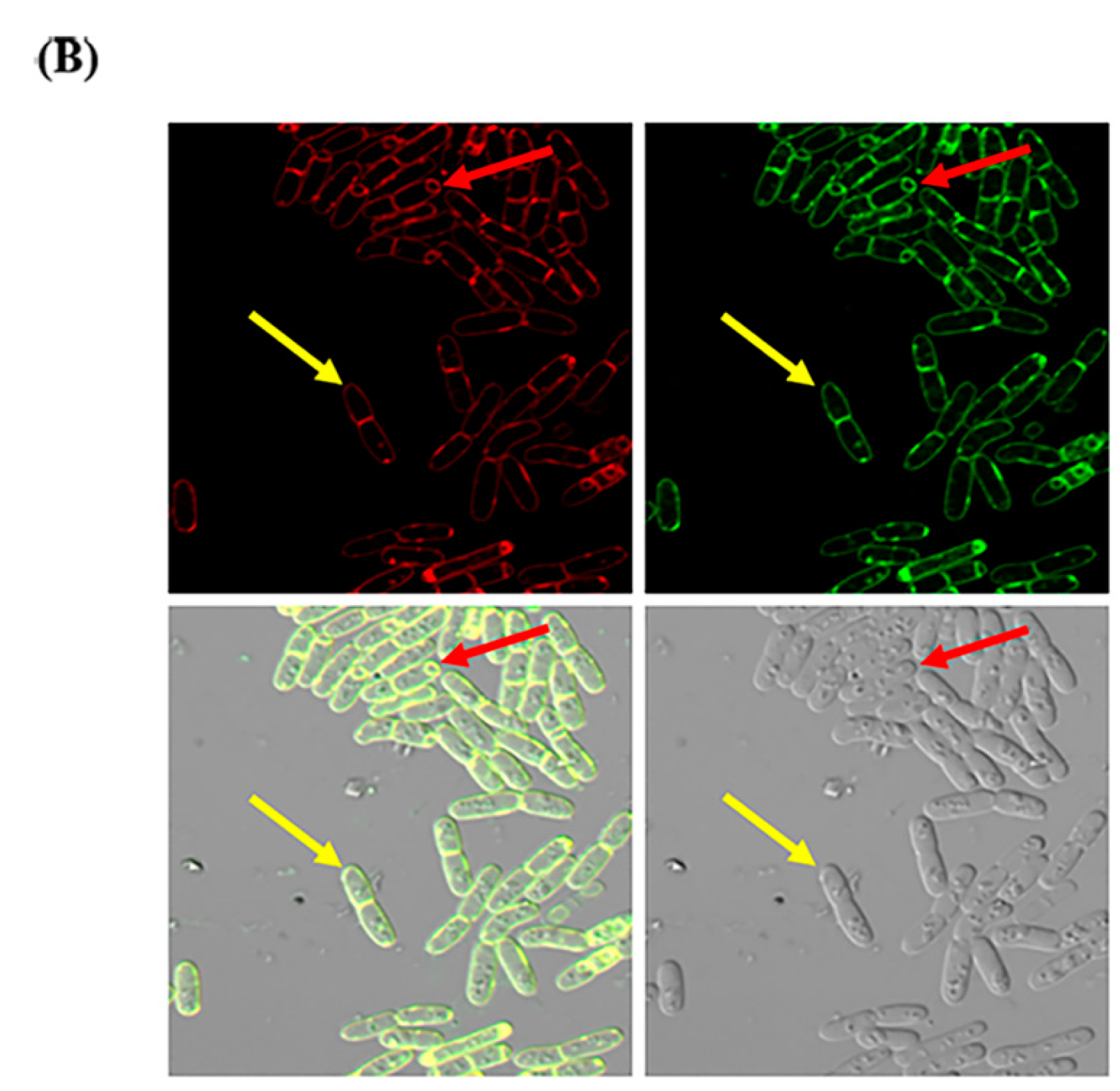
2.3. Localization of CBPA in Bt HD-73
2.4. Analysis of cbpA Promoter Activity under Alkaline Induction
2.5. Chitin Binding Ability and Chitinase Activity of CBPA
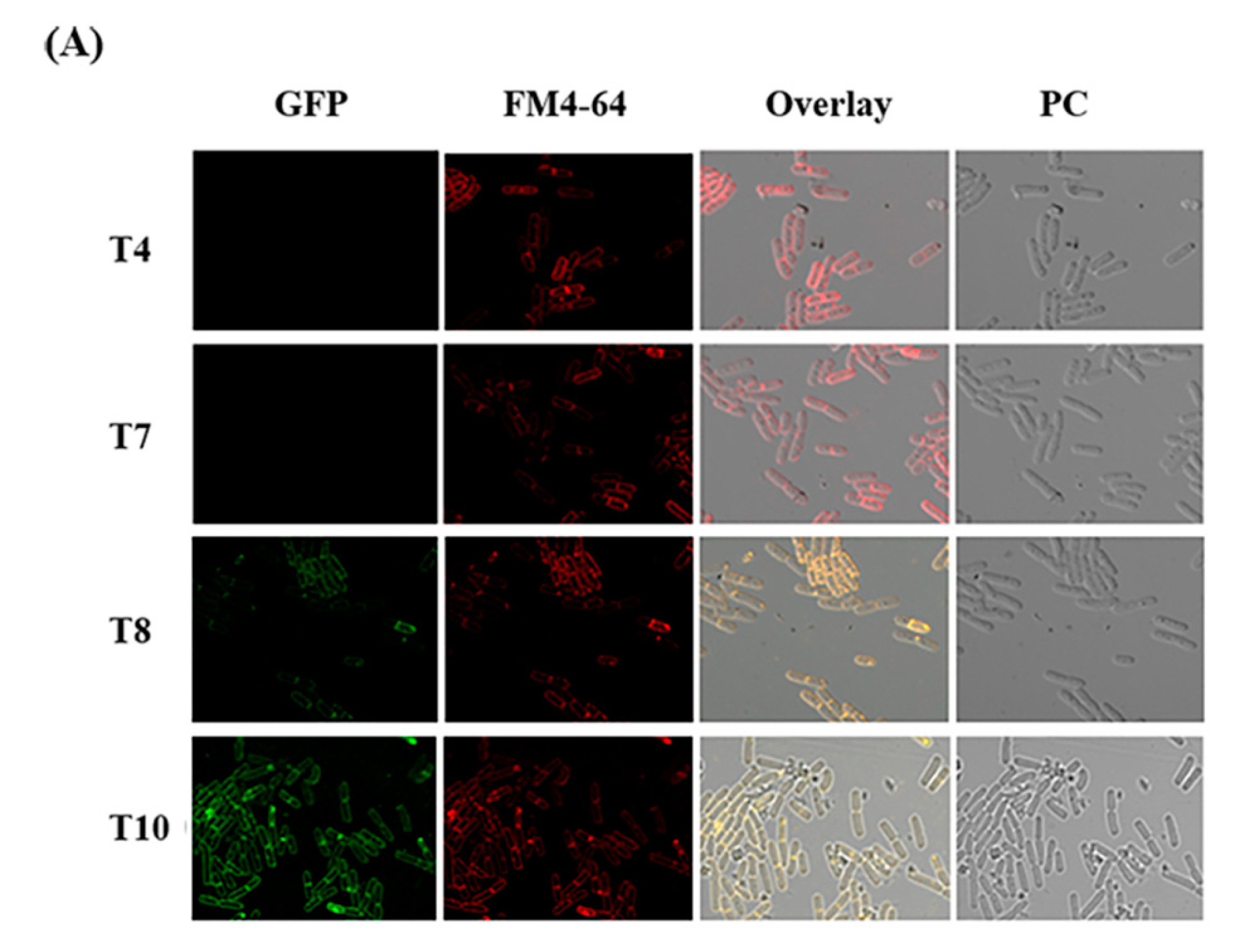
2.6. Expression of CBPA-GFP Fusion in Vivo in G. mellonella
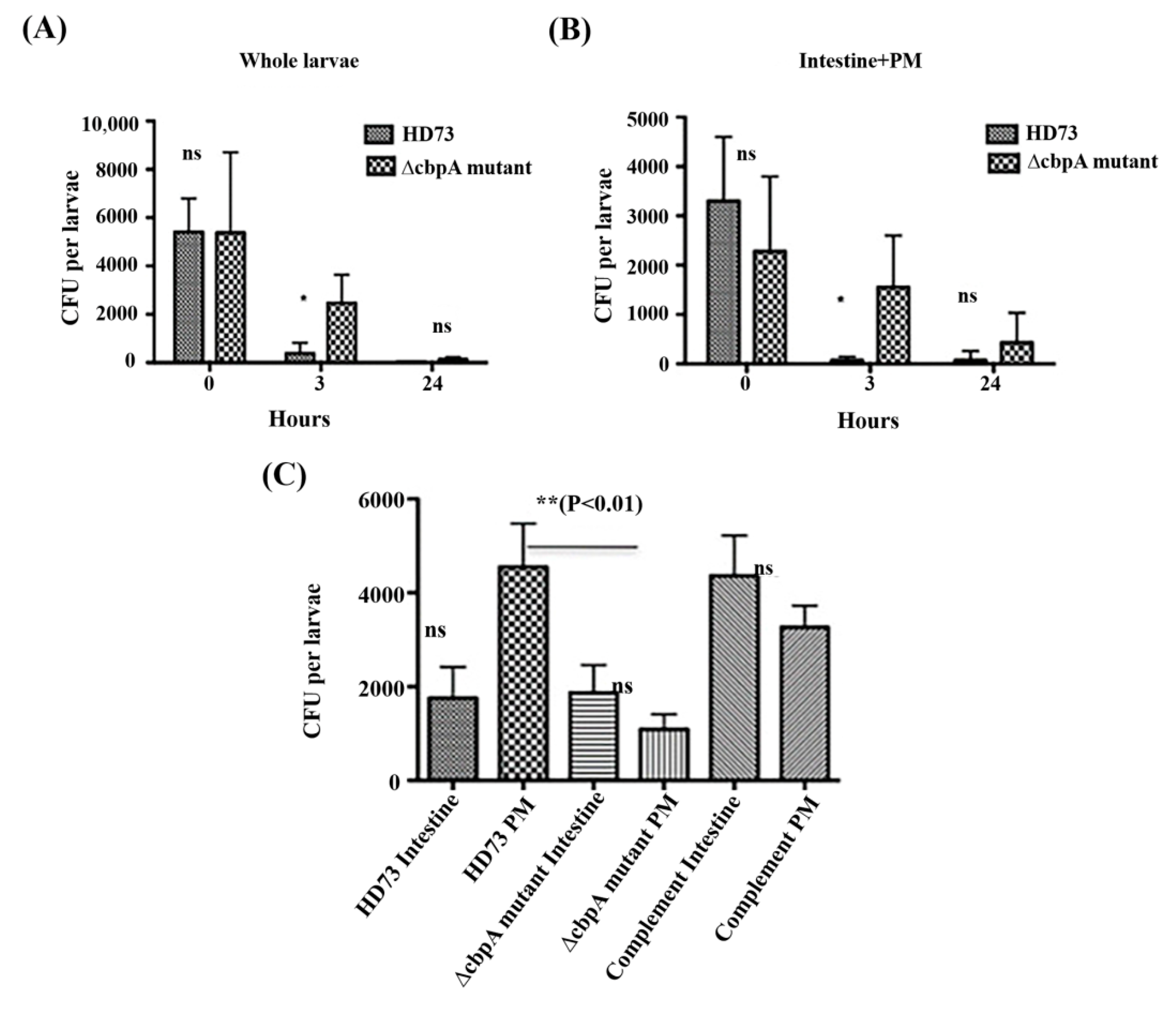
2.7. HD73 and HD73 ∆cbpA Intestinal Transit and Localization Assays
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Insects
4.3. DNA Manipulation and Transformation
4.4. Cloning of the HD73-cbpA Gene
4.5. Expression and Purification of CBPA
4.6. Determination of Chitinase Activity
4.7. Construction and Expression of Recombinant gfp-conjugated cbpA
4.8. Laser-Scanning Confocal Microscopy of CBPA-GFP Fusions
4.9. Construction of the Transcriptional Promoter PcbpA-lacZ Fusion Gene
4.10. β-Galactosidase Assays
4.11. Construction of the HD-73 ∆cbpA Mutant
4.12. Complementation of the HD-73 ∆cbpA Mutant
4.13. Crystal Spore Mixture Preparation for Asian Corn Borer Bioassays
4.14. Spore Preparation for Asian Corn Borer Bioassays
4.15. Spore Counts
4.16. Dose-Mortality Response Bioassays Against Asian Corn Borer
4.17. In Vivo G. mellonella Virulence Assays with HD73 and ∆cbpA Mutant Strains
4.18. Expression of the CBPA-GFP Fusion Protein in Vivo in G. mellonella
4.19. Intestinal Particle Transit Time for Final-Instar G. mellonella
4.20. HD73 and HD-73 ∆cbpA Intestinal Transit and Localization Assays
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423. [Google Scholar] [CrossRef] [PubMed]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Chapter Two–diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv. Insect Physiol. 2014, 47, 39–87. [Google Scholar]
- Ben, R.; Johnston, P.R.; Lereclus, D.; Nielsen-Leroux, C.; Crickmore, N. Bacillus thuringiensis: An impotent pathogen? Trends Microbiol. 2010, 18, 189. [Google Scholar]
- Stones, D.H.; Krachler, A.M. Against the tide: The role of bacterial adhesion in host colonization. Biochem. Soc. Trans. 2016, 44, 1571–1580. [Google Scholar] [CrossRef]
- Pizarrocerdá, J.; Cossart, P. Bacterial adhesion and entry into host cells. Cell 2006, 124, 715. [Google Scholar] [CrossRef]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Toprak, U. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285. [Google Scholar] [CrossRef]
- Weiss, B.L.; Savage, A.F.; Griffith, B.C.; Wu, Y.; Aksoy, S. The peritrophic matrix mediates differential infection outcomes in the tsetse fly gut following challenge with commensal, pathogenic, and parasitic microbes. J. Immunol. 2014, 193, 773–782. [Google Scholar] [CrossRef]
- Nielsen-Leroux, C.; Gaudriault, S.; Ramarao, N.; Lereclus, D.; Givaudan, A. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr. Opin. Microbiol. 2012, 15, 220–231. [Google Scholar] [CrossRef]
- Dubois, T.; Faegri, K.; Perchat, S.; Lemy, C.; Buisson, C.; Nielsen-LeRoux, C.; Lereclus, D. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog. 2012, 8, e1002629. [Google Scholar] [CrossRef]
- Kuraishi, T.; Binggeli, O.; Opota, O.; Buchon, N.; Lemaitre, B. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2011, 108, 15966–15971. [Google Scholar] [CrossRef]
- Lehane, M.J. Peritrophic matrix structure and function. Annu. Rev. Entomol. 1997, 42, 525–550. [Google Scholar] [CrossRef]
- Huber, M.; Cabib, E.; Miller, L.H. Malaria Parasite Chitinase and Penetration of the Mosquito Peritrophic Membrane. Proc. Natl. Acad. Sci. USA 1991, 88, 2807. [Google Scholar] [CrossRef]
- Sirichotpakorn, N.; Rongnoparut, P.; Choosang, K.; Panbangred, W. Coexpression of Chitinase and the cry11Aa1 toxin genes in Bacillus thuringiensis serovar israelensis. J. Invertebr. Pathol. 2001, 78, 160. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, W.A. Three years of aerial field experiments with Bacillus thuringiensis plus chitinase formulation against the spruce budworm. J. Invertebr. Pathol. 1974, 24, 344–348. [Google Scholar] [CrossRef]
- Tantimavanich, S.; Pantuwatana, S.; Bhumiratana, A.; Panbangred, W. Cloning of a chitinase gene into Bacillus thuringiensis subsp. aizawai for enhanced insecticidal activity. J. Gen. Appl. Microbiol. 1997, 43, 341–347. [Google Scholar] [CrossRef]
- Thamthiankul, S.; Moar, W.J.; Miller, M.E.; Panbangred, W. Improving the insecticidal activity of Bacillus thuringiensis subsp. aizawai against Spodoptera exigua by chromosomal expression of a chitinase gene. Appl. Microbiol. Biotechnol. 2004, 65, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Sampson, M.N.; Gooday, G.W. Involvement of chitinases of Bacillus thuringiensis during pathogenesis in insects. Microbiology 1998, 144 Pt 8, 2189. [Google Scholar] [CrossRef]
- Hashimoto, M.; Ikegami, T.; Seino, S.; Ohuchi, N.; Fukada, H.; Sugiyama, J.; Watanabe, T. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J. Bacteriol. 2000, 182, 3045–3054. [Google Scholar] [CrossRef]
- Vaajekolstad-Kolstad, G.; Horn, S.J.; van Aalten, D.M.; Synstad, B.; Eijsink, V.G. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 2005, 280, 28492–28497. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Anne, A.C.; Mathiesen, G.; Eijsink, V.G.H. The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of alpha-and beta-chitin. FEBS J. 2009, 276, 2402–2415. [Google Scholar] [CrossRef]
- Bayer, E.A.; Shoham, Y.; Lamed, R. The cellulosome. Glycomicrobiology 2002, 387–439. [Google Scholar]
- Carrard, G.; Koivula, A.; Soderlund, H.; Beguin, P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 2000, 97, 10342–10347. [Google Scholar] [CrossRef] [PubMed]
- Lehtiö, J.; Sugiyama, J.; Gustavsson, M.; Fransson, L.; Linder, M.; Teeri, T.T. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. USA 2003, 100, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Katouno, F.; Nikaidou, N.; Nonaka, T.; Sugiyama, J.; Watanabe, T. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 2001, 276, 41343. [Google Scholar] [CrossRef]
- Van Aalten, D.M.; Synstad, B.; Brurberg, M.B.; Hough, E.; Riise, B.W.; Eijsink, V.G.; Wierenga, R.K. Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9-A resolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5842–5847. [Google Scholar] [CrossRef]
- Xu, H.; Xie, W.; Gong, Z. Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett. 2000, 478, 123. [Google Scholar]
- Kamakura, T.; Yamaguchi, S.; Saitoh, K.; Teraoka, T.; Yamaguchi, I. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant Microbe Interact. MPMI 2002, 15, 437. [Google Scholar] [CrossRef]
- Kao, G.W.; Qiu, L.L.; Peng, Q.; Zhang, J.; Li, J.; Song, F.P. The analysis of major metabolic pathways in Bacillus thuringiensis under alkaline stress. Acta Microbiol. Sin. 2016, 56, 485–495. [Google Scholar]
- Zhan, Y.L.; Guo, S.Y. Three-dimensional (3D) structure prediction and function analysis of the chitin-binding domain 3 protein HD73_3189 from Bacillus thuringiensis HD73. Bio Med Mater. Eng. 2015, 26, S2019–S2024. [Google Scholar] [CrossRef]
- Li, R.S.; Jarrett, P.; Burges, H.D. Importance of spores, crystals, and δ-endotoxins in the pathogenicity of different varieties of Bacillus thuringiensis in Galleria mellonella and Pieris brassicae. J. Invertebr. Pathol. 1987, 50, 277–284. [Google Scholar] [CrossRef]
- Aamer Mehmood, M.; Hussain, K.; Latif, F.; Rizwan Tabassum, M.; Gull, M.; Shahid Gill, S.; Iqbal, Z. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 2000, 146 Pt 11, 2825. [Google Scholar]
- Bouillaut, L.; Ramarao, N.C.; Gilois, N.; Gohar, M.; Lereclus, D.; Nielsen-Leroux, C. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 2005, 71, 8903. [Google Scholar] [CrossRef] [PubMed]
- Vachon, V.; Laprade, R.; Schwartz, J.L. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: A critical review. J. Invertebr. Pathol. 2012, 111, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.S.; Yen, D.F.; Peng, W.K. Germination and proliferation of Bacillus thuringiensis in the gut of rice moth larva, Corcyra cephalonica. J. Invertebr. Pathol. 1986, 48, 96–99. [Google Scholar] [CrossRef]
- Bauer, L.S.; Pankratz, H.S. Ultrastructural effects of Bacillus thuringiensis var. san diego on midgut cells of the cottonwood leaf beetle. Jinvertebrpathol 1991, 60, 15–25. [Google Scholar] [CrossRef]
- Arora, N.; Sachdev, B.; Gupta, R.; Vimala, Y.; Bhatnagar, R.K. Characterization of a Chitin-Binding Protein from Bacillus thuringiensis HD-1. PLoS ONE 2013, 8, e66603. [Google Scholar] [CrossRef]
- Mehmood, M.A.; Xiao, X.; Hafeez, F.Y.; Gai, Y.B.; Wang, F.P. Molecular characterization of the modular chitin binding protein Cbp50 from Bacillus thuringiensis serovar konkukian. Antonie Van Leeuwenhoek 2011, 100, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Schrempf, H. Characteristics of chitin-binding proteins from Streptomycetes. Exs 1999, 87, 99–108. [Google Scholar]
- Chu, H.H.; Hoang, V.; Hofemeister, J.; Schrempf, H. A Bacillus amyloliquefaciens ChbB protein binds beta- and alpha-chitin and has homologues in related strains. Microbiology 2001, 147, 1793–1803. [Google Scholar] [CrossRef]
- Mehmood, M.A.; Latif, M.; Hussain, K.; Gull, M.; Latif, F.; Rajoka, M.I. Heterologous expression of the antifungal β-chitin binding protein CBP24 from Bacillus thuringiensis and its synergistic action with bacterial chitinases. Protein Pept. Lett. 2015, 22, 39–44. [Google Scholar] [CrossRef]
- Aamer Mehmood, M.; Hussain, K.; Latif, F.; Rizwan Tabassum, M.; Gull, M.; Shahid Gill, S.; Iqbal, Z. Synergistic Action of the Antifungal β-chitin Binding Protein CBP50 from Bacillus thuringiensis with Bacterial Chitinases. Curr. Proteom. 2014, 11, 23–26. [Google Scholar] [CrossRef]
- Du, C.; Nickerson, K.W. Bacillus thuringiensis HD-73 Spores have surface-localized Cry1Ac toxin: Physiological and pathogenic consequences. Appl. Environ. Microbiol. 1996, 62, 3722–3726. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Ahmad, T.; Rajagopal, R.; Bhatnagar, R.K. A constitutively expressed 36 kDa exochitinase from Bacillus thuringiensis HD-1. Biochem. Biophys. Res. Commun. 2003, 307, 620–625. [Google Scholar] [CrossRef]
- Fang, S.; Wang, L.; Guo, W.; Zhang, X.; Peng, D.; Luo, C.; Sun, M. Bacillus thuringiensis Bel Protein Enhances the Toxicity of Cry1Ac Protein to Helicoverpa armigera Larvae by DegradingInsect Intestinal Mucin. Appl. Environ. Microbiol. 2009, 75, 5237–5243. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004, 186, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Song, L.; Shu, C.; Wang, P.; Deng, C.; Peng, Q.; Lereclus, D.; Wang, X.; Huang, D.; Zhang, J.; et al. Complete Genome Sequence of Bacillus thuringiensis subsp. kurstaki Strain HD73. Genome Announcements. 2013, 1, e0008013. [Google Scholar] [CrossRef] [PubMed]
- Jr, R.S.M.; Sasaki, K. Protein D, a putative immunoglobulin D-binding protein produced by Haemophilus influenzae, is glycerophosphodiester phosphodiesterase. J. Bacteriol. 1993, 175, 4569–4571. [Google Scholar]
- Yang, J.; Peng, Q.; Chen, Z.; Deng, C.; Shu, C.; Zhang, J.; Song, F. Transcriptional regulation and characteristics of a novel N-acetylmuramoyl-l-alanine amidase gene involved in Bacillus thuringiensis mother cell lysis. J. Bacteriol. 2013, 195, 2887. [Google Scholar] [CrossRef]
- Arantes, O.; Lereclus, D. Construction of cloning vectors for Bacillus thuringiensis. Gene 1991, 108, 115–119. [Google Scholar] [CrossRef]
- Agaisse, H.; Lereclus, D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 1994, 13, 97–107. [Google Scholar] [CrossRef]
- Gennaro, M.L.; Iordanescu, S.; Novick, R.P.; Murray, R.W.; Steck, T.R.; Khan, S.A. Functional organization of the plasmid pT181 replication origin. J. Mol. Biology 1989, 205, 355–362. [Google Scholar] [CrossRef]
- Wang, G.J.; Zhang, J.; Song, F.P.; Wu, J.; Feng, S.L.; Huang, D.F. Engineered Bacillus thuringiensis GO33A with broad insecticidal activity against lepidopteran and coleopteran pests. Appl. Microbiol. Biotechnol. 2006, 72, 924. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, Volume 1; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Lereclus, D.; Arantès, O.; Chaufaux, J.; Lecadet, M. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 1989, 51, 211–217. [Google Scholar] [CrossRef]
- Yang, H.; Wang, P.; Peng, Q.; Rong, R.; Liu, C.; Lereclus, D.; Huang, D. Weak transcription of the cry1Ac gene in nonsporulating Bacillus thuringiensis Cells. Appl. Environ. Microbiol. 2012, 78, 6466–6474. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.J.H. Assay of β galactosidase. In Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2015; pp. 352–359. [Google Scholar]
- Clements, M.O.; Moir, A. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriology 1998, 180, 6729. [Google Scholar] [CrossRef]
- Van Cuyk, S.; Deshpande, A.; Hollander, A.; Duval, N.; Ticknor, L.; Layshock, J.; Omberg, K.M. Persistence of Bacillus thuringiensis subsp. kurstaki in urban environments following spraying. Appl. Environ. Microbiol. 2011, 77, 7954–7961. [Google Scholar] [CrossRef]
- Zhang, T.; He, M.; Gatehouse, A.M.; Wang, Z.; Edwards, M.G.; Li, Q.; He, K. Inheritance patterns, dominance and cross-resistance of Cry1Ab-and Cry1Ac-selected (Guenée). Bautechnik 2014, 74, 395–400. [Google Scholar]
- Zhou, Z.; Wang, Z.; Liu, Y.; Liang, G.; Shu, C.; Song, F.; Zhang, J. Identification of ABCC2 as a binding protein of Cry1Ac on brush border membrane vesicles from Helicoverpa armigera by an improved pull-down assay. Microbiologyopen 2016, 5, 659–669. [Google Scholar] [CrossRef]
- Ramarao, N.; Nielsen-Leroux, C.; Lereclus, D. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis. Exp. Jov. 2012, 70, e4392. [Google Scholar] [CrossRef]




| No. | Concentration of Cry Protein (μg/g) | Number of Spore (Numbers/g) | Mortality (%) | Significance | |
|---|---|---|---|---|---|
| Wild-Type | ∆cbpA Mutant | ||||
| 1 | 0.005 | 5.4 × 106 | 0.0 ± 0.0 | 0.7 ± 0.7 | 0.374 |
| 2 | 0.025 | 2.7 × 107 | 4.7 ± 0.7 | 2.7 ± 1.3 | 0.251 |
| 3 | 0.050 | 5.4 × 107 | 17.0 ± 1.2 | 8.7 ± 0.7* | 0.003 |
| 4 | 0.250 | 2.7 × 108 | 35.3 ± 1.5 | 19.7 ± 0.7 * | 0.001 |
| 5 | 0.500 | 5.4 × 108 | 51.3 ± 1.8 | 33.7 ± 1.3* | 0.001 |
| 6 | 1.000 | 1.1 × 109 | 76.3 ± 1.3 | 54.7 ± 0.7* | << 0.001** |
| 7 | 2.500 | 2.7 × 109 | 90.3 ± 2.7 | 58.7 ± 1.3 * | <<0.001*** |
| LC50(Spore CFU) | 95% Confidence Interval | |
|---|---|---|
| BtHD73 | 6.59 × 105 | 3.41 × 105–1.04 × 106 |
| △cbpA mutant | 4.85 × 106 | 2.25 × 106–7.61 × 106 |
| △cbpA::cbpA | 5.72 × 105 | 3.18 × 105–8.98 × 105 |
| Chitinase Substrate | Chitinase Activity (Substrate Degradation, μmol/min) | ||
|---|---|---|---|
| Negative Control | CPBA | Positive Control | |
| 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide | 4.79 × 104 | 4.88 ×1 04 | 3.47 × 106 |
| 4-Methylumbelliferyl Β-D-N,N’-diacetylchitobioside hydrate | 6.35 × 104 | 5.88 × 104 | 4.81 × 106 |
| 4-Methylumbelliferyl Β-D-N,N,N″-triacetylchitotriose | 5.01 × 104 | 4.79 × 104 | 2.21 × 106 |
| Strain or Plasmid | Characeristics | Reference or Source |
|---|---|---|
| E. coli strains | ||
| JM110 | rpsL(strr),thr,leu,thi-1,lacY,galK,galT,ara,tonA,tsx,dam,dcm,supE44,Δ(lac-proAB), [F’,traD36,proAB,laclqZΔM15] | This laboratory |
| BL21/DE3 | E. coli B, F-,dcm,ompT, hsdS(rB-mB-), gal, λ(DE3) | [47] |
| ET | ∆(lac-proAB) RpsL(strr), thr, leu, endA, thi-1, lacY, galK, galT, ara, tonA, tsx, dam, dcm, supE44, (F’ traD36proABlacIqZ∆M15) | This laboratory |
| B. thuringiensis subsp. kurstaki strains | ||
| HD73 | Contains cry1Ac gene | [46] |
| HD73(pHT-gfp) | HD73 strain containing plasmid pHT-gfp | [48] |
| HD73(pHT-cbpA-gfp) | HD73 strain containing the translational fusion plasmid pHT-cbpA-gfp | This study |
| HD (PcbpA-lacZ) | HD73 strain containing plasmid pHTPcbpA | This study |
| HD73(pRN5101ΩcbpA) | HD73 strain containing plasmid pRN5101ΩcbpA | This study |
| HD73(ΔcbpA) | HD73 mutant, ΔcbpA | This study |
| HD73(ΔcbpA::cbpA) | HD73(ΔcbpA) containing plasmid pHTCcbpA | This study |
| Plasmids | ||
| pET-21b | Expression vector, Ampr, 5.4 kb | Novagen |
| BL21 (pETcbpA) | BL21(DE3) with pETcbpA plasmid | This study |
| pHT315 | B. thuringiensis-E. coli shuttle vector, 6.5kb | [49] |
| pHT304-18Z | Promoterless lacZ vector, Eryr Ampr, 9.7 kb | [50] |
| pHTPcbpA | pHT304-18Z carrying PcbpA, Ampr Ermr | This study |
| pETcbpA | pET-21b containing cbpA gene, Ampr | This study |
| pETCcbpA | pHT304 carrying cbpA gene,Ampr | This study |
| pHT PcbpA-3189-gfp | pHT315 containing PcbpA-cbpA-gfp gene | This study |
| pRN5101 | Temperature-sensitive plasmid, 8.0 kb | [51] |
| pRN5101ΩcbpA | pRN5101 carrying partial cbpA deletion gene | This study |
| pDG780 | Containing a kanamycin resistance gene | [52] |
| Primer | Sequence | Restriction Site |
|---|---|---|
| cbpA-a | CGCGGATCCGATGAACATGAATAATCGAT | BamH I |
| cbpA-b | ACGCGTCGACTTACACTGTTTTCCATAAT | Sal I |
| cbpA-c | GCATGCCTGCAGGTCGACTCTAGAGGGCTGCTTTGAATTTGAAGGAAT | |
| cbpA-d | TGTAAAACGACGGCCAGTGAATTAAAGCCCATCATCTCTTAGTTCAT | |
| gfp-1 | CGGGATCCAAGAGGCTGCTTTGAATTTGAAGG | BamH I |
| gfp-2 | CCGCCTCCACCTGACACTGTTTTCCATAATG | |
| gfp-3 | CATTATGGAAAACAGTGTCAGGTGGAGGCGG | |
| gfp-4 | CATGCATGCTTATTTGTATAGTTCATCCATGCC | SphI(Pae I) |
| PcbpA-F | TGCACTGCAGGGCTGCTTTGAATTTGAAGGAATC | Pst I |
| PcbpA-R | CGGGATCCGTTCATGTCCCCTTCTTGTTATAC | BamH I |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, J.; Tong, Z.; Zhan, Y.; Buisson, C.; Song, F.; He, K.; Nielsen-LeRoux, C.; Guo, S. A Bacillus thuringiensis Chitin-Binding Protein is Involved in Insect Peritrophic Matrix Adhesion and Takes Part in the Infection Process. Toxins 2020, 12, 252. https://doi.org/10.3390/toxins12040252
Qin J, Tong Z, Zhan Y, Buisson C, Song F, He K, Nielsen-LeRoux C, Guo S. A Bacillus thuringiensis Chitin-Binding Protein is Involved in Insect Peritrophic Matrix Adhesion and Takes Part in the Infection Process. Toxins. 2020; 12(4):252. https://doi.org/10.3390/toxins12040252
Chicago/Turabian StyleQin, Jiaxin, Zongxing Tong, Yiling Zhan, Christophe Buisson, Fuping Song, Kanglai He, Christina Nielsen-LeRoux, and Shuyuan Guo. 2020. "A Bacillus thuringiensis Chitin-Binding Protein is Involved in Insect Peritrophic Matrix Adhesion and Takes Part in the Infection Process" Toxins 12, no. 4: 252. https://doi.org/10.3390/toxins12040252
APA StyleQin, J., Tong, Z., Zhan, Y., Buisson, C., Song, F., He, K., Nielsen-LeRoux, C., & Guo, S. (2020). A Bacillus thuringiensis Chitin-Binding Protein is Involved in Insect Peritrophic Matrix Adhesion and Takes Part in the Infection Process. Toxins, 12(4), 252. https://doi.org/10.3390/toxins12040252






