PGN and LTA from Staphylococcus aureus Induced Inflammation and Decreased Lactation through Regulating DNA Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells
Abstract
1. Introduction
2. Results
2.1. Effects of PGN and LTA on Transcriptome
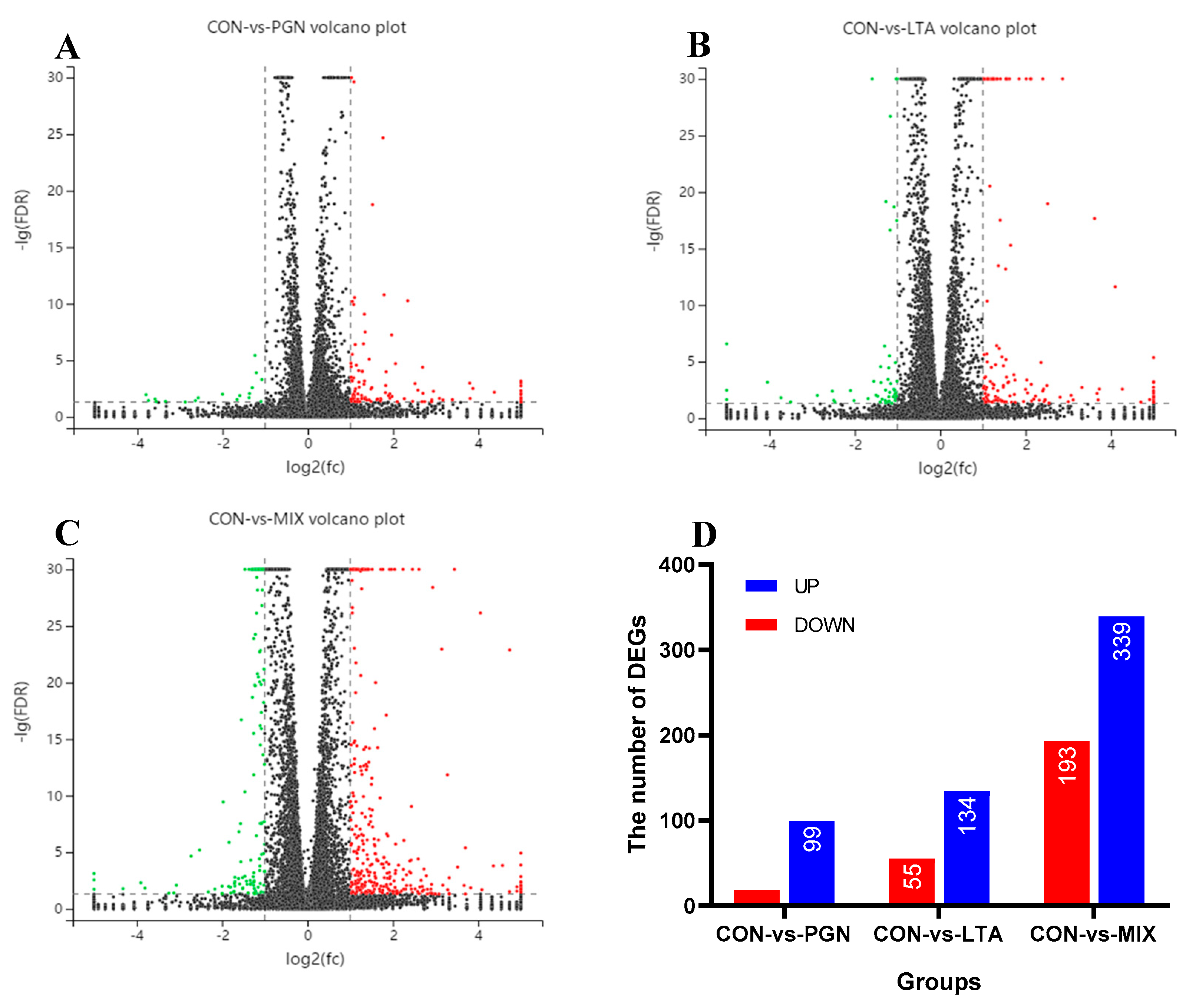
2.1.1. Overview of RNA Sequencing Data
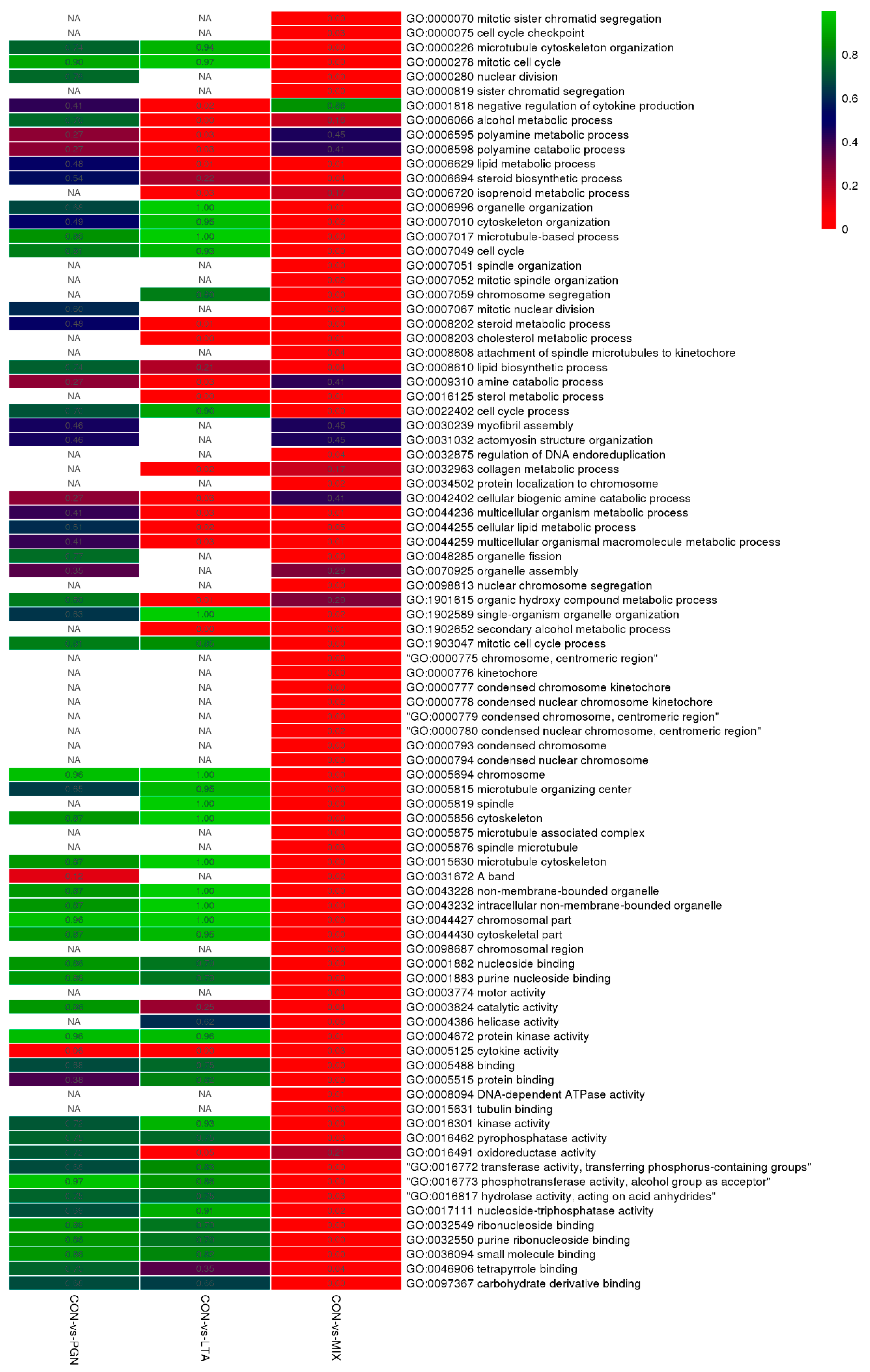
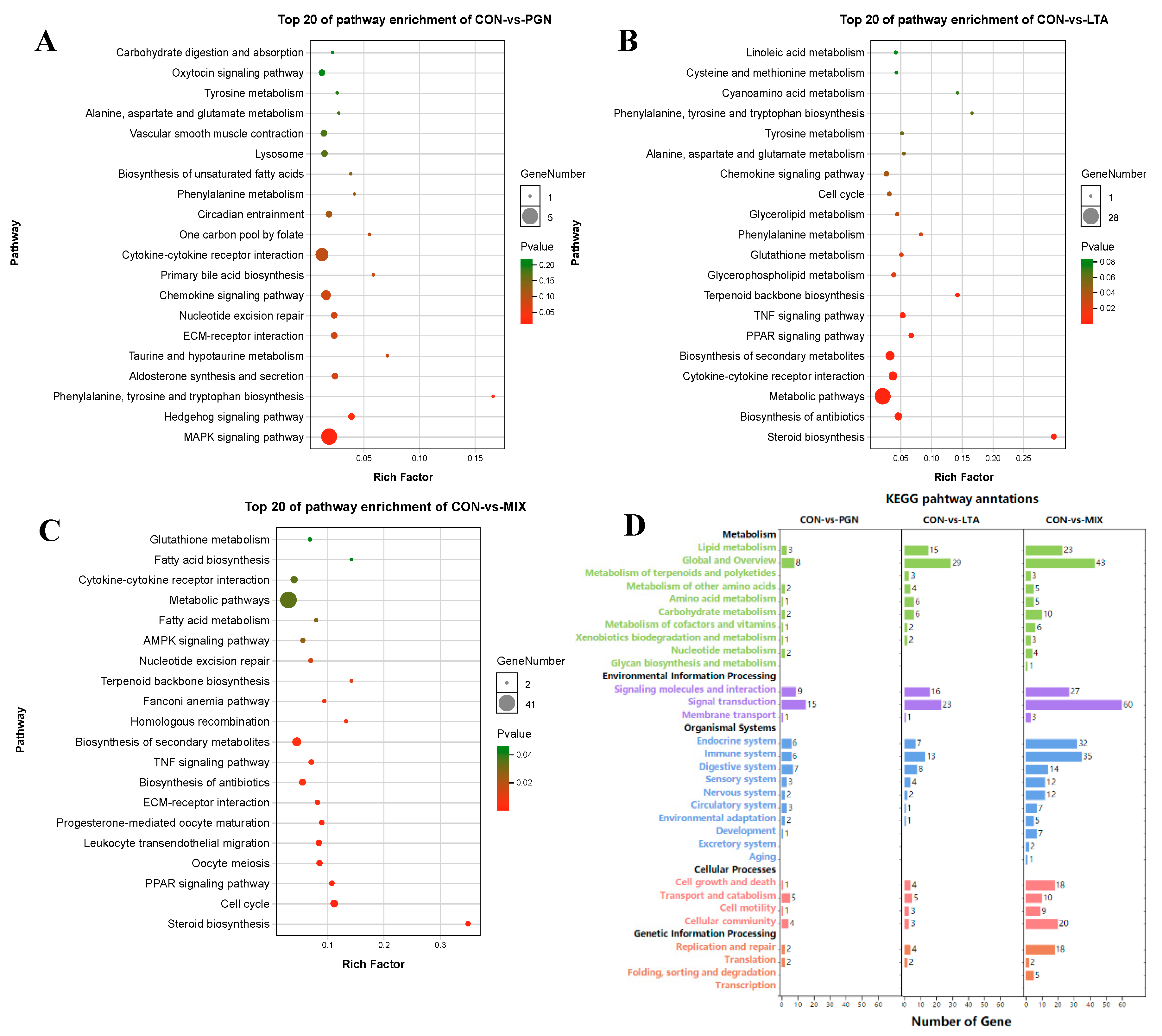
2.1.2. GO Function Annotation and KEGG Pathway Enrichment Analyses of DEGs
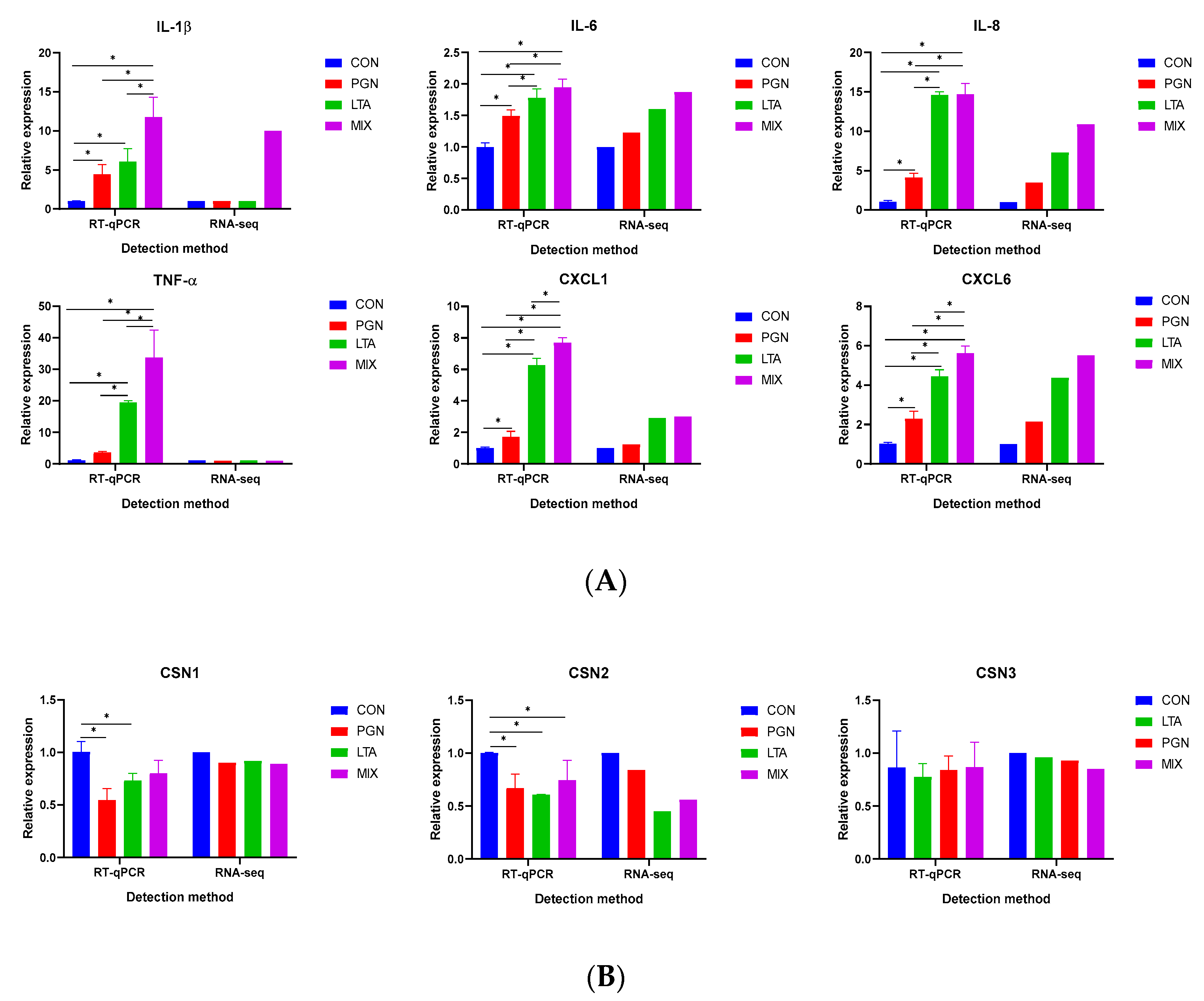
2.2. Validation of Selected DEGs by RT-qPCR Analyses
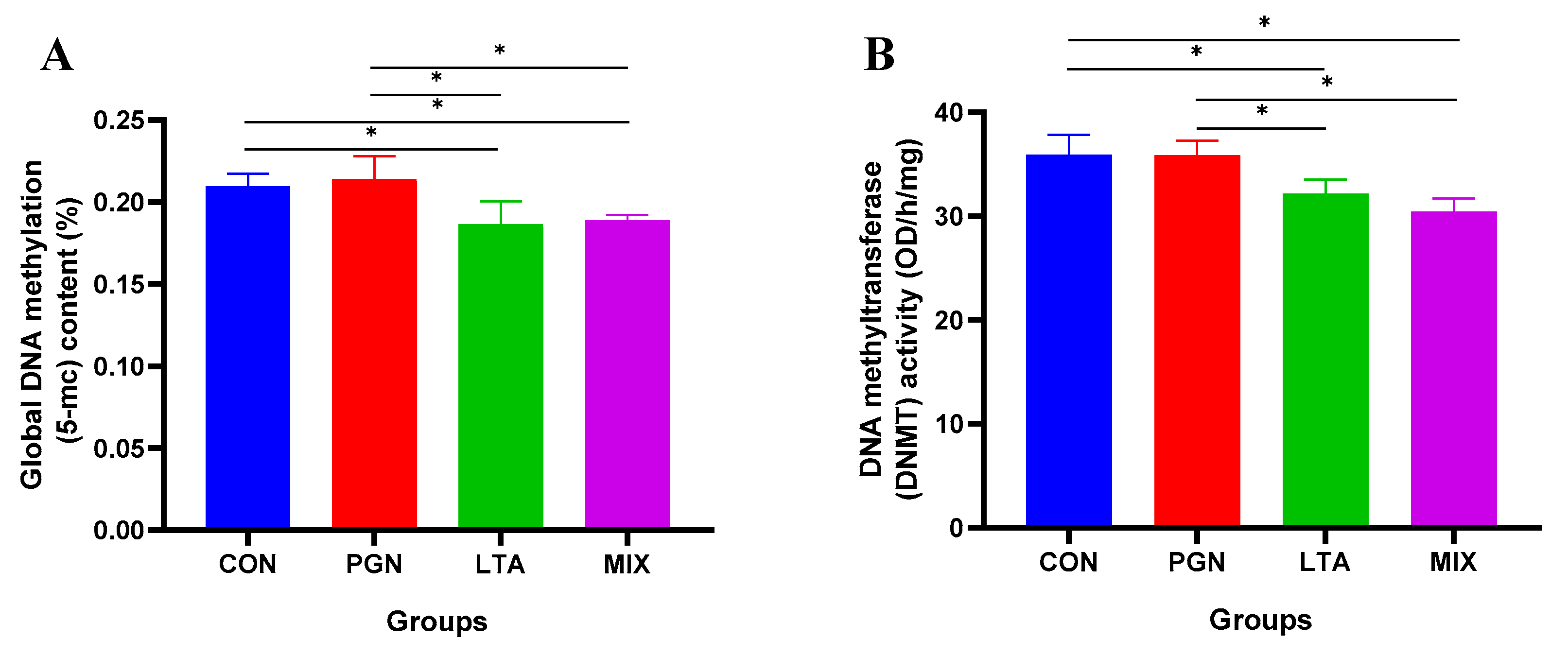
2.3. LTA and PGN + LTA Reduced Global DNA Methylation Levels by Suppressing DNMT Activity
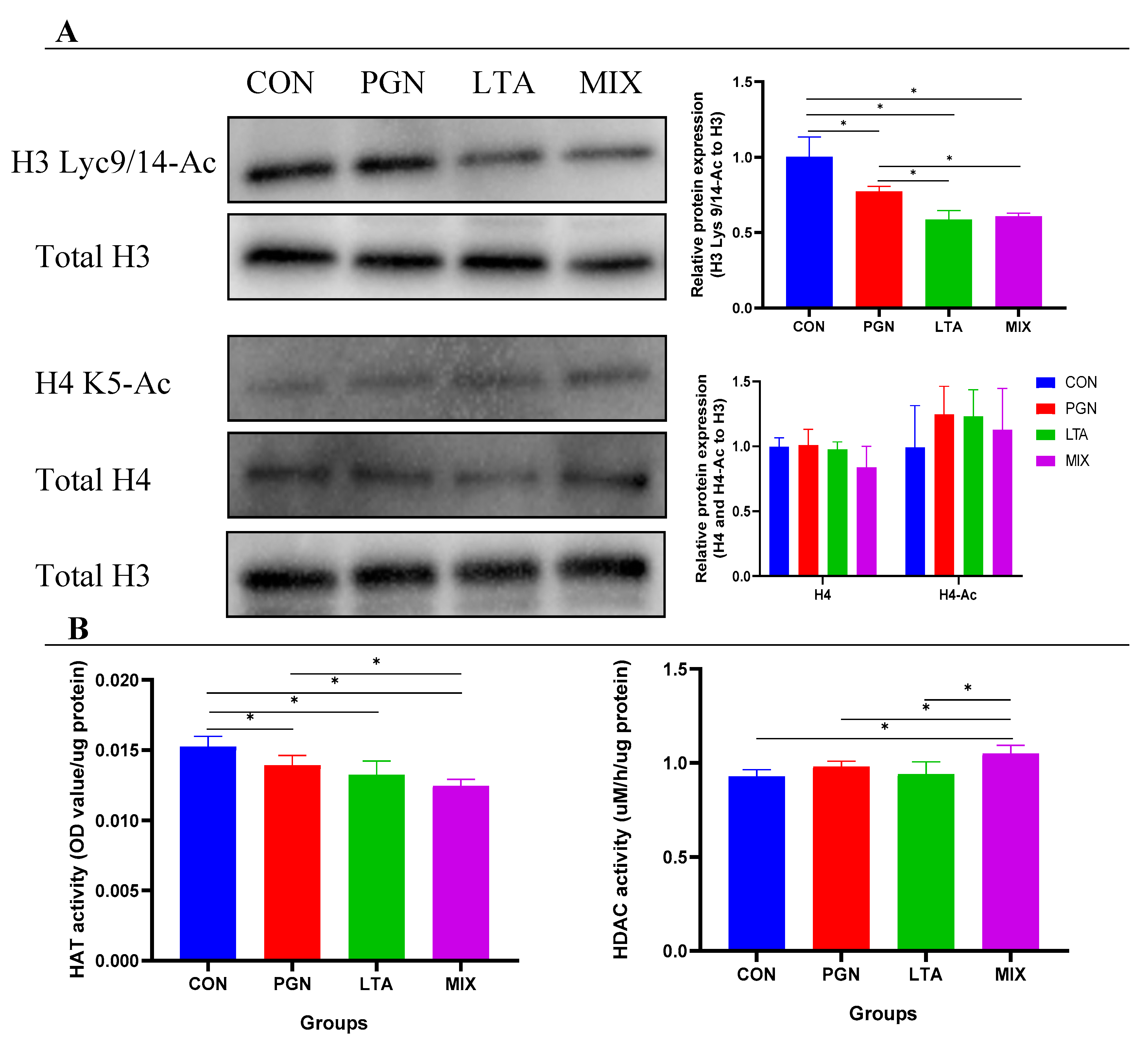
2.4. PGN and LTA Reduced Histone H3 Acetylation Levels by Suppressing HAT Activity
2.5. PGN and LTA Promoted Inflammation But Reduced Lactation
2.5.1. PGN and LTA Increased Proinflammatory Factor Secretion
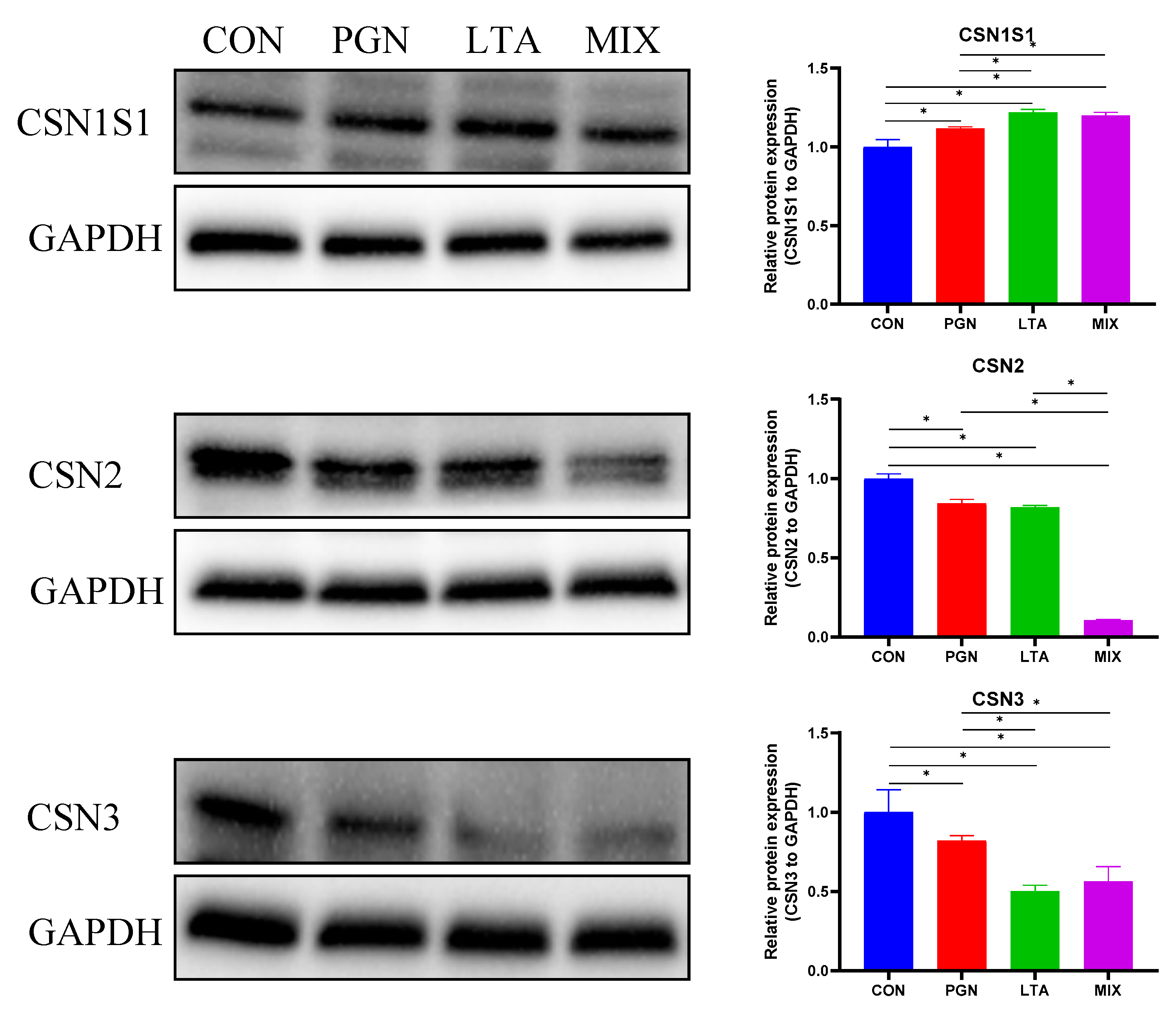
2.5.2. PGN and LTA Decreased Casein Synthesis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Culture and Treatments
5.2. RNA-Seq Data Analysis
5.3. Total RNA Isolation, Reverse Transcription, and RT-qPCR
5.4. Analysis of Total DNA Methylation, Histone Acetylation, and Activity of DNMT, HAT and HDAC
5.5. ELISA and Western Blot Assays
5.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Szyda, J.; Mielczarek, M.; Fraszczak, M.; Minozzi, G.; Williams, J.L.; Wojdak-Maksymiec, K. The genetic background of clinical mastitis in Holstein-Friesian cattle. Animal 2019, 13, 2156–2163. [Google Scholar] [CrossRef]
- Yu, C.; Shi, Z.R.; Chu, C.Y.; Lee, K.H.; Zhao, X.; Lee, J.W. Expression of bovine granulocyte chemotactic protein-2 (GCP-2) in neutrophils and a mammary epithelial cell line (MAC-T) in response to various bacterial cell wall components. Vet. J. 2010, 186, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Strandberg, Y.; Gray, C.; Vuocolo, T.; Donaldson, L.; Broadway, M.; Tellam, R. Lipopolysaccharide and lipoteichoic acid induce different innate immune responses in bovine mammary epithelial cells. Cytokine 2005, 31, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Eckel, E.F.; Ametaj, B.N. Invited review: Role of bacterial endotoxins in the etiopathogenesis of periparturient diseases of transition dairy cows. J. Dairy Sci. 2016, 99, 5967–5990. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Deng, R.; Li, X.; Zhang, Y.; Gao, M. RNA-seq analysis of different inflammatory reactions induced by lipopolysaccharide and lipoteichoic acid in bovine mammary epithelial cells. Microb. Pathog. 2019, 130, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Im, J.; Lee, T.; Jeon, J.H.; Baik, J.E.; Kim, K.W.; Kang, S.-S.; Yun, C.-H.; Kim, H.; Han, S.H. Gene expression profiling of bovine mammary gland epithelial cells stimulated with lipoteichoic acid plus peptidoglycan from Staphylococcus aureus. Int. Immunopharmacol. 2014, 21, 231–240. [Google Scholar] [CrossRef]
- Kiku, Y.; Nagasawa, Y.; Tanabe, F.; Sugawara, K.; Watanabe, A.; Hata, E.; Ozawa, T.; Nakajima, K.; Arai, T.; Hayashi, T. The cell wall component lipoteichoic acid of Staphylococcus aureus induces chemokine gene expression in bovine mammary epithelial cells. J. Vet. Med. Sci. 2016, 78, 1505–1510. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, S.J.; Maksim, S.; Yao, Y.F.; Liu, H.M.; Du, J. Epigenetic modification in macrophages: A promising target for tumor and inflammation-associated disease therapy. Curr. Top. Med. Chem. 2019, 19, 1350–1362. [Google Scholar] [CrossRef]
- Zong, D.D.; Ouyang, R.Y.; Chen, P. Epigenetic mechanisms in chronic obstructive pulmonary disease. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 844–856. [Google Scholar]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Sajjanar, B.; Trakooljul, N.; Wimmers, K.; Ponsuksili, S. DNA methylation analysis of porcine mammary epithelial cells reveals differentially methylated loci associated with immune response against Escherichia coli challenge. BMC Genom. 2019, 20, 623. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Li, Q.; Cai, L.; Zhan, M.; Xu, Q. The effect of DNA methylation on the miRNA expression pattern in lipopolysaccharide-induced inflammatory responses in human dental pulp cells. Mol. Immunol. 2019, 111, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhan, M.; Meng, R.; Wang, X.; Xu, Q. 5-Aza-2’-deoxycytidine enhances lipopolysaccharide-induced inflammatory cytokine expression in human dental pulp cells by regulating TRAF6 methylation. Bioengineered 2019, 10, 197–206. [Google Scholar] [CrossRef]
- Korkmaz, F.T.; Kerr, D.E. Genome-wide methylation analysis reveals differentially methylated loci that are associated with an age-dependent increase in bovine fibroblast response to LPS. BMC Genom. 2017, 18, 405. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Sun, Y.; Dong, X.; Wang, Z.; Zhang, Z.; Xiao, Y.; Dong, G. Bacterial lipopolysaccharide induced alterations of genome-wide DNA methylation and promoter methylation of lactation-related genes in bovine mammary epithelial cells. Toxins 2019, 11, 298. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Y.; Sun, Y.; Dong, X.; Wang, Z.; Zhang, Z.; Xiao, Y.; Dong, G. Bacterial endotoxin decreased histone H3 acetylation of bovine mammary epithelial cells and the adverse effect was suppressed by sodium butyrate. BMC Vet. Res. 2019, 15, 267. [Google Scholar] [CrossRef]
- Song, M.; He, Y.; Zhou, H.; Zhang, Y.; Li, X.; Yu, Y. Combined analysis of DNA methylome and transcriptome reveal novel candidate genes with susceptibility to bovine Staphylococcus aureus subclinical mastitis. Sci. Rep. 2016, 6, 29390. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Jiang, Q.; Hao, H.; Ju, Z.; Yang, C.; Sun, Y.; Wang, C.; Zhong, J.; Huang, J.; et al. DNA methylation rather than single nucleotide polymorphisms regulates the production of an aberrant splice variant of IL6R in mastitic cows. Cell Stress Chaperones 2018, 23, 617–628. [Google Scholar] [CrossRef]
- Wang, D.; Wei, Y.; Shi, L.; Khan, M.Z.; Fan, L.; Wang, Y.; Yu, Y. Genome-wide DNA methylation pattern in a mouse model reveals two novel genes associated with Staphylococcus aureus mastitis. Asian-Australas. J. Anim. Sci. 2019, 15, 203–211. [Google Scholar] [CrossRef]
- Tesfaye, G.Y.; Regassa, F.G.; Kelay, B. Milk yield and associated economic losses in quarters with subclinical mastitis due to Staphylococcus aureus in Ethiopian crossbred dairy cows. Trop. Anim. Health Prod. 2010, 42, 925–931. [Google Scholar] [CrossRef]
- Choudhury, A.; Solanki, B.; Singh, S.; Sahu, U.; Parvez, S.; Kar, S.; Ganguly, S. Persistent peripheral presence of Staphylococcus aureus promotes histone H3 hypoacetylation and decreases tyrosine hydroxylase protein level in rat brain tissues. Neuroreport 2019, 30, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Su, L.D.; Lu, Z.Q. Expression of DNA methylation and histone acetylation related genes in response to bacterial infection in the silkworm, Bombyx mori. Acta Entomol. Sin. 2015, 58, 941–949. [Google Scholar]
- Barkema, H.W.; Schukken, Y.H.; Lam, T.; Beiboer, M.L.; Wilmink, H.; Benedictus, G.; Brand, A. Incidence of clinical mastitis in dairy herds grouped in three categories by bulk milk somatic cell counts. J. Dairy Sci. 1998, 81, 411–419. [Google Scholar] [CrossRef]
- Riollet, C.; Rainard, P.; Poutrel, B. Cell subpopulations and cytokine expression in cow milk in response to chronic Staphylococcus aureus infection. J. Dairy Sci. 2001, 84, 1077–1084. [Google Scholar] [CrossRef]
- Barkema, H.W.; Schukken, Y.H.; Zadoks, R.N. Invited review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 2006, 89, 1877–1895. [Google Scholar] [CrossRef]
- Zaatout, N.; Ayachi, A.; Kecha, M.; Kadlec, K. Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J. Appl. Microbiol. 2019, 127, 1305–1314. [Google Scholar] [CrossRef]
- Pidgeon, S.E.; Pires, M.M. Cell wall remodeling of staphylococcus aureus in live caenorhabditis elegans. Bioconj. Chem. 2017, 28, 2310–2315. [Google Scholar] [CrossRef] [PubMed]
- Steven, B.; Maria, J.P.S., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Rajagopal, M.; Walker, S. Envelope structures of Gram-positive bacteria. In Protein and Sugar Export and Assembly in Gram-Positive Bacteria; Bagnoli, F., Rappuoli, R., Eds.; Springer International Publishing: Basel, Switzerland, 2017; Volume 404, pp. 1–44. [Google Scholar]
- Kratochvilova, L.; Kharkevich, K.; Slama, P. TNF-alpha and IL-10 are Produced by Leukocytes During the Experimental Inflammatory Response of Bovine Mammary Gland Induced by Peptidoglycan; Mendel University: Brno, Czech Republic, 2018; pp. 376–379. [Google Scholar]
- Kharkevich, K.; Kratochvilova, L.; Slama, P. Transforming Growth Factor Beta 1 Production During Inflammatory Response of Mammary Gland Induced by Peptidoglycan; Mendel University: Brno, Czech Republic, 2018; pp. 366–369. [Google Scholar]
- Zhang, W.Y.; Li, X.Z.; Xu, T.; Ma, M.R.; Zhang, Y.; Gao, M.Q. Inflammatory responses of stromal fibroblasts to inflammatory epithelial cells are involved in the pathogenesis of bovine mastitis. Exp. Cell Res. 2016, 349, 45–52. [Google Scholar] [CrossRef]
- Sartori, C.; Boss, R.; Bodmer, M.; Leuenberger, A.; Ivanovic, I.; Graber, H.U. Sanitation of Staphylococcus aureus genotype B-positive dairy herds: A field study. J. Dairy Sci. 2018, 101, 6897–6914. [Google Scholar] [CrossRef]
- Vangan, N.; Cao, Y.F.; Jia, X.Y.; Bao, W.L.; Wang, Y.F.; He, Q.; Binderiya, U.; Feng, X.; Li, T.T.; Hao, H.F.; et al. mTORC1 mediates peptidoglycan induced inflammatory cytokines expression and NF-kappa B activation in macrophages. Microb. Pathog. 2016, 99, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Zingarelli, B. Peptidoglycan is an important pathogenic factor of the inflammatory response in sepsis. Crit. Care Med. 2004, 32, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Iyer, J.K.; Khurana, T.; Langer, M.; West, C.M.; Ballard, J.D.; Metcalf, J.P.; Merkel, T.J.; Coggeshall, K.M. Inflammatory cytokine response to bacillus anthracis peptidoglycan requires phagocytosis and lysosomal trafficking. Infect. Immun. 2010, 78, 2418–2428. [Google Scholar] [CrossRef] [PubMed]
- Boveri, M.; Kinsner, A.; Berezowski, V.; Lenfant, A.M.; Draing, C.; Cecchelli, R.; Dehouck, M.P.; Hartung, T.; Prieto, P.; Bal-Price, A. Highly purified lipoteichoic acid from gram-positive bacteria induces In Vitro blood-brain barrier disruption through glia activation: Role of pro-inflammatory cytokines and nitric oxide. Neuroscience 2006, 137, 1193–1209. [Google Scholar] [CrossRef]
- Wang, J.E.; Dahle, M.K.; McDonald, M.; Foster, S.J.; Aasen, A.O.; Thiemermann, C. Peptidoglycan and lipoteichoic acid in gram-positive bacterial sepsis: Receptors, signal transduction, biological effects, and synergism. Shock 2003, 20, 402–414. [Google Scholar] [CrossRef]
- Kang, S.S.; Sim, J.R.; Yun, C.H.; Han, S.H. Lipoteichoic acids as a major virulence factor causing inflammatory responses via Toll-like receptor 2. Arch. Pharmacal Res. 2016, 39, 1519–1529. [Google Scholar] [CrossRef]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Chen, T.P.; Li, E. Structure and function of eukaryotic DNA methyltransferases. In Stem Cells in Development and Disease; Schatten, G.P., Ed.; Elsevier: Amsterdam, The Netherlands, 2004; Volume 60, pp. 55–89. [Google Scholar]
- Yen, C.; Huang, H.; Shu, C.; Hou, M.; Yuan, S.F.; Wang, H.; Chang, Y.; Farooqi, A.A.; Tang, J.; Chang, H. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 2016, 373, 185–192. [Google Scholar] [CrossRef]
- Xu, F.; Mao, C.; Ding, Y.; Rui, C.; Wu, L.; Shi, A.; Zhang, H.; Zhang, L.; Xu, Z. Molecular and enzymatic profiles of mammalian DNA methyltransferases: Structures and targets for drugs. Curr. Med. Chem. 2010, 17, 4052–4071. [Google Scholar] [CrossRef]
- Low, D.; Mizoguchi, A.; Mizoguchi, E. DNA methylation in inflammatory bowel disease and beyond. World J. Gastroenterol. 2013, 19, 5238–5249. [Google Scholar] [CrossRef]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer t cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, Y.L.; Yang, T.; Sahay, B.; Mohamadzadeh, M. Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus. Gut Microbes 2013, 4, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Shuto, T.; Furuta, T.; Oba, M.; Xu, H.D.; Li, J.D.; Cheung, J.; Gruenert, D.C.; Uehara, A.; Suico, M.A.; Okiyoneda, T.; et al. Promoter hypomethylation of Toll-like receptor-2 gene is associated with increased proinflammatory response toward bacterial peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB J. 2006, 20, 782. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Deckert, J.; Struhl, K. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 2001, 21, 2726–2735. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Tagai, C.; Shiraishi, T.; Miyaji, K.; Iwamuro, S. Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against Gram-positive Staphylococcus aureus. Peptides 2013, 48, 75–82. [Google Scholar] [CrossRef]
- Kim, T.; Yoon, J.; Cho, H.S.; Lee, W.B.; Kim, J.; Song, Y.H.; Kim, S.N.; Yoon, J.H.; Kim-Ha, J.; Kim, Y.J. Downregulation of lipopolysaccharide response in drosophila by negative crosstalk between the AP1 and NF-kappa B signaling modules. Nat. Immunol. 2005, 6, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.N.; Jeong, C.H.; Seo, H.G.; Han, S.G. Moringa extract attenuates inflammatory responses and increases gene expression of casein in bovine mammary epithelial cells. Animals 2019, 9, 391. [Google Scholar] [CrossRef]
- Wu, T.; Wang, C.; Ding, L.; Shen, Y.; Cui, H.; Wang, M.; Wang, H. Arginine relieves the inflammatory response and enhances the casein expression in bovine mammary epithelial cells induced by lipopolysaccharide. Mediators Inflamm. 2016, 9, 9618795. [Google Scholar] [CrossRef]
- Zhang, K.; Chang, G.; Xu, T.; Xu, L.; Guo, J.; Jin, D.; Shen, X. Lipopolysaccharide derived from the digestive tract activates inflammatory gene expression and inhibits casein synthesis in the mammary glands of lactating dairy cows. Oncotarget 2016, 7, 9652–9665. [Google Scholar] [CrossRef]
- Huynh, H.T.; Robitaille, G.; Turner, J.D. Establishment of bovine mammary epithelial-cells (mac-t)—An In Vitro model for bovine lactation. Exp. Cell Res. 1991, 197, 191–199. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Chen, J.; Sun, Y.; Dong, X.; Wang, Z.; Chen, J.; Dong, G. PGN and LTA from Staphylococcus aureus Induced Inflammation and Decreased Lactation through Regulating DNA Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells. Toxins 2020, 12, 238. https://doi.org/10.3390/toxins12040238
Wu Y, Chen J, Sun Y, Dong X, Wang Z, Chen J, Dong G. PGN and LTA from Staphylococcus aureus Induced Inflammation and Decreased Lactation through Regulating DNA Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells. Toxins. 2020; 12(4):238. https://doi.org/10.3390/toxins12040238
Chicago/Turabian StyleWu, Yongjiang, Jingbo Chen, Yawang Sun, Xianwen Dong, Zili Wang, Juncai Chen, and Guozhong Dong. 2020. "PGN and LTA from Staphylococcus aureus Induced Inflammation and Decreased Lactation through Regulating DNA Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells" Toxins 12, no. 4: 238. https://doi.org/10.3390/toxins12040238
APA StyleWu, Y., Chen, J., Sun, Y., Dong, X., Wang, Z., Chen, J., & Dong, G. (2020). PGN and LTA from Staphylococcus aureus Induced Inflammation and Decreased Lactation through Regulating DNA Methylation and Histone H3 Acetylation in Bovine Mammary Epithelial Cells. Toxins, 12(4), 238. https://doi.org/10.3390/toxins12040238




