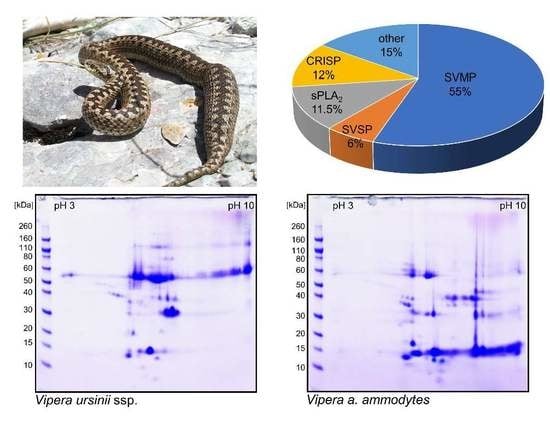
Biological Activities and Proteomic Profile of the Venom of Vipera ursinii ssp., a very Rare Karst Viper from Croatia
Abstract
1. Introduction
2. Results
2.1. Toxinological Characterisation of V. ursinii ssp. Venom
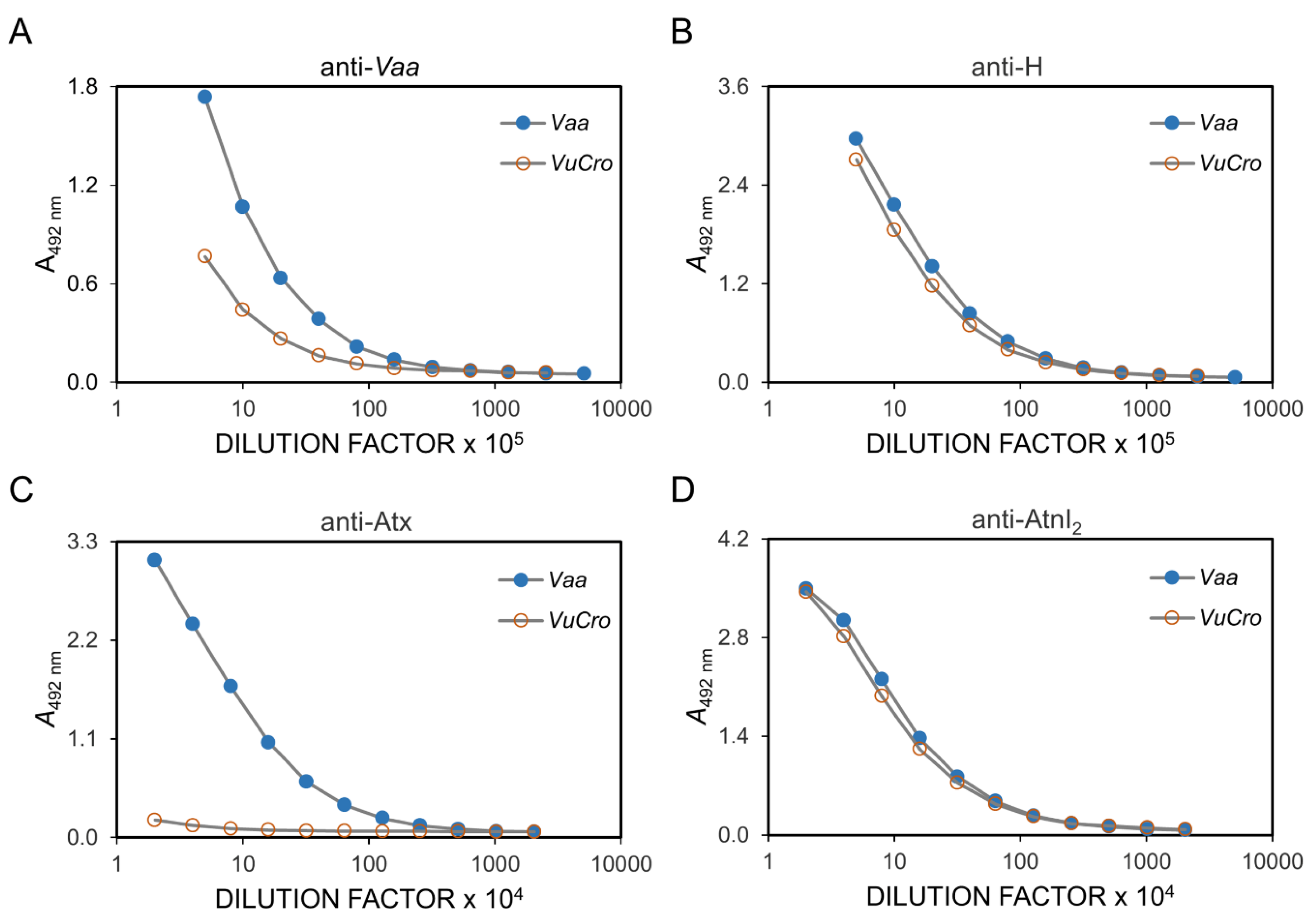
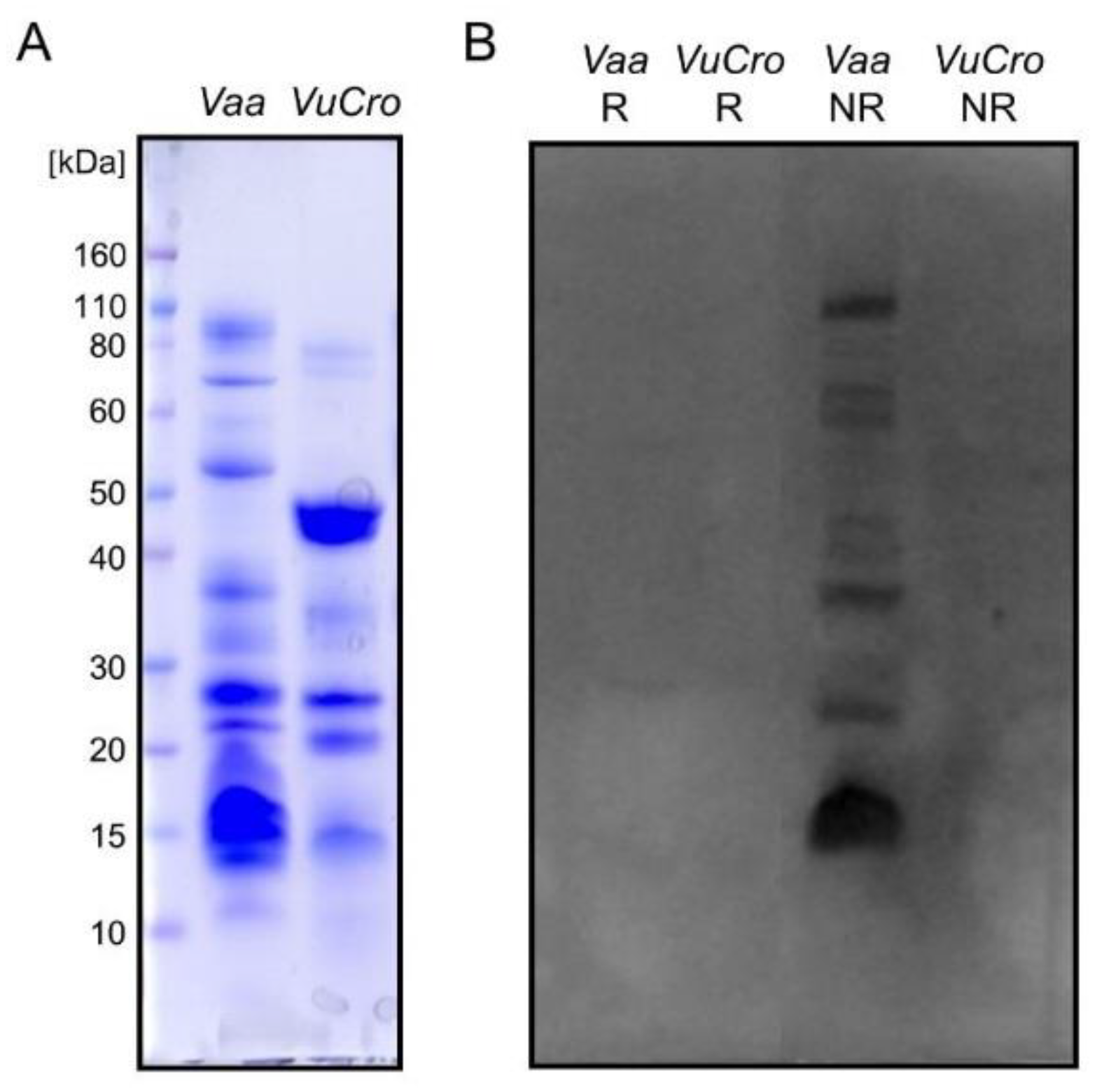
2.2. Immunological Cross-Reactivity between V. ursinii ssp. and V. a. ammodytes Venoms
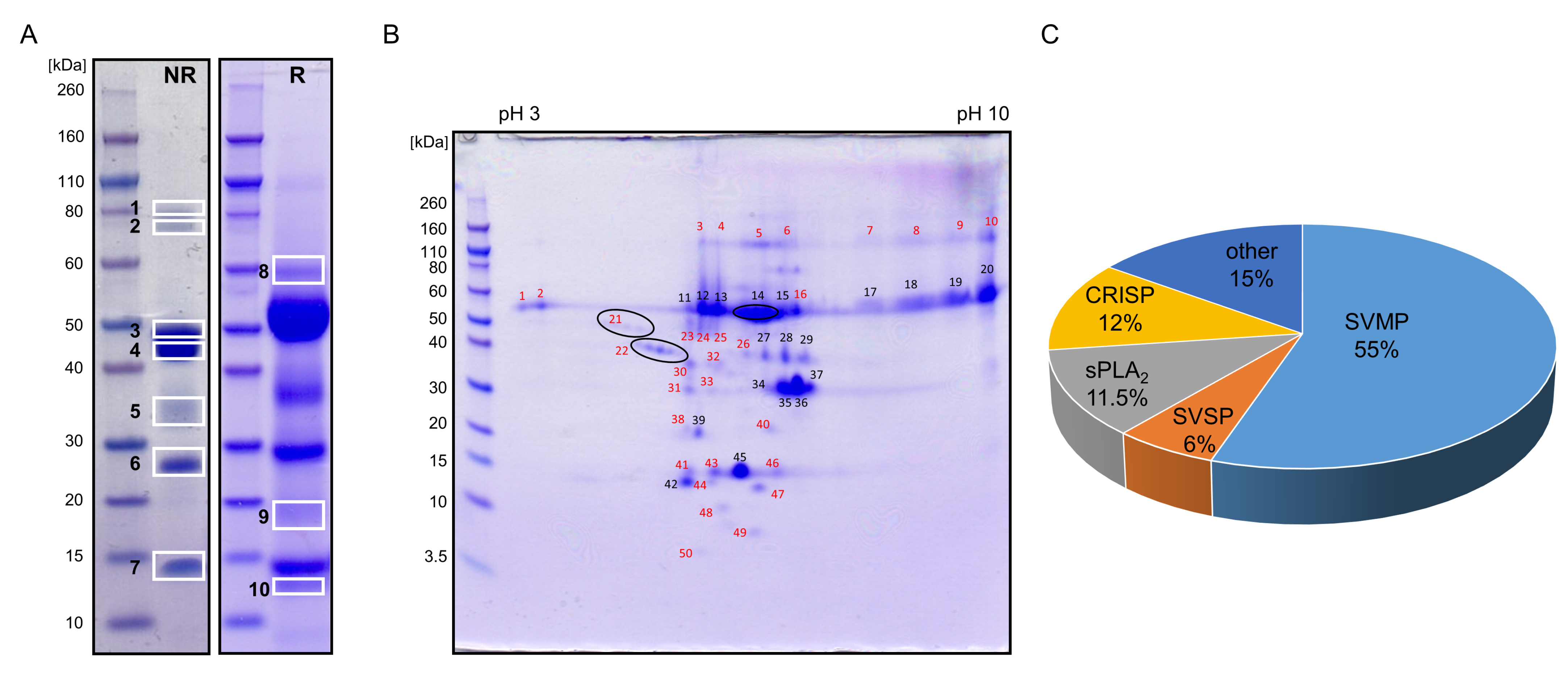
2.3. Proteomics of V. ursinii ssp. Venom and Its Comparison with V. a. ammodytes Venom
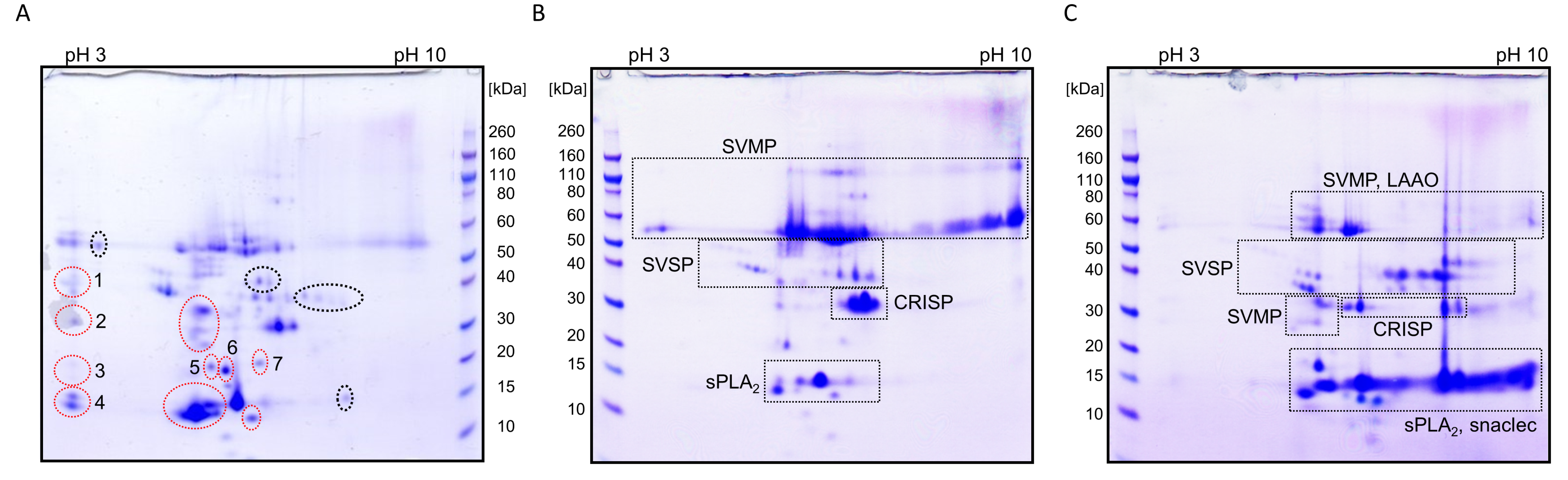
2.3.1. Proteome of V. ursinii ssp. Venom
2.3.2. Comparative Analysis of V. ursinii ssp. and V. a. ammodytes Venoms
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagents and Chemicals
5.2. Snake Venoms and Antivenoms
5.3. Animals for In Vivo Assays
5.4. Assay of Lethal Toxicity in Mice
5.5. Assay of Lethal Toxicity in Crickets
5.6. Assay for Assessing Haemorrhagic Activity
5.7. Immunochemical Assays
5.7.1. ELISA Assay
5.7.2. One-Dimensional SDS-PAGE (1-DE) and Western Blot
5.8. Two-Dimensional Gel Electrophoresis (2-DE)
5.9. Mass Spectrometry (MS) Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uetz, P.; Freed, P.; Hošek, J. The Reptile Database. Available online: http://www.reptile-database.org/ (accessed on 2 December 2019).
- Mizsei, E.; Jablonski, D.; Roussos, S.A.; Dimaki, M.; Ioannidis, Y.; Nilson, G.; Nagy, Z.T. Nuclear markers support the mitochondrial phylogeny of Vipera ursinii-renardi complex (Squamata: Viperidae) and species status for the Greek meadow viper. Zootaxa 2017, 4227. [Google Scholar] [CrossRef] [PubMed]
- Council of Europe, Convention on the Conservation of European Wildlife and Natural Habitats: Action Plan for the Conservation of the Meadow Viper (Vipera ursinii) in Europe, T-PVS/Inf, 2005 1 Revised. Available online: https://rm.coe.int/16807462b0 (accessed on 2 December 2019).
- Ferchaud, A.L.; Ursenbacher, S.; Cheylan, M.; Luiselli, L.; Jelić, D.; Halpern, B.; Major, Á.; Kotenko, T.; Keyan, N.; Behrooz, R.; et al. Phylogeography of the Vipera ursinii complex (Viperidae): Mitochondrial markers reveal an east-west disjunction in the Palaearctic region. J. Biogeogr. 2012, 39, 1836–1847. [Google Scholar] [CrossRef]
- Jelić, D.; Ajtić, R.; Sterijovski, B.; Crnobrnia-Isailović, J.; Lelo, S.; Tomović, L. Distribution of the genus Vipera in the western and central Balkans (Squamata: Serpentes: Viperidae). Herpetozoa 2013, 25, 109–132. [Google Scholar]
- Jelić, D.; Ajtić, R.; Sterijovski, B.; Crnobrnja-Isailović, J.; Lelo, S.; Tomović, L. Legal status and assessment of conservation threats to vipers (Reptilia: Squamata: Viperidae) of the Western and Central Balkans. Herpetol. Conserv. Biol. 2013, 8, 764–770. [Google Scholar]
- Starkov, V.G.; Osipov, A.V.; Utkin, Y.N. Toxicity of venoms from vipers of Pelias group to crickets Gryllus assimilis and its relation to snake entomophagy. Toxicon 2007, 49, 995–1001. [Google Scholar] [CrossRef]
- Krescák, L.; Zacher, G.; Malina, Z. Clinical picture of envenoming with the Meadow Viper (Vipera (Acridophaga) ursinii). Clin. Toxicol. 2011, 49, 13–20. [Google Scholar]
- Gvoždík, V.; Jandzik, D.; Cordos, B.; Rehák, I.; Kotlík, P. A mitochondrial DNA phylogeny of the endangered vipers of the Vipera ursinii complex. Mol. Phylogenet. Evol. 2012, 62, 1019–1024. [Google Scholar] [CrossRef]
- Ferchaud, A.-L.; Lyet, A.; Cheylan, M.; Arnal, V.; Baron, J.-P.; Montgelard, C.; Ursenbacher, S. High genetic differentiation among French populations of the Orsini’s viper (Vipera ursinii ursinii) based on mitochondrial and microsatellite data: Implications for conservation management. J. Hered. 2011, 102, 67–78. [Google Scholar] [CrossRef]
- Mebs, D.; Langelüddeke, T. European viper venoms: Haemorrhagic and myotoxic activities. Toxicon 1992, 30, 1303–1306. [Google Scholar] [CrossRef]
- Jan, V.M.; Guillemin, I.; Robbe-Vincent, A.; Choumet, V. Phospholipase A2 diversity and polymorphism in European viper venoms: Paradoxical molecular evolution in Viperinae. Toxicon 2007, 50, 1140–1161. [Google Scholar] [CrossRef]
- Halassy, B.; Brgles, M.; Habjanec, L.; Balija, M.L.; Kurtović, T.; Marchetti-Deschmann, M.; Križaj, I.; Allmaier, G. Intraspecies variability in Vipera ammodytes ammodytes venom related to its toxicity and immunogenic potential. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Halassy, B.; Habjanec, L.; Balija, M.L.; Kurtović, T.; Brgles, M.; Križaj, I. Ammodytoxin content of Vipera ammodytes ammodytes venom as a prognostic factor for venom immunogenicity. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010, 151, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, A.; Sajevic, T.; Pungerčar, J.; Križaj, I. Comprehensive study of the proteome and transcriptome of the venom of the most venomous european viper: Discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019, 18, 2287–2309. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.P.; Barlow, A.; Wüster, W. Venom lethality and diet: Differential responses of natural prey and model organisms to the venom of the saw-scaled vipers (Echis). Toxicon 2012, 59, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Sixberry, N.M.; Heyborne, W.H.; Fritts, T. Venom of the brown treesnake, Boiga irregularis: Ontogenetic shifts and taxa-specific toxicity. Toxicon 2006, 47, 537–548. [Google Scholar] [CrossRef]
- Calvete, J.J.; Sanz, L.; Angulo, Y.; Lomonte, B.; Gutiérrez, J.M. Venoms, venomics, antivenomics. FEBS Lett. 2009, 583, 1736–1743. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Latinović, Z.; Leonardi, A.; Šribar, J.; Sajevic, T.; Žužek, M.C.; Frangež, R.; Halassy, B.; Trampuš-Bakija, A.; Pungerčar, J.; Križaj, I.; et al. Venomics of Vipera berus berus to explain differences in pathology elicited by Vipera ammodytes ammodytes envenomation: Therapeutic implications. J. Proteom. 2016, 146, 34–47. [Google Scholar] [CrossRef]
- Sajevic, T.; Leonardi, A.; Križaj, I. Haemostatically active proteins in snake venoms. Toxicon 2011, 57, 627–645. [Google Scholar] [CrossRef]
- Chellapandi, P. Structural, functional and therapeutic aspects of snake venom metalloproteinases. Mini Rev. Org. Chem. 2014, 11, 28–44. [Google Scholar] [CrossRef]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins 2016, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Sajevic, T.; Leonardi, A.; Kovačič, L.; Lang-Balija, M.; Kurtović, T.; Pungerčar, J.; Halassy, B.; Trampuš-Bakija, A.; Križaj, I. VaH3, one of the principal hemorrhagins in Vipera ammodytes ammodytes venom, is a homodimeric P-IIIc metalloproteinase. Biochimie 2013, 95, 1158–1170. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, J.; Kool, J.; Harrison, R.A.; Casewell, N.R. Haemotoxic snake venoms: Their functional activity, impact on snakebite victims and pharmaceutical promise. Br. J. Haematol. 2017, 177, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, A.; Fox, J.W.; Trampuš-Bakija, A.; Križaj, I. Two coagulation factor X activators from Vipera a. ammodytes venom with potential to treat patients with dysfunctional factors IXa or VIIa. Toxicon 2008, 52, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Takeya, H.; Nishida, S.; Miyata, T.; Kawada, S.; Saisaka, Y.; Morita, T.; Iwanaga, S. Coagulation factor X activating enzyme from Russell’s viper venom (RVV-X). A novel metalloproteinase with disintegrin (platelet aggregation inhibitor)-like and C-type lectin-like domains. J. Biol. Chem. 1992, 267, 14109–14117. [Google Scholar]
- Tokunaga, F.; Nagasawa, K.; Tamura, S.; Miyata, T.; Iwanaga, S.; Kisiel, W. The factor V-activating enzyme (RVV-V) from Russell’s viper venom. Identification of isoproteins RVV-V alpha, -V beta, and -V gamma and their complete amino acid sequences. J. Biol. Chem. 1988, 263, 17471–17481. [Google Scholar]
- Segers, K.; Rosing, J.; Nicolaes, G.A.F. Structural models of the snake venom factor V activators from Daboia russelli and Daboia lebetina. Proteins 2006, 64, 968–984. [Google Scholar] [CrossRef]
- Latinović, Z.; Leonardi, A.; Kovačič, L.; Koh, C.; Šribar, J.; Bakija, A.; Venkateswarlu, D.; Kini, R.; Križaj, I. The first intrinsic tenase complex inhibitor with serine protease structure offers a new perspective in anticoagulant therapy. Thromb. Haemost. 2018, 118, 1713–1728. [Google Scholar] [CrossRef]
- de Queiroz, M.R.; de Sousa, B.B.; da Cunha Pereira, D.F.; Mamede, C.C.N.; Matias, M.S.; de Morais, N.C.G.; de Oliveira Costa, J.; de Oliveira, F. The role of platelets in hemostasis and the effects of snake venom toxins on platelet function. Toxicon 2017, 133, 33–47. [Google Scholar] [CrossRef]
- Komori, Y.; Nikai, T.; Sugihara, H. Comparative study of three phospholipase A2s from the venom of Vipera aspis. Comp. Biochem. Physiol. B 1990, 97, 507–514. [Google Scholar] [CrossRef]
- Tsai, I.-H.; Wang, Y.-M.; Cheng, A.C.; Starkov, V.; Osipov, A.; Nikitin, I.; Makarova, Y.; Ziganshin, R.; Utkin, Y. cDNA cloning, structural, and functional analyses of venom phospholipases A₂ and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon 2011, 57, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Križaj, I. Ammodytoxin: A window into understanding presynaptic toxicity of secreted phospholipases A2 and more. Toxicon 2011, 58, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.; Yamazaki, Y.; Hyodo, F.; Sugiyama, Y.; Nozaki, M.; Morita, T. Structural divergence of cysteine-rich secretory proteins in snake venoms. J. Biochem. 2009, 145, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Lodovicho, M.E.; Costa, T.R.; Bernardes, C.P.; Menaldo, D.L.; Zoccal, K.F.; Carone, S.E.; Rosa, J.C.; Pucca, M.B.; Cerni, F.A.; Arantes, E.C.; et al. Investigating possible biological targets of Bj-CRP, the first cysteine-rich secretory protein (CRISP) isolated from Bothrops jararaca snake venom. Toxicol. Lett. 2016, 265, 156–169. [Google Scholar] [CrossRef]
- Wang, Y.L.; Kuo, J.H.; Lee, S.C.; Liu, J.S.; Hsieh, Y.C.; Shih, Y.T.; Chen, C.J.; Chiu, J.J.; Wu, W.G. Cobra CRISP functions as an inflammatory modulator via a novel Zn 2+- and heparan sulfate-dependent transcriptional regulation of endothelial cell adhesion molecules. J. Biol. Chem. 2010, 285, 37872–37883. [Google Scholar] [CrossRef]
- Adade, C.M.; Carvalho, A.L.O.; Tomaz, M.A.; Costa, T.F.R.; Godinho, J.L.; Melo, P.A.; Lima, A.P.C.A.; Rodrigues, J.C.F.; Zingali, R.B.; Souto-Padrón, T.; et al. Crovirin, a snake venom cysteine-rich secretory protein (CRISP) with promising activity against Trypanosomes and Leishmania. PLoS Negl. Trop. Dis. 2014, 8, e3252. [Google Scholar] [CrossRef]
- Lecht, S.; Chiaverelli, R.A.; Gerstenhaber, J.; Calvete, J.J.; Lazarovici, P.; Casewell, N.R.; Harrison, R.; Lelkes, P.I.; Marcinkiewicz, C. Anti-angiogenic activities of snake venom CRISP isolated from Echis carinatus sochureki. Biochim. Biophys. Acta 2015, 1850, 1169–1179. [Google Scholar] [CrossRef]
- Trummal, K.; Tõnismägi, K.; Paalme, V.; Järvekülg, L.; Siigur, J.; Siigur, E. Molecular diversity of snake venom nerve growth factors. Toxicon 2011, 58, 363–368. [Google Scholar] [CrossRef]
- Ritonja, A.; Turk, V.; Gubenšek, F. Serine proteinase inhibitors from Vipera ammodytes venom. Isolation and kinetic studies. Eur. J. Biochem. 1983, 133, 427–432. [Google Scholar] [CrossRef]
- Thakur, R.; Mukherjee, A.K. Pathophysiological significance and therapeutic applications of snake venom protease inhibitors. Toxicon 2017, 131, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Brgles, M.; Kurtović, T.; Kovačič, L.; Križaj, I.; Barut, M.; Balija, M.L.; Allmaier, G.; Marchetti-Deschmann, M.; Halassy, B. Identification of proteins interacting with ammodytoxins in Vipera ammodytes ammodytes venom by immuno-affinity chromatography. Anal. Bioanal. Chem. 2014, 406, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.L.; Sanz, L.; Chiucchi, J.E.; Farrell, T.M.; Calvete, J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri). J. Proteom. 2011, 74, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Nakashima, K.; Nobuhisa, I.; Deshimaru, M.; Shimohigashi, Y.; Fukumaki, Y.; Sakaki, Y.; Hattori, S.; Ohno, M. Accelerated evolution of snake venom phospholipase A2 isozymes for acquisition of diverse physiological functions. Toxicon 1996, 34, 1229–1236. [Google Scholar] [CrossRef]
- Aird, S.D.; Aggarwal, S.; Villar-Briones, A.; Tin, M.M.-Y.; Terada, K.; Mikheyev, A.S. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 2015, 16, 647. [Google Scholar] [CrossRef]
- Halassy, B.; Habjanec, L.; Brgles, M.; Balija, M.L.; Leonardi, A.; Kovačič, L.; Prijatelj, P.; Tomašić, J.; Križaj, I. The role of antibodies specific for toxic sPLA2s and haemorrhagins in neutralizing potential of antisera raised against Vipera ammodytes ammodytes venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 148, 178–183. [Google Scholar] [CrossRef]
- Kurtović, T.; Leonardi, A.; Lang Balija, M.; Brgles, M.; Habjanec, L.; Križaj, I.; Halassy, B. The standard mouse assay of anti-venom quality does not measure antibodies neutralising the haemorrhagic activity of Vipera ammodytes venom. Toxicon 2012, 59, 709–717. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Theakston, R.D.G.; Reid, H.A. Development of simple standard assay procedures for the characterization of snake venoms. Bull. World Health Organ. 1983, 61, 949–956. [Google Scholar]
- Lang Balija, M.; Vrdoljak, A.; Habjanec, L.; Dojnović, B.; Halassy, B.; Vranešić, B.; Tomašić, J. The variability of Vipera ammodytes ammodytes venoms from Croatia—Biochemical properties and biological activity. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2005, 140, 257–263. [Google Scholar] [CrossRef]
- Kurtović, T.; Lang Balija, M.; Ayvazyan, N.; Halassy, B. Paraspecificity of Vipera a. ammodytes-specific antivenom towards Montivipera raddei and Macrovipera lebetina obtusa venoms. Toxicon 2014, 78, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, A.; Biass, D.; Kordiš, D.; Stöcklin, R.; Favreau, P.; Križaj, I. Conus consors snail venom proteomics proposes functions, pathways, and novel families involved in its venomic system. J. Proteome Res. 2012, 11, 5046–5058. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, A.; Sajevic, T.; Kovačič, L.; Pungerčar, J.; Lang Balija, M.; Halassy, B.; Trampuš Bakija, A.; Križaj, I. Hemorrhagin VaH4, a covalent heterodimeric P-III metalloproteinase from Vipera ammodytes ammodytes with a potential antitumour activity. Toxicon 2014, 77, 141–155. [Google Scholar] [CrossRef] [PubMed]

| VuCro Venom | Vaa Venom | |
|---|---|---|
| Lethal Toxicity | ||
| LD50 in mice (µg) | 37.0 ± 0.1 (n = 2) | 4.4–13.7 * |
| ** Mass-normalized LD50 in mice (µg/g) | 1.94 | 0.47 |
| LD50 in crickets (µg) | 7.1 ± 0.4 (n = 2) | 38.7 ± 3.3 (n = 2) |
| ** Mass-normalized LD50 in crickets (µg/g) | 9.8 | 53.4 |
| Haemorrhagic Activity | ||
| MHD in rats (µg) | 34.1 ± 4.8 (n = 4) | 21.6–42.8* |
| Higher Abundance | Protein Family |
|---|---|
| AtnI1 (B) isoform (Vu) | sPLA2 |
| Vur-PL2 | sPLA2 |
| AtnI2 (D) isoform (Vu) | sPLA2 |
| RVV-X light chain 2 (Daboia siamensis) | snaclec |
| Vaa-SP-3 * | SVSP |
| Vaa-SP-4 | SVSP |
| Vaa-SPH-1 (Vaa) | SVSP |
| VNGF (Vu) | VNGF |
| Vur-KIn (V. renardi) | KSPI |
| Lower Abundance | |
| Metalloproteinases | SVMP |
| Vaa-CRISP-1 (Vaa) | CRISP |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang Balija, M.; Leonardi, A.; Brgles, M.; Sviben, D.; Kurtović, T.; Halassy, B.; Križaj, I. Biological Activities and Proteomic Profile of the Venom of Vipera ursinii ssp., a very Rare Karst Viper from Croatia. Toxins 2020, 12, 187. https://doi.org/10.3390/toxins12030187
Lang Balija M, Leonardi A, Brgles M, Sviben D, Kurtović T, Halassy B, Križaj I. Biological Activities and Proteomic Profile of the Venom of Vipera ursinii ssp., a very Rare Karst Viper from Croatia. Toxins. 2020; 12(3):187. https://doi.org/10.3390/toxins12030187
Chicago/Turabian StyleLang Balija, Maja, Adrijana Leonardi, Marija Brgles, Dora Sviben, Tihana Kurtović, Beata Halassy, and Igor Križaj. 2020. "Biological Activities and Proteomic Profile of the Venom of Vipera ursinii ssp., a very Rare Karst Viper from Croatia" Toxins 12, no. 3: 187. https://doi.org/10.3390/toxins12030187
APA StyleLang Balija, M., Leonardi, A., Brgles, M., Sviben, D., Kurtović, T., Halassy, B., & Križaj, I. (2020). Biological Activities and Proteomic Profile of the Venom of Vipera ursinii ssp., a very Rare Karst Viper from Croatia. Toxins, 12(3), 187. https://doi.org/10.3390/toxins12030187