Occurrence of Methicillin-Resistant Staphylococcus spp. on Brazilian Dairy Farms that Produce Unpasteurized Cheese
Abstract
1. Introduction
2. Results
3. Discussion
4. Material and Methods
4.1. Staphylococcus spp. Sampling, Isolation and Identification
4.2. Detection of Enterotoxin Genes
4.3. Antimicrobial Susceptibility Test of MRS Isolates
4.4. Sequencing
4.5. RAPD Markers
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Duijkeren, E.; Box, A.; Heck, M.; Wannet, W.; Fluit, A. Methicillin-resistant staphylococci isolated from animals. Veter. Microbiol. 2004, 103, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Juhász-Kaszanyitzky, É.; Jánosi, S.; Somogyi, P.; Dán, Á.; Van Bloois, L.V.; Van Duijkeren, E.; Wagenaar, J.A. MRSA Transmission between Cows and Humans. Emerg. Infect. Dis. 2007, 13, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Fisher, E.A.; Paterson, G.K. Prevalence and characterization of methicillin-resistant staphylococci from bovine bulk tank milk in England and Wales. J. Glob. Antimicrob. Resist. 2020, 22, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Klibi, A.; Maaroufi, A.; Torres, C.; Jouini, A. Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Schnitt, A.; Tenhagen, B.A. Risk Factors for the Occurrence of Methicillin-Resistant Staphylococcus aureus in Dairy Herds: An Update. Foodborne Pathog. Dis. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans, J.; Van Leeuwen, W.; Goessens, W.; Hollis, R.; Messer, S.; Herwaldt, L.A.; Bruining, H.; Heck, M.; Rost, J. Food-initiated outbreak of methicillin-resistant Staphylococcus aureus analyzed by pheno- and genotyping. J. Clin. Microbiol. 1995, 33, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Aires-De-Sousa, M. Methicillin-resistant Staphylococcus aureus among animals: Current overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Siiriken, B.; Yildirim, T.; Güney, A.K.; Erol, I.; Durupinar, B. Prevalence and Molecular Characterization of Methicillin-Resistant Staphylococcus aureus in Foods of Animal Origin, Turkey. J. Food Prot. 2016, 79, 1990–1994. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y.; Djoko, C.F. Mannitol-fermenting methicillin-resistant staphylococci (MRS) in pig abattoirs in Cameroon and South Africa: A serious food safety threat. Int. J. Food Microbiol. 2018, 285, 50–60. [Google Scholar] [CrossRef]
- Soares, M.J.S.; Tokumaru-Miyazaki, N.H.; Noleto, A.L.S.; Figueiredo, A.M.S.; John, S.F.; Hillier, V.F.; Handley, P.S.; Derrick, M.R. Enterotoxin production by Staphylococcus aureus clones and detection of Brazilian epidemic MRSA clone (III::B:A) among isolates from food handlers. J. Med. Microbiol. 1997, 46, 214–221. [Google Scholar] [CrossRef]
- Adame-Gómez, R.; Toribio-Jimenez, J.; Vences-Velázquez, A.; Rodríguez-Bataz, E.; Dionisio, M.C.S.; Ramírez-Peralta, A. Methicillin-Resistant Staphylococcus aureus (MRSA) in Artisanal Cheeses in México. Int. J. Microbiol. 2018, 2018, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Saka, E.; Gulel, G.T. Detection of Enterotoxin Genes and Methicillin-Resistance in Staphylococcus aureus Isolated from Water Buffalo Milk and Dairy Products. J. Food Sci. 2018, 83, 1716–1722. [Google Scholar] [CrossRef]
- Sergelidis, D.; Angelidis, A.S. Methicillin-resistant Staphylococcus aureus (MRSA): A controversial food-borne pathogen. Lett. Appl. Microbiol. 2017, 64, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Podkowik, M.; Park, J.; Seo, K.S.; Bystroń, J.; Bania, J. Enterotoxigenic potential of coagulase-negative staphylococci. Int. J. Food Microbiol. 2013, 163, 34–40. [Google Scholar] [CrossRef]
- Guimarães, F.; Manzi, M.; Joaquim, S.F.; Richini-Pereira, V.B.; Langoni, H. Short communication: Outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-associated mastitis in a closed dairy herd. J. Dairy Sci. 2017, 100, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Oppegaard, H.; Steinum, T.M.; Wasteson, Y. Horizontal Transfer of a Multi-Drug Resistance Plasmid between Coliform Bacteria of Human and Bovine Origin in a Farm Environment. Appl. Environ. Microbiol. 2001, 67, 3732–3734. [Google Scholar] [CrossRef]
- Yoon, Y.; Lee, S.; Choi, K.H. Microbial benefits and risks of raw milk cheese. Food Control 2016, 63, 201–215. [Google Scholar] [CrossRef]
- Normanno, G.; Corrente, M.; La Salandra, G.; Dambrosio, A.; Quaglia, N.C.; Parisi, A.; Greco, G.; Bellacicco, A.; Virgilio, S.; Celano, G.V. Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol. 2007, 117, 219–222. [Google Scholar] [CrossRef]
- Vanderhaeghen, W.; Cerpentier, T.; Adriaensen, C.; Vicca, J.; Hermans, K.; Butaye, P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Veter. Microbiol. 2010, 144, 166–171. [Google Scholar] [CrossRef]
- Helak, I.; Daczkowska-Kozon, E.; Dłubała, A. Short communication: Enterotoxigenic potential of coagulase-negative staphylococci isolated from bovine milk in Poland. J. Dairy Sci. 2020, 103, 3076–3081. [Google Scholar] [CrossRef]
- Kim, S.J.; Moon, D.C.; Park, S.C.; Kang, H.Y.; Na, S.H.; Lim, S.K. Antimicrobial resistance and genetic characterization of coagulase-negative staphylococci from bovine mastitis milk samples in Korea. J. Dairy Sci. 2019, 102, 11439–11448. [Google Scholar] [CrossRef] [PubMed]
- Goetz, C.; Tremblay, Y.D.; Lamarche, D.; Blondeau, A.; Gaudreau, A.M.; Labrie, J.; Malouin, F.; Jacques, M. Coagulase-negative staphylococci species affect biofilm formation of other coagulase-negative and coagulase-positive staphylococci. J. Dairy Sci. 2017, 100, 6454–6464. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.; Júnior, W.D.L.; Abley, M.; Gebreyes, W.A.; Oliveira, C.J. Antimicrobial resistance and genotypic relatedness of environmental staphylococci in semi-extensive dairy farms. Veter Anim. Sci. 2018, 6, 103–106. [Google Scholar] [CrossRef]
- Dhaouadi, S.; Soufi, L.; Campanile, F.; Dhaouadi, F.; Sociale, M.; Lazzaro, L.; Cherif, M.; Stefani, S.; Elandoulsi, R.B. Prevalence of methicillin-resistant and susceptible coagulase-negative staphylococci with the first detection of mecC gene among cows, humans and manure in Tunisia. Int. J. Antimicrob. Agents 2020, 55, 105826. [Google Scholar] [CrossRef] [PubMed]
- Pizauro, L.J.L.; de Almeida, C.C.; Soltes, G.A.; Slavic, D.; De Ávila, F.A.; Zafalon, L.F.; MacInnes, J.I. Short communication: Detection of antibiotic resistance, mecA, and virulence genes in coagulase-negative Staphylococcus spp. from buffalo milk and the milking environment. J. Dairy Sci. 2019, 102, 11459–11464. [Google Scholar] [CrossRef]
- Zafalon, L.F.; Cunha, M.L.R.S.; Brandão, H.D.M.; Mosqueira, V.C.F.; Santana, R.C.; Júnior, W.B.; Martins, K.B.; Pilon, L.E. Relationship between virulence factor genes in coagulase-negative Staphylococcus spp. and failure of antimicrobial treatment of subclinical mastitis in sheep. Pesquisa Veterinária Bras. 2018, 38, 579–585. [Google Scholar] [CrossRef]
- Moon, J.S.; Lee, A.R.; Kang, H.M.; Lee, E.S.; Kim, M.N.; Paik, Y.; Park, Y.; Joo, Y.S.; Koo, H.C. Phenotypic and Genetic Antibiogram of Methicillin-Resistant Staphylococci Isolated from Bovine Mastitis in Korea. J. Dairy Sci. 2007, 90, 1176–1185. [Google Scholar] [CrossRef]
- Berge, A.C.; Baars, T. Raw milk producers with high levels of hygiene and safety. Epidemiol. Infect. 2020, 148, e14. [Google Scholar] [CrossRef]
- Al-Ashmawy, M.A.M.; Sallam, K.I.; Abd-ElGhany, S.M.; Elhadidy, M.; Tamura, T. Prevalence, Molecular Characterization, and Antimicrobial Susceptibility of Methicillin-Resistant Staphylococcus aureus Isolated from Milk and Dairy Products. Foodborne Pathog. Dis. 2016, 13, 156–162. [Google Scholar] [CrossRef]
- Riva, A.; Borghi, E.; Cirasola, D.; Colmegna, S.; Borgo, F.C.; Amato, E.; Pontello, M.; Morace, G. Methicillin-Resistant Staphylococcus aureus in Raw Milk: Prevalence, SCCmec Typing, Enterotoxin Characterization, and Antimicrobial Resistance Patterns. J. Food Prot. 2015, 78, 1142–1146. [Google Scholar] [CrossRef]
- Shanehbandi, D.; Baradaran, B.; Sadigh-Eteghad, S.; Zarredar, H. Occurrence of Methicillin Resistant and Enterotoxigenic Staphylococcus aureusin Traditional Cheeses in the North West of Iran. ISRN Microbiol. 2014, 129580. [Google Scholar] [CrossRef]
- Momtaz, H.; Hafezi, L. Meticillin-resistant Staphylococcus aureus isolated from Iranian hospitals: Virulence factors and antibiotic resistance properties. Bosn. J. Basic Med Sci. 2014, 14, 219–226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kousta, M.; Mataragas, M.; Skandamis, P.; Drosinos, E.H. Prevalence and sources of cheese contamination with pathogens at farm and processing levels. Food Control 2010, 21, 805–815. [Google Scholar] [CrossRef]
- Aquino, G.D.V.; Maluta, R.P.; Avila, F.A. Prevalence of Methicillin-Resistant Staphylococci on a Farm: Staff can Harbour MRS When Animals Do Not. Zoonoses Public Health 2012, 59, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Keskimäki, M.; Eklund, M.; Pesonen, H.; Heiskanen, T.; Siitonen, A. EPEC, EAEC and STEC in stool specimens: Prevalence and molecular epidemiology of isolates. Diagn. Microbiol. Infect. Dis. 2001, 40, 151–156. [Google Scholar] [CrossRef]
- Pereira, V.; Lopes, C.; Castro, A.I.; Silva, J.; Gibbs, P.; Teixeira, P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009, 26, 278–282. [Google Scholar] [CrossRef]
- Morot-Bizot, S.C.; Talon, R.; Leroy, S. Development of a multiplex PCR for the identification of Staphylococcus genus and four staphylococcal species isolated from food. J. Appl. Microbiol. 2004, 97, 1087–1094. [Google Scholar] [CrossRef]
- Gortel, K.; Campbell, K.L.; Kakoma, I.; Whittem, T.; Schaeffer, D.J.; Weisiger, R.M. Methicillin resistance among staphylococci isolated from dogs. Am. J. Vet. Res. 1999, 60, 1526–1530. [Google Scholar]
- Paniagua-Contreras, G.; Sáinz-Espuñes, T.; Monroy-Pérez, E.; Rodríguez-Moctezuma, J.R.; Arenas-Aranda, D.; Negrete-Abascal, E.; Vaca, S. Virulence Markers in Staphylococcus aureus Strains Isolated from Hemodialysis Catheters of Mexican Patients. Adv. Microbiol. 2012, 2, 476–487. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- NCCLS. Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard. NCCLS Document M2-A8, 8th ed.; NCCLS: Wayne, PA, USA, 2000; ISBN 1-56238-485-6. [Google Scholar]
- Bag, S.; Saha, B.; Mehta, O.; Anbumani, D.; Kumar, N.; Dayal, M.; Pant, A.; Kumar, P.; Saxena, S.; Allin, K.H.; et al. An Improved Method for High Quality Metagenomics DNA Extraction from Human and Environmental Samples. Nature 2016, 6, 26775. [Google Scholar] [CrossRef] [PubMed]
- Euzeby, J.P. List of Bacterial Names with Standing in Nomenclature: A Folder Available on the Internet. Int. J. Syst. Microbiol. 1997, 47, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Yugueros, J.; Temprano, A.; Sánchez, M.; Luengo, J.M.; Naharro, G. Identification of Staphylococcus spp. by PCR-Restriction Fragment Length Polymorphism of gap Gene. J. Clin. Microbiol. 2001, 39, 3693–3695. [Google Scholar] [CrossRef][Green Version]
- Bergeron, M.; Dauwalder, O.; Gouy, M.; Freydière, A.-M.; Bes, M.; Meugnier, H.; Benito, Y.; Etienne, J.; Lina, G.; Vandenesch, F.; et al. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 343–354. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using PHRED II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Green, P. PHRAP Documentation. 1996. Available online: http://bozeman.mbt.washington.edu/phrap.docs/phrap.html (accessed on 10 August 2016).
- Gordon, D.; Abajian, C.; Green, P. Consed:A Graphical Tool for Sequence Finishing. Genome Res. 1998, 8, 195–202. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony. v.4.; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
| Farm | Sample | IsolateS (N) | Antimicrobial Drug | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CPM | CLO | ERI | PEN | OXA | SUT | CIP | |||
| B | Cheese handlers‘ hands | 3 | 3 (100) | 0 | 1 (33,33) | 3 (100) | 3 (100) | 3 (100) | 0 |
| Artisanal cheese | 4 | 2 (50) | 0 | 0 | 4 (100) | 4 (100) | 2 (50) | 0 | |
| C | Cheese handlers‘ hands | 5 | 2 (40) | 0 | 0 | 5 (100) | 5 (100) | 5 (100) | 5 (100) |
| Artisanal cheese | 4 | 0 | 3 (75) | 1 (25) | 4 (100) | 4 (100) | 3 (75) | 4 (100) | |
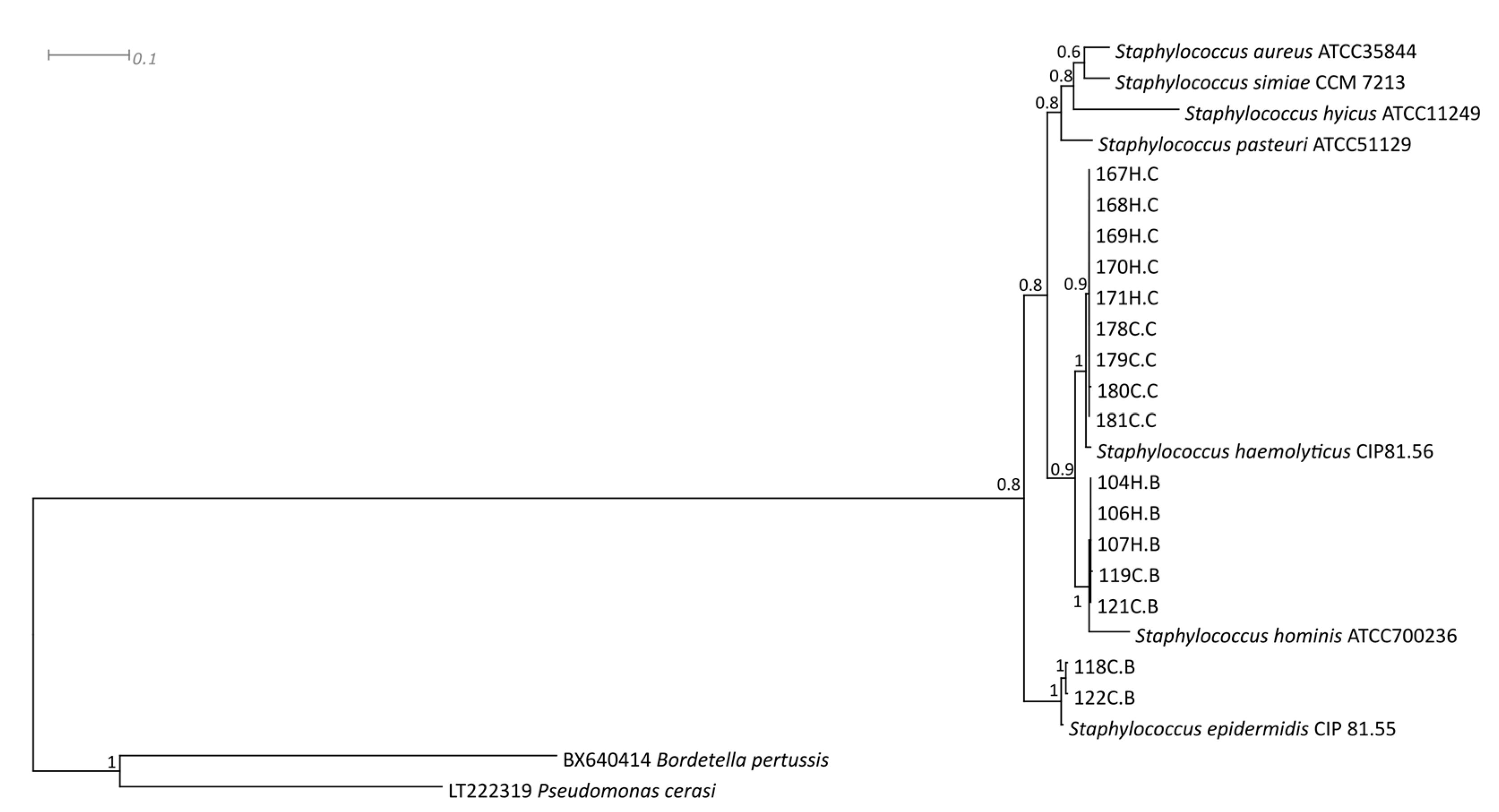
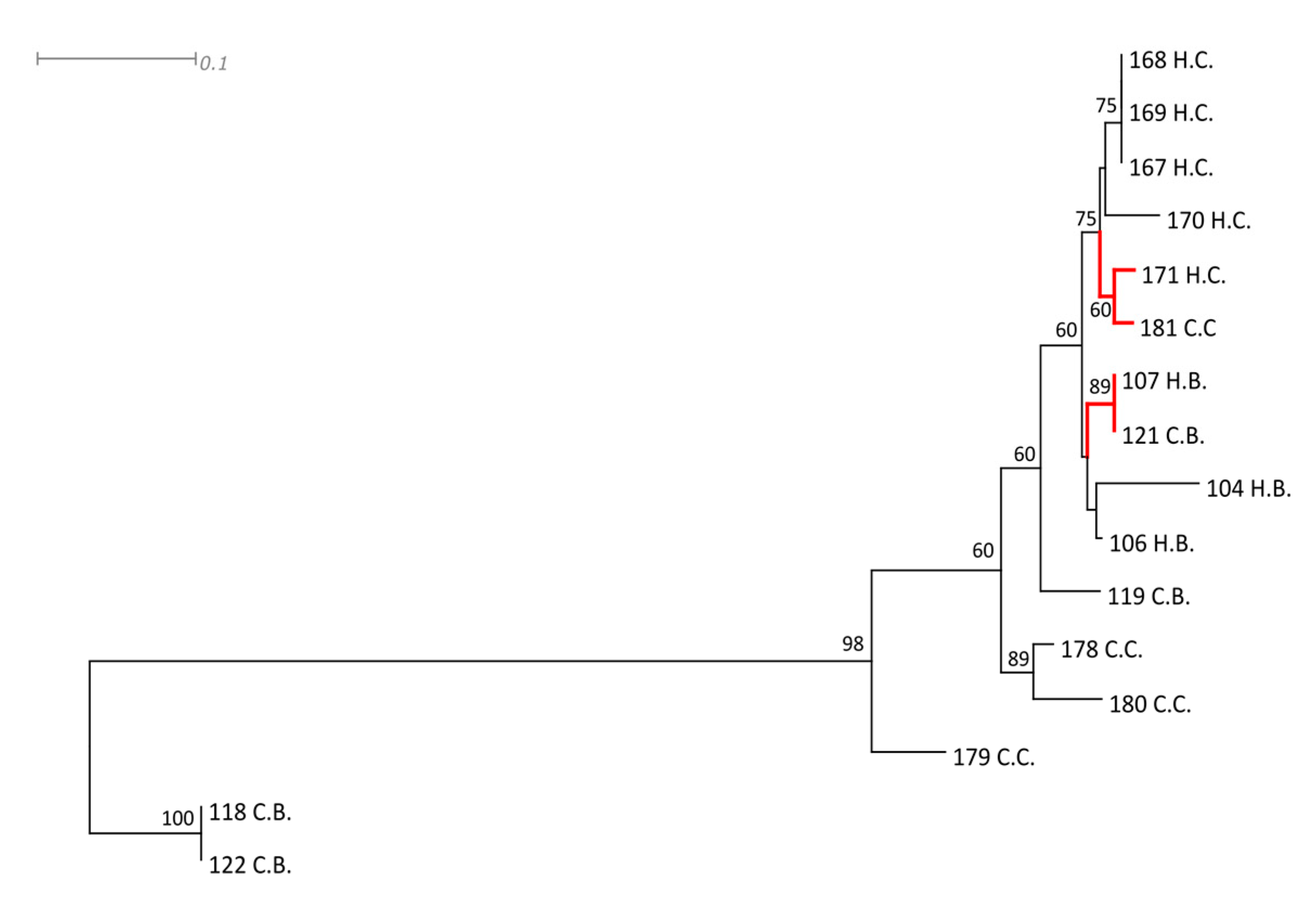
| Staphylococcus Species | Strain | rpoB | gap | tuf |
|---|---|---|---|---|
| Staphylococcus aureus | ATCC 35844 | AF325894 | HM352968 | HM352930 |
| Staphylococcus epidermidis | CIP 81.55 | EU659944 | EU659906 | EU652794 |
| Staphylococcus haemolyticus | CIP81.56 | EU659950 | EU659920 | EU652800 |
| Staphylococcus hominis | ATCC700236 | MF679108 | HM352973 | HM352925 |
| Staphylococcus hyicus | ATCC 11249 | AF325876 | FJ578002 | CP008747 |
| Staphylococcus pasteuri | ATCC 51129 | EU659961 | HM352972 | HM352929 |
| Staphylococcus simiae | CCM 7213 | EU888127 | HM352970 | HM352931 |
| Staphylococcus hominis | 104H.B | MT832255 | MT832271 | MT832239 |
| Staphylococcus hominis | 106H.B | MT832256 | MT832272 | MT832240 |
| Staphylococcus hominis | 107H.B | MT832257 | MT832273 | MT832241 |
| Staphylococcus epidermidis | 118C.B | MT832258 | MT832274 | MT832242 |
| Staphylococcus hominis | 119C.B | MT832259 | MT832275 | MT832243 |
| Staphylococcus hominis | 121C.B | MT832260 | MT832276 | MT832244 |
| Staphylococcus epidermidis | 122C.B | MT832261 | MT832277 | MT832245 |
| Staphylococcus haemolyticus | 167H.C | MT832262 | MT832278 | MT832246 |
| Staphylococcus haemolyticus | 168H.C | MT832263 | MT832279 | MT832247 |
| Staphylococcus haemolyticus | 169H.C | MT832264 | MT832280 | MT832248 |
| Staphylococcus haemolyticus | 170H.C | MT832265 | MT832281 | MT832249 |
| Staphylococcus haemolyticus | 171H.C | MT832266 | MT832282 | MT832250 |
| Staphylococcus haemolyticus | 178C.C | MT832267 | MT832283 | MT832251 |
| Staphylococcus haemolyticus | 179C.C | MT832268 | MT832284 | MT832252 |
| Staphylococcus haemolyticus | 180C.C | MT832269 | MT832285 | MT832253 |
| Staphylococcus haemolyticus | 181C.C | MT832270 | MT832286 | MT832254 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freitas Ribeiro, L.; Akira Sato, R.; de Souza Pollo, A.; Marques Rossi, G.A.; do Amaral, L.A. Occurrence of Methicillin-Resistant Staphylococcus spp. on Brazilian Dairy Farms that Produce Unpasteurized Cheese. Toxins 2020, 12, 779. https://doi.org/10.3390/toxins12120779
Freitas Ribeiro L, Akira Sato R, de Souza Pollo A, Marques Rossi GA, do Amaral LA. Occurrence of Methicillin-Resistant Staphylococcus spp. on Brazilian Dairy Farms that Produce Unpasteurized Cheese. Toxins. 2020; 12(12):779. https://doi.org/10.3390/toxins12120779
Chicago/Turabian StyleFreitas Ribeiro, Laryssa, Rafael Akira Sato, Andressa de Souza Pollo, Gabriel Augusto Marques Rossi, and Luiz Augusto do Amaral. 2020. "Occurrence of Methicillin-Resistant Staphylococcus spp. on Brazilian Dairy Farms that Produce Unpasteurized Cheese" Toxins 12, no. 12: 779. https://doi.org/10.3390/toxins12120779
APA StyleFreitas Ribeiro, L., Akira Sato, R., de Souza Pollo, A., Marques Rossi, G. A., & do Amaral, L. A. (2020). Occurrence of Methicillin-Resistant Staphylococcus spp. on Brazilian Dairy Farms that Produce Unpasteurized Cheese. Toxins, 12(12), 779. https://doi.org/10.3390/toxins12120779







