Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin
Abstract
1. Introduction
2. Results
2.1. ENN and BEA Possess Antibacterial and Antifungal Activities
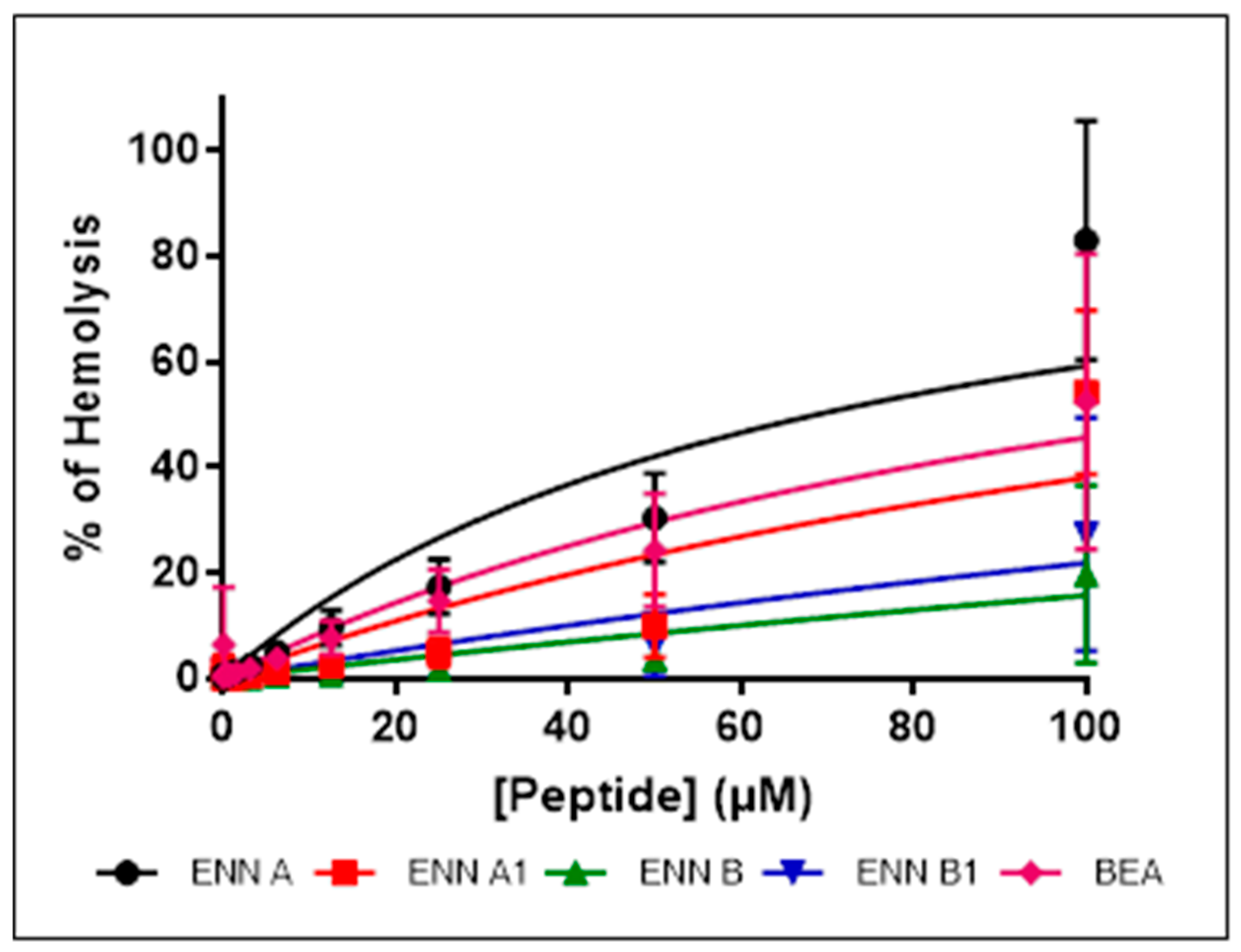
2.2. ENN and BEA Have Low Hemolytic Activity
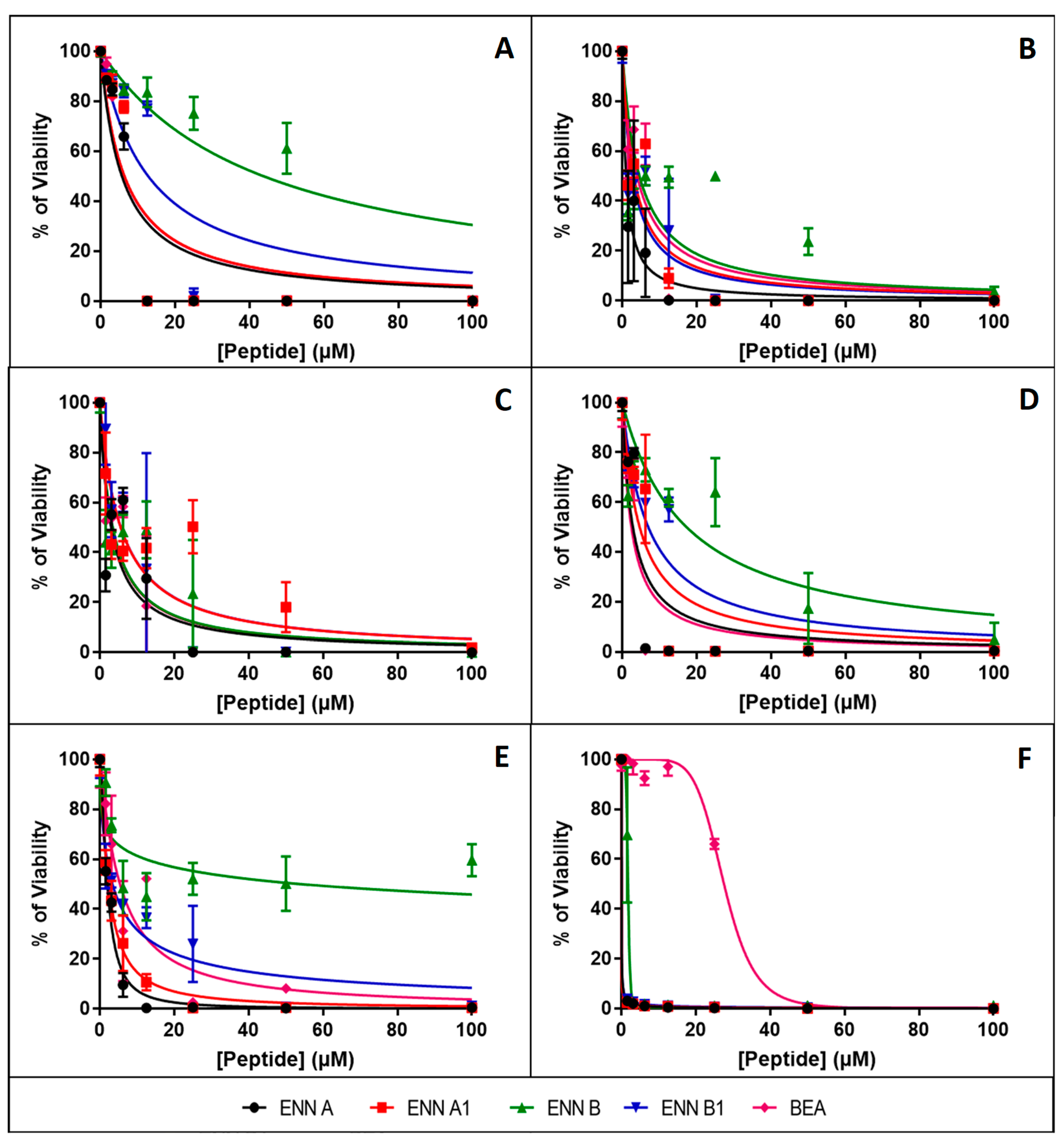
2.3. ENN and BEA are Toxic to Nucleated Human Cells
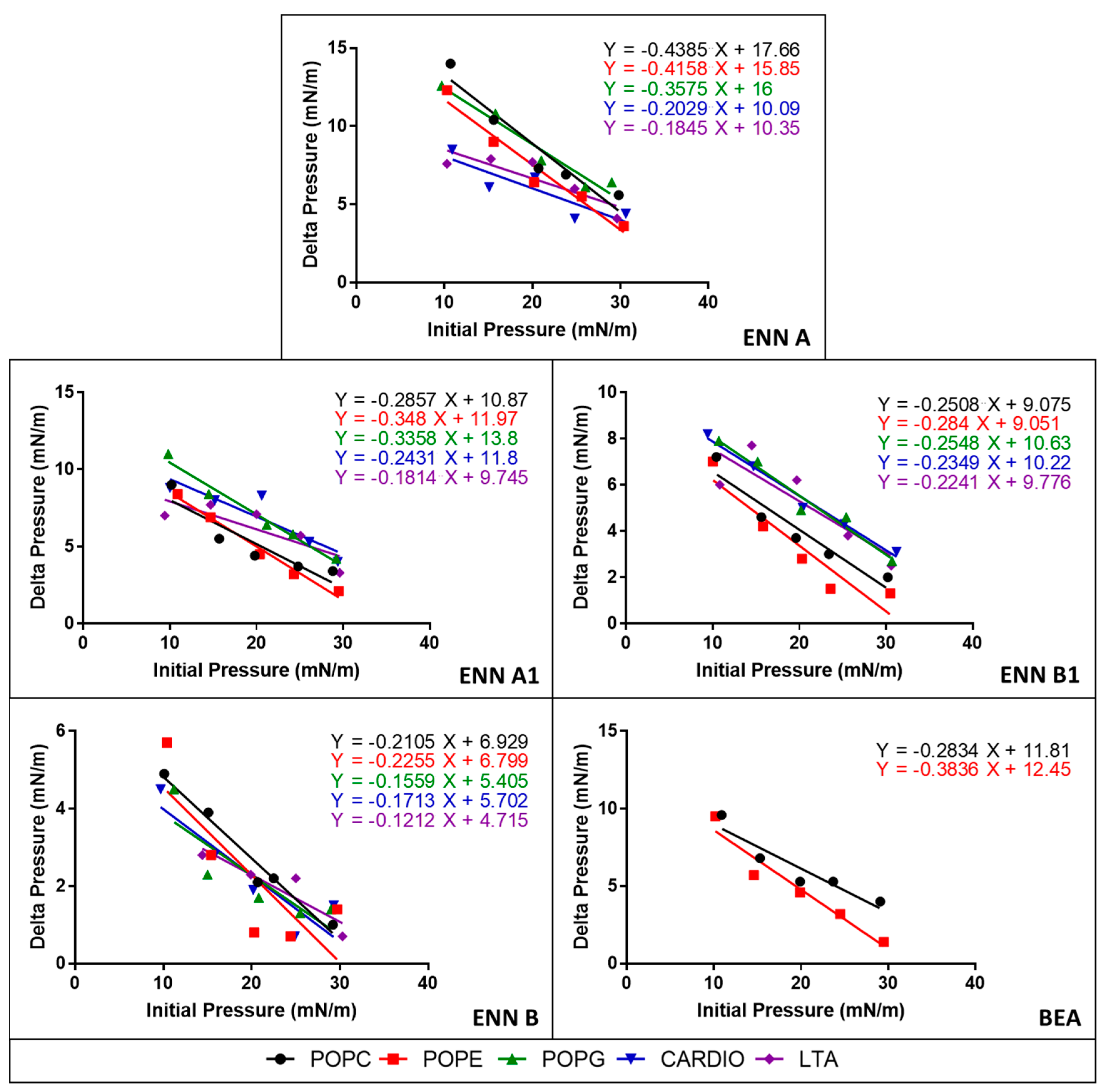
2.4. ENN and BEA Interact and Insert into Bacterial Lipids
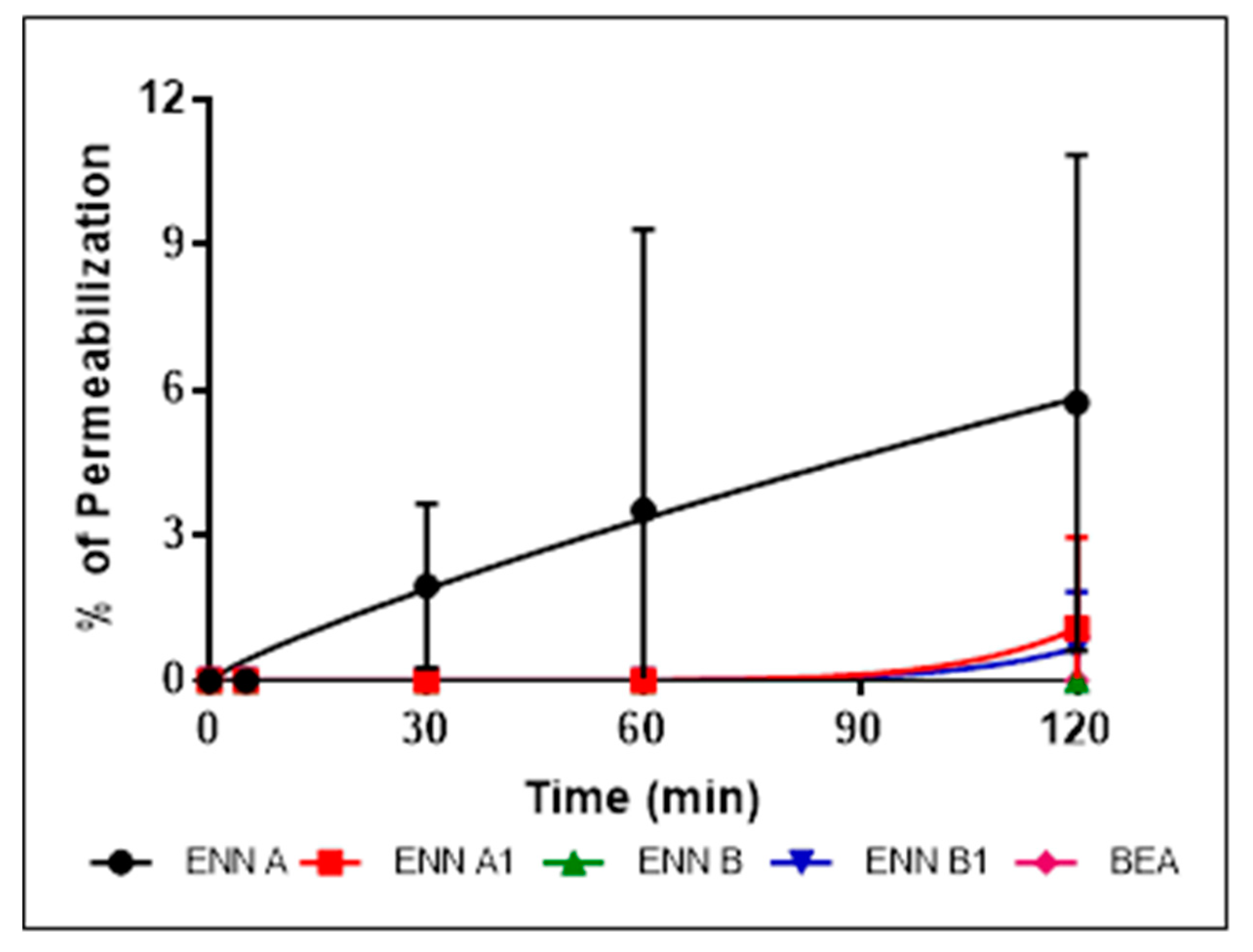
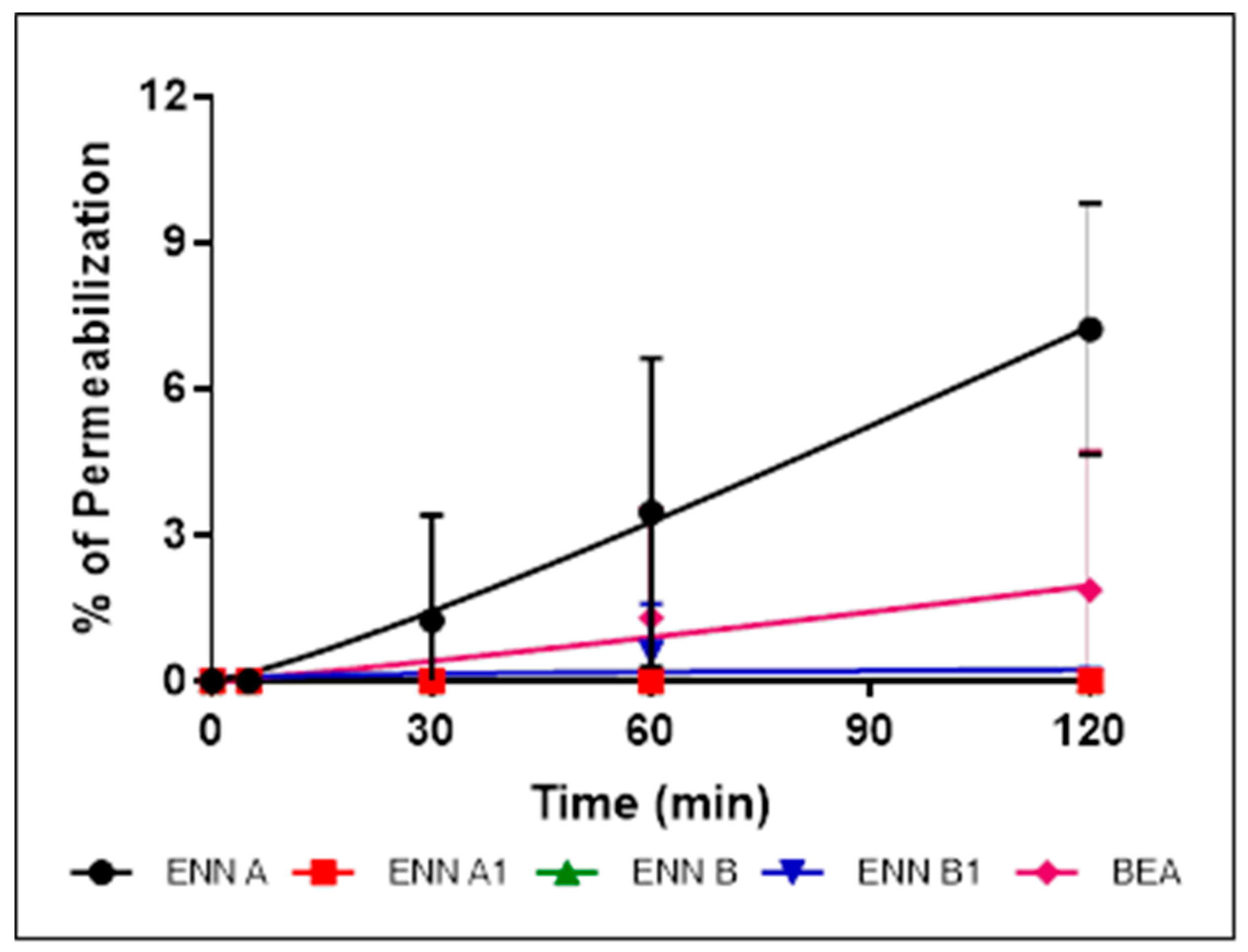
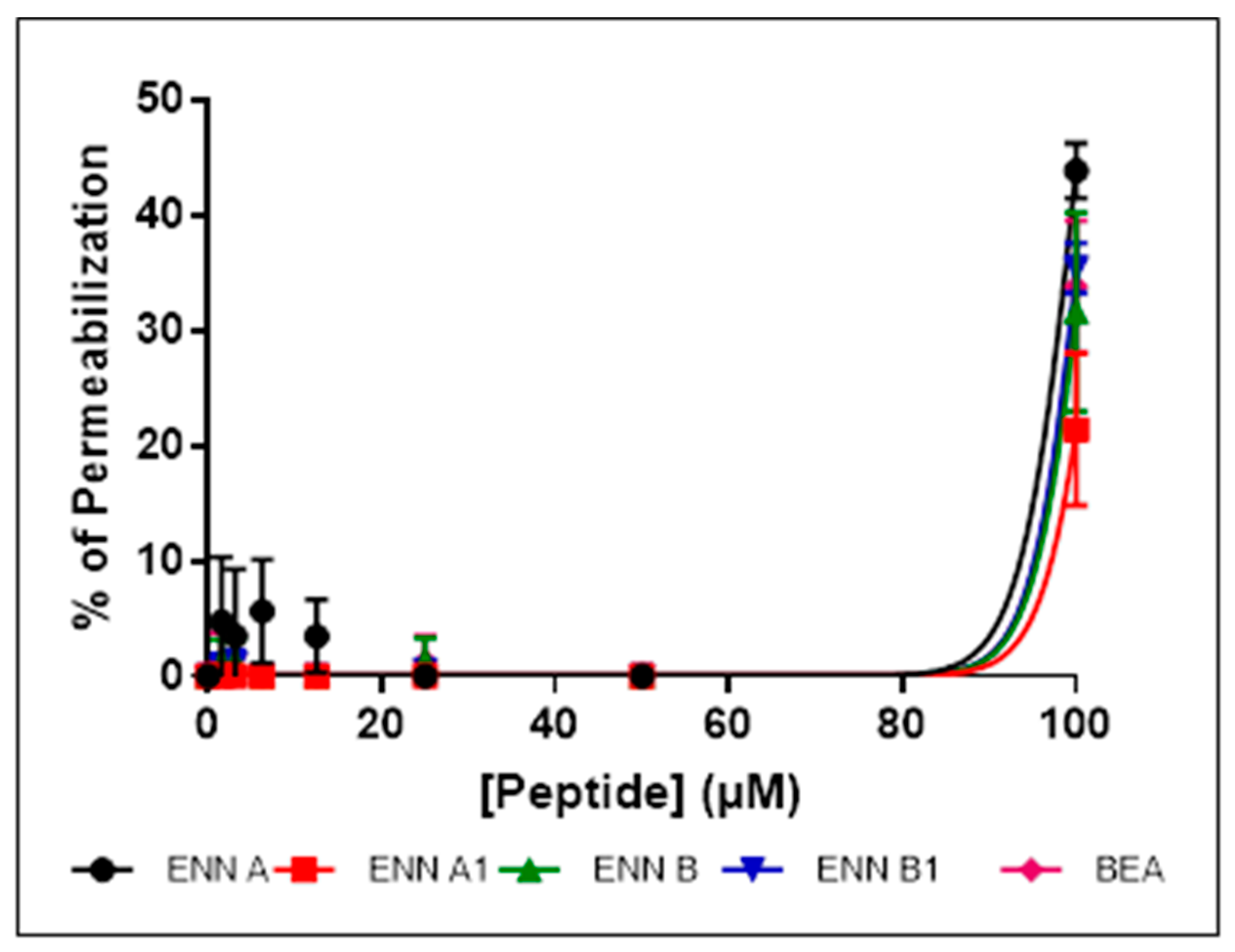
2.5. ENN and BEA Permeabilize the Bacterial Membrane but Only at High Doses
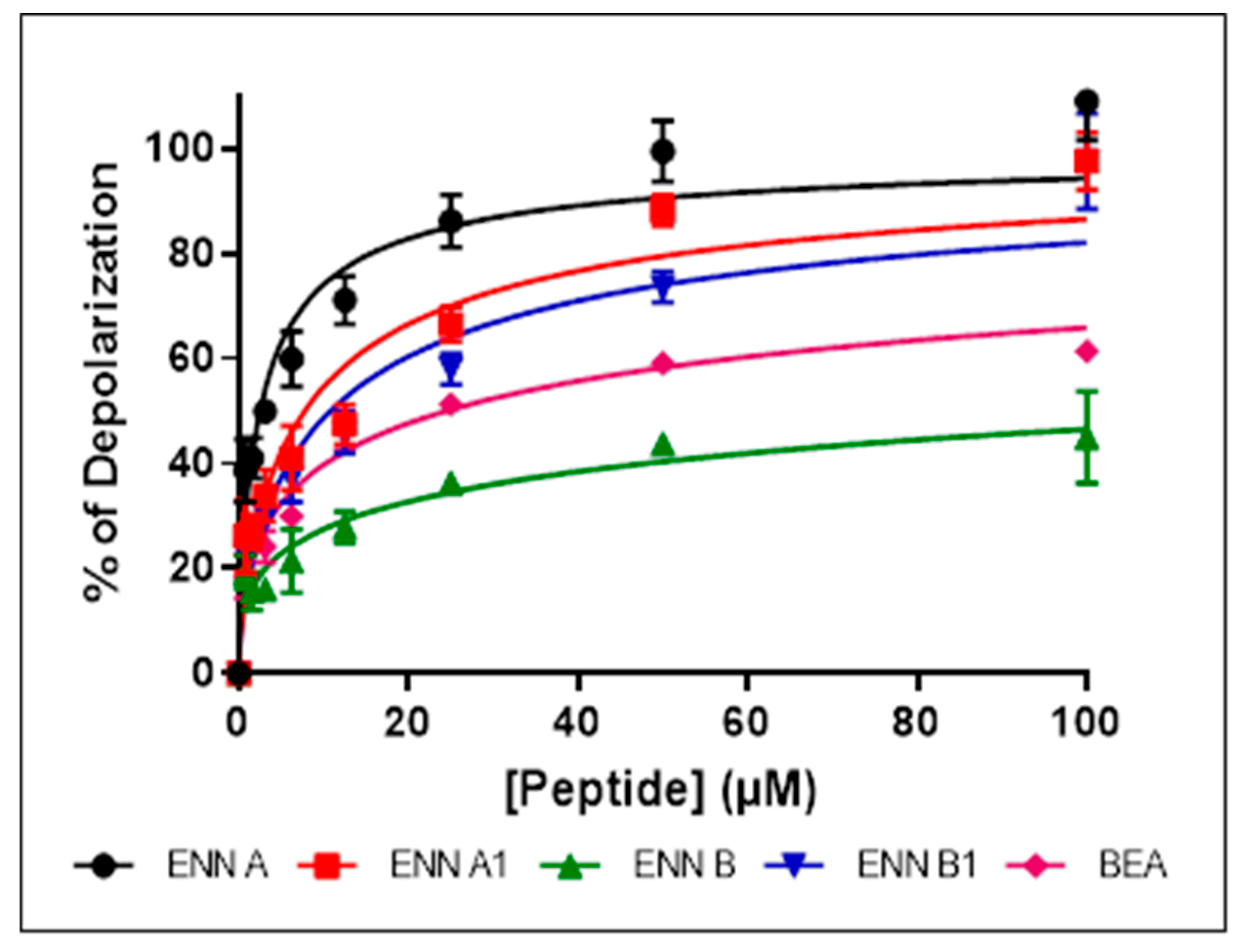
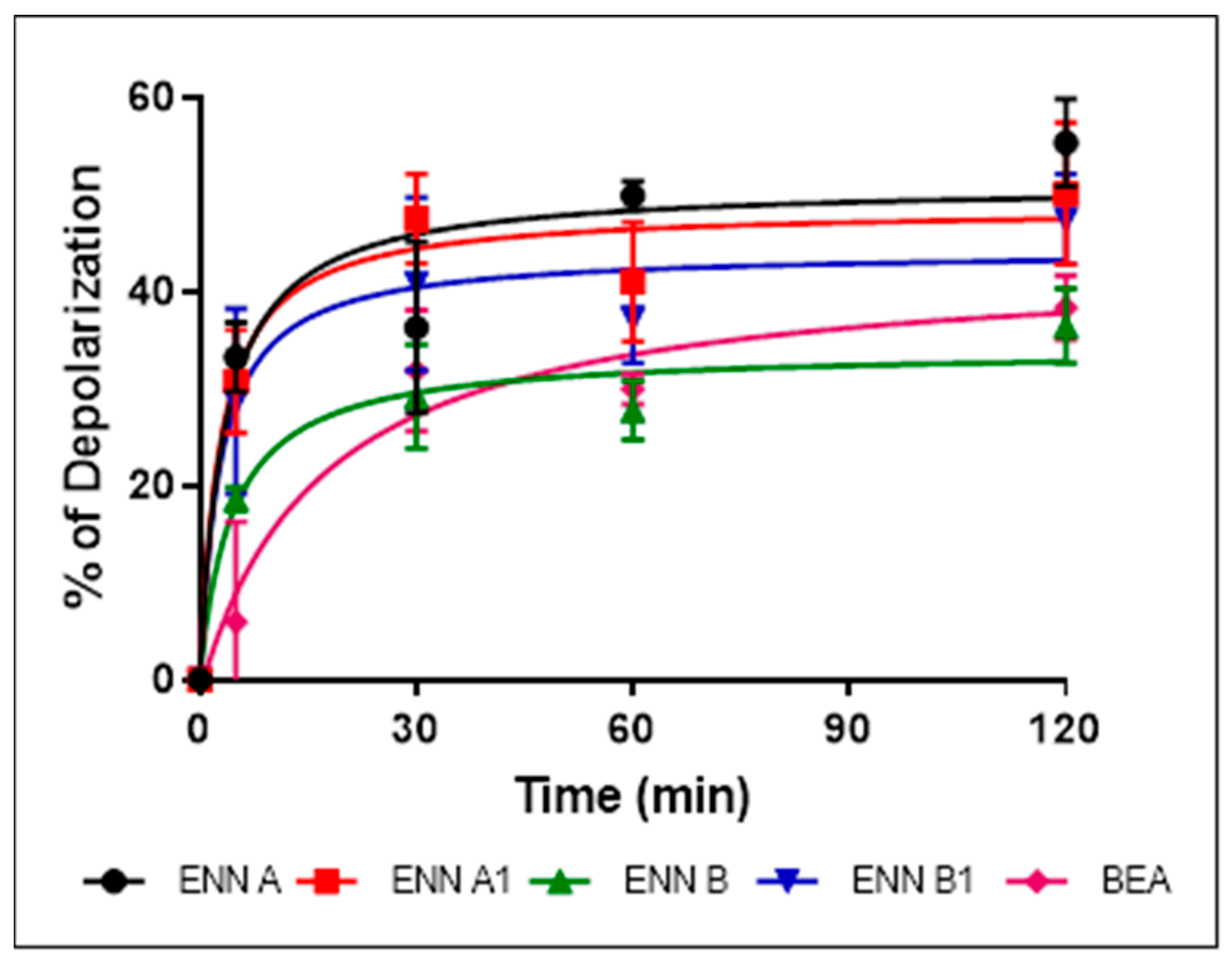
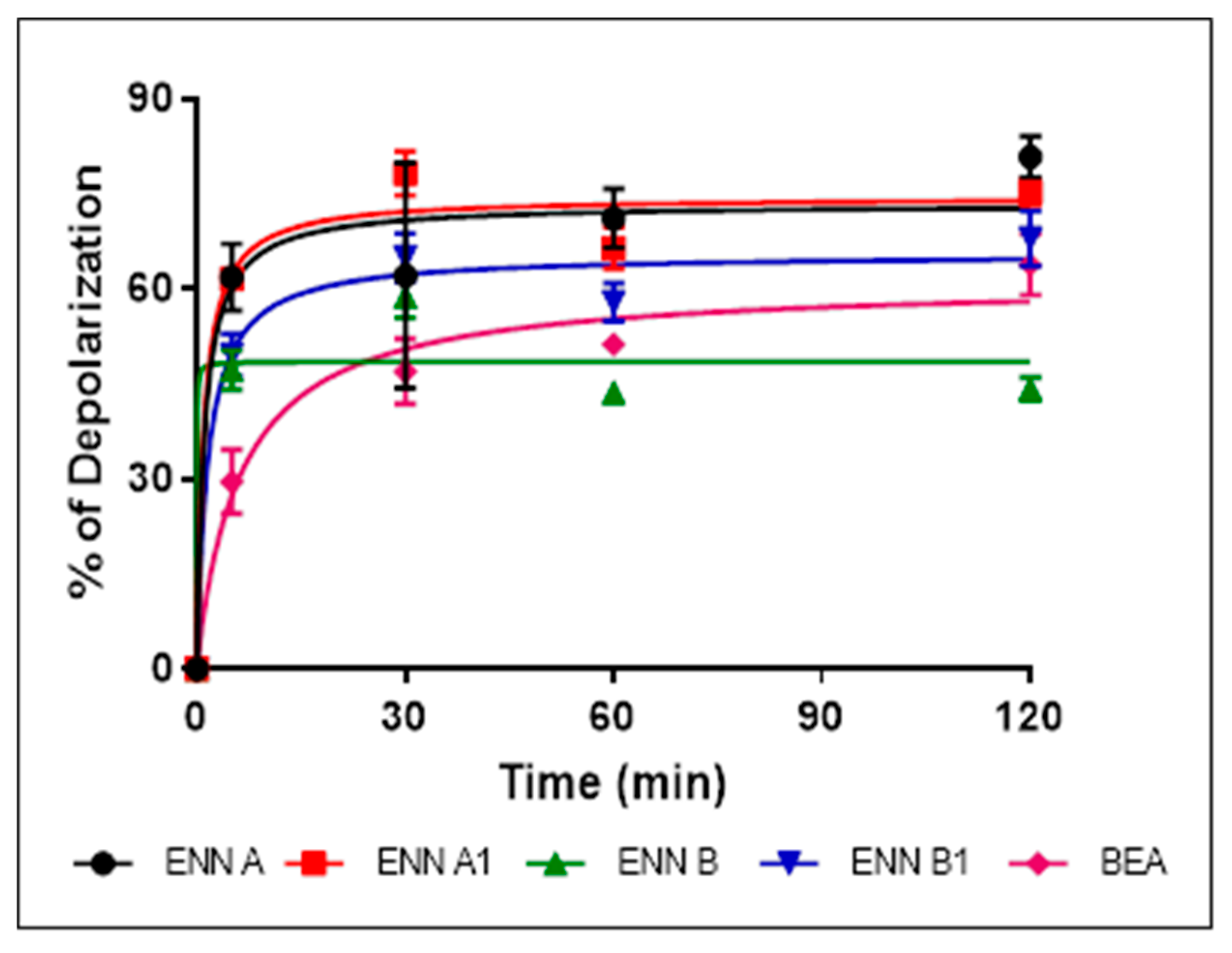
2.6. ENN and BEA at Low Doses Cause the Depolarization of the Bacterial Membrane
2.7. ENN and BEA at Low Doses Inhibit the Synthesis of Bacterial Macromolecules
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. Microorganism Strains and Growth Conditions
5.3. Antimicrobial Activity Assay
5.4. Hemolytic Activity Assay
5.5. Cytotoxic Assays
5.6. Peptid–eLipid Interaction Assay
5.7. Bacterial Membrane Permeabilization Assay
5.8. Membrane Depolarization Assay
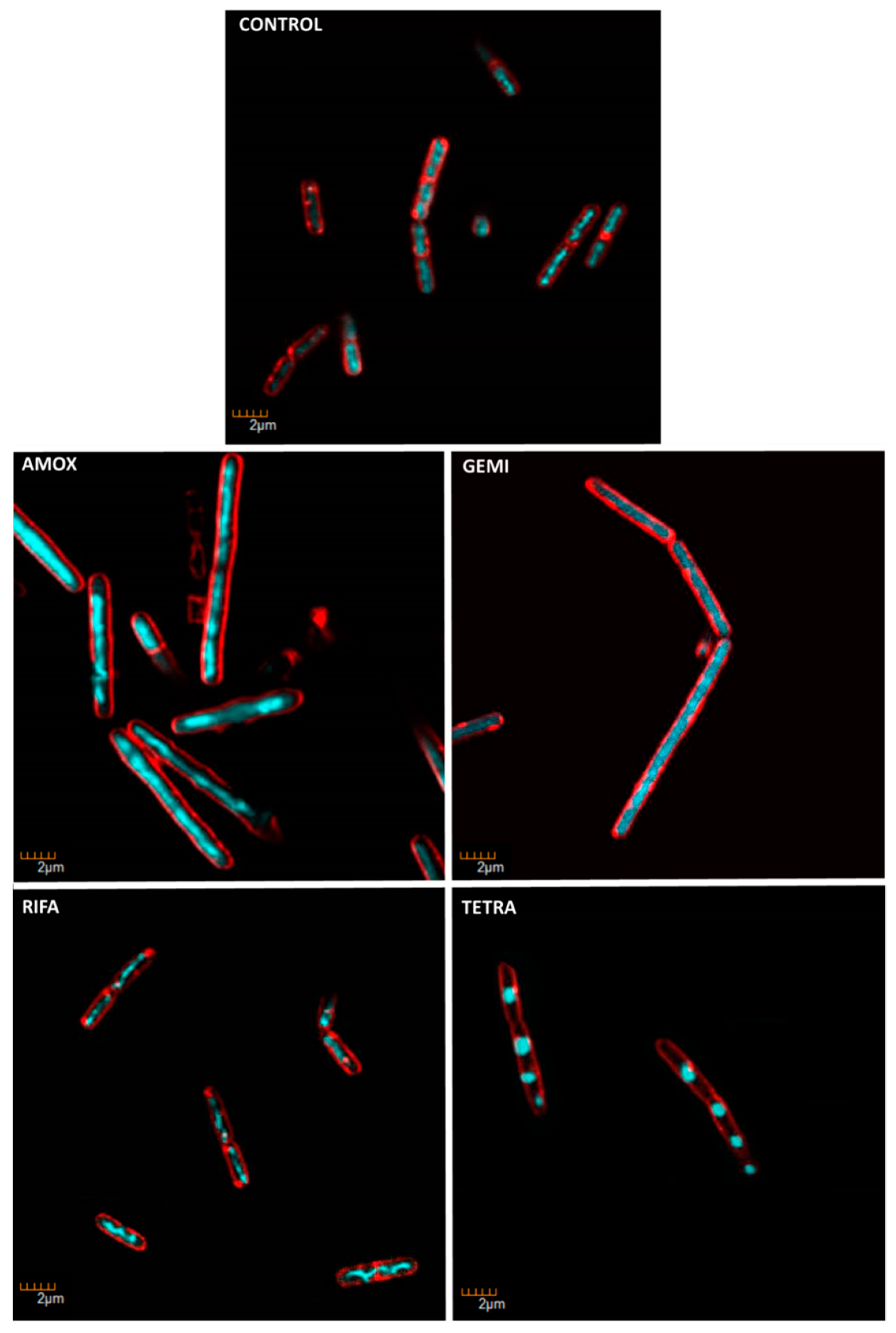
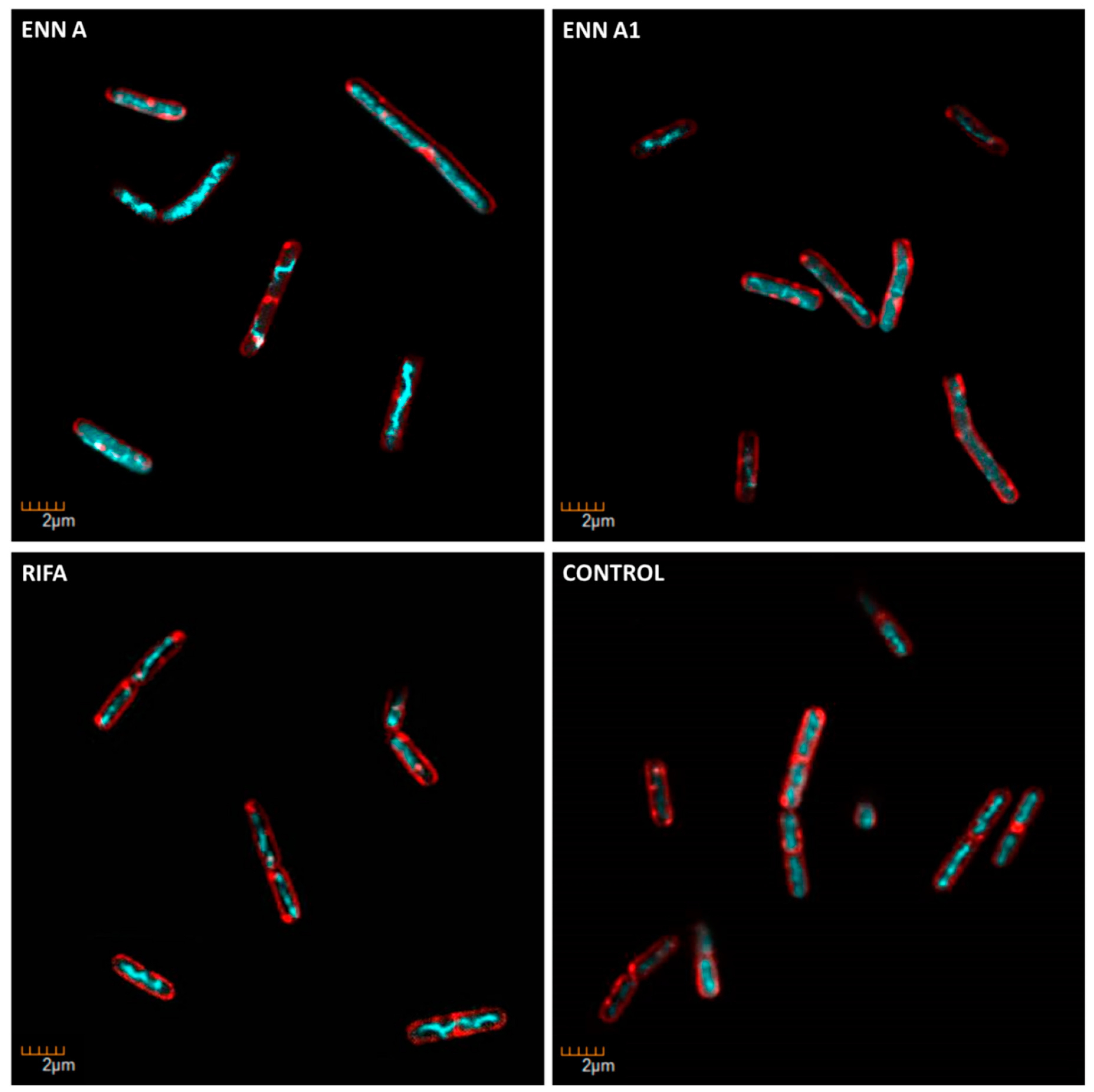
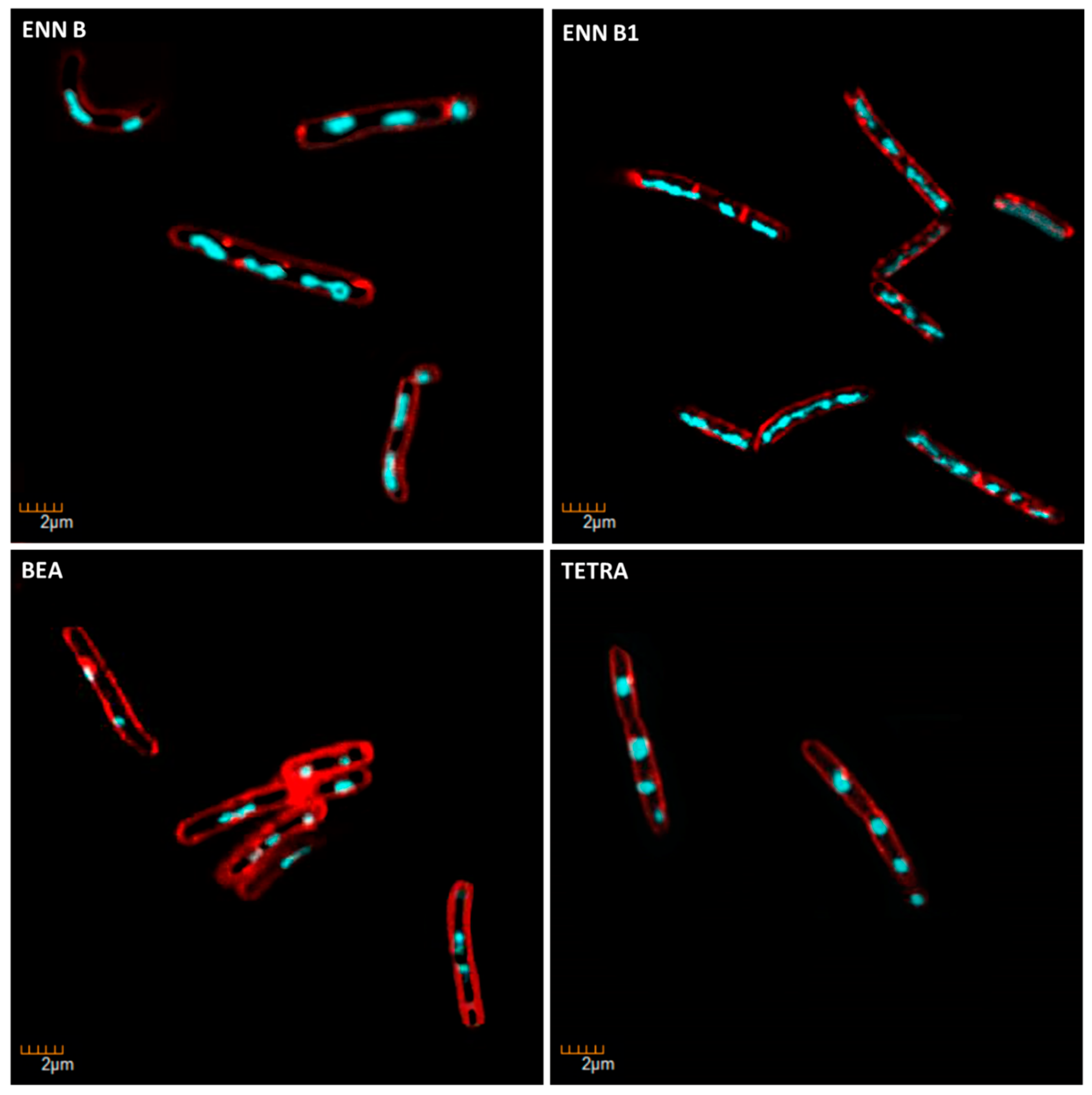
5.9. Determination of the Mechanism of Action on Bacteria Using Microscopic Observation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benkhaled, B.T.; Hadiouch, S.; Olleik, H.; Perrier, J.; Ysacco, C.; Guillaneuf, Y.; Gigmes, D.; Maresca, M.; Lefay, C. Elaboration of antimicrobial polymeric materials by dispersion of well-defined amphiphilic methacrylic SG1-based copolymers. Polym. Chem. 2018, 9, 3127–3141. [Google Scholar] [CrossRef]
- Kouchak, F.; Askarian, M. Nosocomial Infections: The Definition Criteria. Iran. J. Med. Sci. 2012, 37, 72–73. [Google Scholar] [PubMed]
- Wang, X.; Gong, X.; Li, P.; Lai, D.; Zhou, L. Structural Diversity and Biological Activities of Cyclic Depsipeptides from Fungi. Molecules 2018, 23, 169. [Google Scholar] [CrossRef] [PubMed]
- Sy-Cordero, A.A.; Pearce, C.J.; Oberlies, N.H. Revisiting the enniatins: A review of their isolation, biosynthesis, structure determination and biological activities. J. Antibiot. (Tokyo) 2012, 65, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Jestoi, M. Emerging Fusarium-Mycotoxins Fusaproliferin, Beauvericin, Enniatins, And Moniliformin—A Review. Crit. Rev. Food Sci. Nutr. 2008, 48, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Bertero, A.; Moretti, A.; Spicer, L.J.; Caloni, F. Fusarium Molds and Mycotoxins: Potential Species-Specific Effects. Toxins 2018, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Fraeyman, S.; Croubels, S.; Devreese, M.; Antonissen, G. Emerging Fusarium and Alternaria Mycotoxins: Occurrence, Toxicity and Toxicokinetics. Toxins 2017, 9, 228. [Google Scholar] [CrossRef]
- Hornbogen, T.; Glinski, M.; Zocher, R. Biosynthesis of Depsipeptide Mycotoxins in Fusarium. Eur. J. Plant Pathol. 2002, 108, 713–718. [Google Scholar] [CrossRef]
- Prosperini, A.; Berrada, H.; Ruiz, M.J.; Caloni, F.; Coccini, T.; Spicer, L.J.; Perego, M.C.; Lafranconi, A. A Review of the Mycotoxin Enniatin B. Front. Public Health 2017, 5. [Google Scholar] [CrossRef]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 2013, 60, 218–237. [Google Scholar] [CrossRef] [PubMed]
- Maresca, M. From the Gut to the Brain: Journey and Pathophysiological Effects of the Food-Associated Trichothecene Mycotoxin Deoxynivalenol. Toxins 2013, 5, 784–820. [Google Scholar] [CrossRef] [PubMed]
- Terciolo, C.; Maresca, M.; Pinton, P.; Oswald, I.P. Review article: Role of satiety hormones in anorexia induction by Trichothecene mycotoxins. Food Chem. Toxicol. 2018, 121, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Krug, I.; Behrens, M.; Esselen, M.; Humpf, H.-U. Transport of enniatin B and enniatin B1 across the blood-brain barrier and hints for neurotoxic effects in cerebral cells. PLoS ONE 2018, 13, e0197406. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Fernández-Franzón, M.; Font, G.; Ruiz, M.J. Toxicity evaluation of individual and mixed enniatins using an in vitro method with CHO-K1 cells. Toxicol. Vitr. 2013, 27, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Ola, A.R.B.; Thomy, D.; Lai, D.; Brötz-Oesterhelt, H.; Proksch, P. Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 2013, 76, 2094–2099. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Bluzat, A.; Seigneuret, M.; Rigaud, J.-L. Alkali cation transport through liposomes by the antimicrobial fusafungine and its constitutive enniatins. Biochem. Pharmacol. 1995, 50, 2105–2107. [Google Scholar] [CrossRef]
- Jayasinghe, L.; Abbas, H.K.; Jacob, M.R.; Herath, W.H.M.W.; Nanayakkara, N.P.D. N-Methyl-4-hydroxy-2-pyridinone Analogues from Fusarium oxysporum. J. Nat. Prod. 2006, 69, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Firáková, S.; Proksa, B.; Sturdíková, M. Biosynthesis and biological activity of enniatins. Int. J. Pharm. Sci. 2007, 62, 563–568. [Google Scholar]
- Jeschke, P.; Benet-Buchholz, J.; Harder, A.; Etzel, W.; Schindler, M.; Thielking, G. Synthesis and anthelmintic activity of cyclohexadepsipeptides with (S,S,S,R,S,R)-configuration. Bioorg. Med. Chem. Lett. 2003, 13, 3285–3288. [Google Scholar] [CrossRef]
- Tong, Y.; Liu, M.; Zhang, Y.; Liu, X.; Huang, R.; Song, F.; Dai, H.; Ren, B.; Sun, N.; Pei, G.; et al. Beauvericin counteracted multi-drug resistant Candida albicans by blocking ABC transporters. Synth. Syst. Biotechnol. 2016, 1, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ruan, C.; Bai, X.; Zhang, M.; Zhu, S.; Jiang, Y. Isolation and Identification of the Antimicrobial Agent Beauvericin from the Endophytic Fusarium oxysporum 5–19 with NMR and ESI-MS/MS. Biomed. Res. Int. 2016, 2016, 1084670. [Google Scholar]
- Fukuda, T.; Arai, M.; Yamaguchi, Y.; Masuma, R.; Tomoda, H.; Omura, S. New Beauvericins, Potentiators of Antifungal Miconazole Activity, Produced by Beauveria sp. FKI-1366. J. Antibiot. (Tokyo) 2004, 57, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Dornetshuber, R.; Heffeter, P.; Kamyar, M.-R.; Peterbauer, T.; Berger, W.; Lemmens-Gruber, R. Enniatin exerts p53-dependent cytostatic and p53-independent cytotoxic activities against human cancer cells. Chem. Res. Toxicol. 2007, 20, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Sharom, F.J.; Lu, P.; Liu, R.; Yu, X. Linear and cyclic peptides as substrates and modulators of P-glycoprotein: Peptide binding and effects on drug transport and accumulation. Biochem. J. 1998, 333, 621–630. [Google Scholar] [CrossRef]
- Tomoda, H.; Huang, X.H.; Cao, J.; Nishida, H.; Nagao, R.; Okuda, S.; Tanaka, H.; Omura, S.; Arai, H.; Inoue, K. Inhibition of acyl-CoA: Cholesterol acyltransferase activity by cyclodepsipeptide antibiotics. J. Antibiot. (Tokyo) 1992, 45, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- Benz, R. Alkali ion transport through lipid bilayer membranes mediated by enniatin A and B and beauvericin. J. Membr. Biol. 1978, 43, 367–394. [Google Scholar] [CrossRef]
- Lifson, S.; Felder, C.E.; Shanzer, A. Enniatin B and valinomycin as ion carriers: An empirical force field analysis. J. Biomol. Struct. Dyn. 1984, 2, 641–661. [Google Scholar] [CrossRef]
- Ovchinnikov, Y.A.; Ivanov, V.T.; Evstratov, A.V.; Mikhaleva, I.I.; Bystrov, V.F.; Portnova, S.L.; Balashova, T.A.; Meshcheryakova, E.N.; Tulchinsky, V.M. The enniatin ionophores. Conformation and ion binding properties. Int. J. Pept. Protein Res. 1974, 6, 465–498. [Google Scholar] [CrossRef]
- Prince, R.C.; Crofts, A.R.; Steinrauf, L.K. A comparison of beauvericin, enniatin and valinomycin as calcium transporting agents in liposomes and chromatophores. Biochem. Biophys. Res. Commun. 1974, 59, 697–703. [Google Scholar] [CrossRef]
- Doebler, J.A. Effects of neutral ionophores on membrane electrical characteristics of NG108-15 cells. Toxicol. Lett. 2000, 114, 27–38. [Google Scholar] [CrossRef]
- Tonshin, A.A.; Teplova, V.V.; Andersson, M.A.; Salkinoja-Salonen, M.S. The Fusarium mycotoxins enniatins and beauvericin cause mitochondrial dysfunction by affecting the mitochondrial volume regulation, oxidative phosphorylation and ion homeostasis. Toxicology 2010, 276, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Chiumento, S.; Roblin, C.; Kieffer-Jaquinod, S.; Tachon, S.; Basset, C.; Aditiyarini, D.; Olleik, H.; Nicoletti, C.; Iranzo, O.; Maresca, M.; et al. RuminococcinC, a promising antibiotic produced by a human gut symbiont. Sci. Adv. 2019, 22. [Google Scholar]
- Nonejuie, P.; Burkart, M.; Pogliano, K.; Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. 2013, 110, 16169–16174. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.; McGaw, L. Natural Cyclic Peptides as an Attractive Modality for Therapeutics: A Mini Review. Molecules 2018, 23, 2080. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases-a field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Olleik, H.; Yahiaoui, S.; Roulier, B.; Courvoisier-Dezord, E.; Perrier, J.; Pérès, B.; Hijazi, A.; Baydoun, E.; Raymond, J.; Boumendjel, A.; et al. Aurone derivatives as promising antibacterial agents against resistant Gram-positive pathogens. Eur. J. Med. Chem. 2019, 165, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Oyama, L.B.; Girdwood, S.E.; Cookson, A.R.; Fernandez-Fuentes, N.; Privé, F.; Vallin, H.E.; Wilkinson, T.J.; Golyshin, P.N.; Golyshina, O.V.; Mikut, R.; et al. The rumen microbiome: An underexplored resource for novel antimicrobial discovery. Npj Biofilms Microbiomes 2017, 3, 33. [Google Scholar] [CrossRef]
- Rex, J.H.; Pfaller, M.A.; Walsh, T.J.; Chaturvedi, V.; Espinel-Ingroff, A.; Ghannoum, M.A.; Gosey, L.L.; Odds, F.C.; Rinaldi, M.G.; Sheehan, D.J.; et al. Antifungal Susceptibility Testing: Practical Aspects and Current Challenges. Clin. Microbiol. Rev. 2001, 14, 643–658. [Google Scholar] [CrossRef]
- Tardy, A.; Honoré, J.-C.; Tran, J.; Siri, D.; Delplace, V.; Bataille, I.; Letourneur, D.; Perrier, J.; Nicoletti, C.; Maresca, M.; et al. Radical Copolymerization of Vinyl Ethers and Cyclic Ketene Acetals as a Versatile Platform to Design Functional Polyesters. Angew. Chem. Int. Ed. 2017, 56, 16515–16520. [Google Scholar] [CrossRef]
- Borie, C.; Mondal, S.; Arif, T.; Briand, M.; Lingua, H.; Dumur, F.; Gigmes, D.; Stocker, P.; Barbarat, B.; Robert, V.; et al. Enediynes bearing polyfluoroaryl sulfoxide as new antiproliferative agents with dual targeting of microtubules and DNA. Eur. J. Med. Chem. 2018, 148, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Di Pasquale, E.; Salmi-Smail, C.; Brunel, J.-M.; Sanchez, P.; Fantini, J.; Maresca, M. Biophysical studies of the interaction of squalamine and other cationic amphiphilic molecules with bacterial and eukaryotic membranes: Importance of the distribution coefficient in membrane selectivity. Chem. Phys. Lipids 2010, 163, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Mahfoud, R.; Maresca, M.; Santelli, M.; Pfohl-Leszkowicz, A.; Puigserver, A.; Fantini, J. pH-Dependent Interaction of Fumonisin B1 with Cholesterol: Physicochemical and Molecular Modeling Studies at the Air−Water Interface. J. Agric. Food Chem. 2002, 50, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Uppu, D.S.S.M.; Haldar, J. Lipopolysaccharide Neutralization by Cationic-Amphiphilic Polymers through Pseudoaggregate Formation. Biomacromolecules 2016, 17, 862–873. [Google Scholar] [CrossRef] [PubMed]
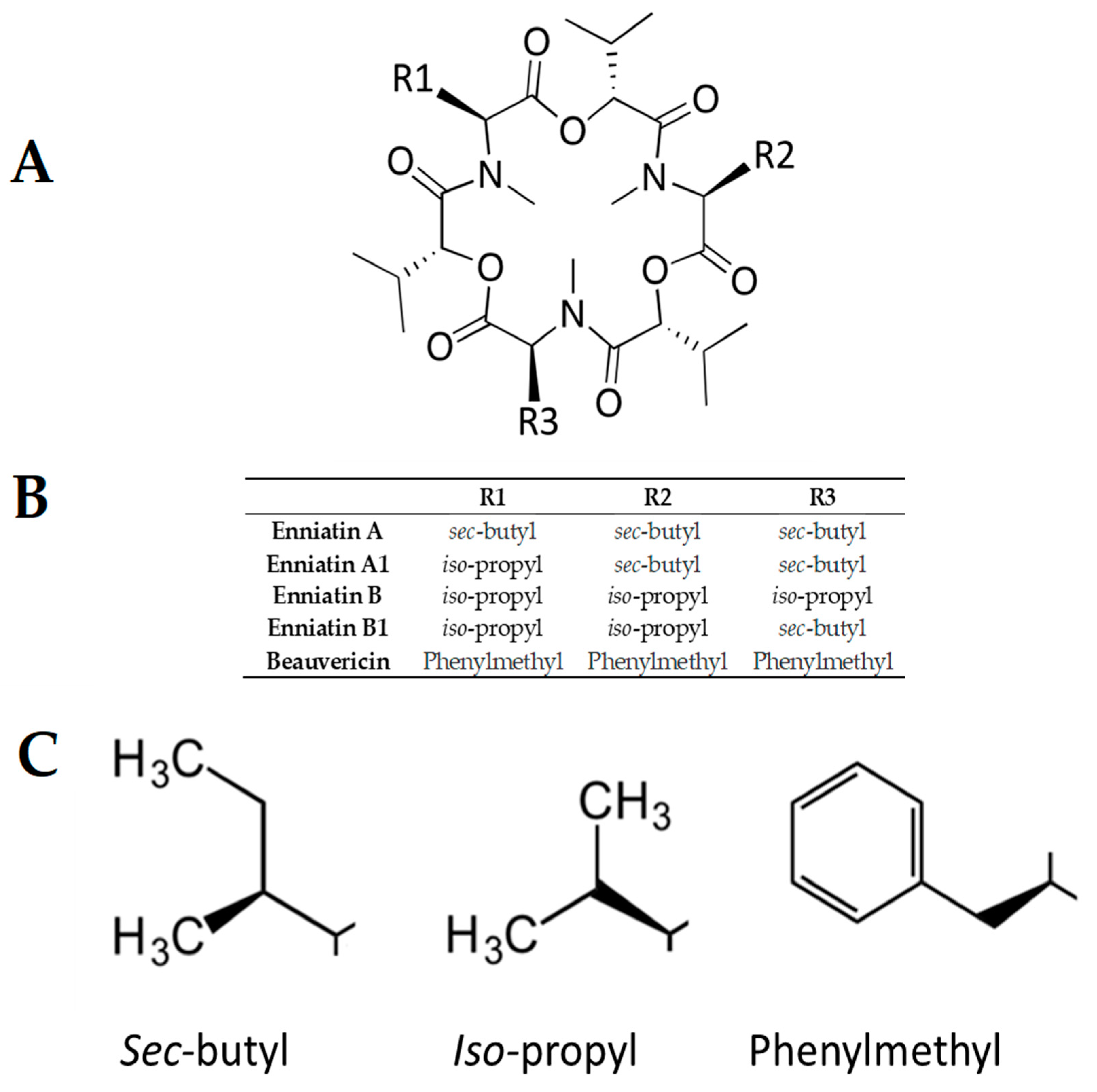
| Microorganisms | ENN A | ENN A1 | ENN B | ENN B1 | BEA | |
|---|---|---|---|---|---|---|
| Gram positive bacteria | B. subtilis | 6.25–12.5 | 12.5–25 | >100 | 25–50 | 6.25–12.5 |
| B. subtilis NR | 6.25–12.5 | 25 | >100 | 50 | 6.25–12.5 | |
| C. perfringens | 3.12 | 3.12–6.25 | 12.5 | 6.25 | 6.25 | |
| E. faecalis | 3.12 | 6.25 | >100 | 12.5 | 12.5 | |
| S. aureus | 6.25–12.5 | 12.5–25 | >100 | 25 | 12.5 | |
| S. aureus MRSA | 6.25–12.5 | 12.5–25 | >100 | 25 | 6.25–12.5 | |
| Gram negative bacteria | A. baumannii | >100 | >100 | >100 | >100 | >100 |
| E. coli | >100 | >100 | >100 | >100 | >100 | |
| H. pylori | >100 | >100 | >100 | >100 | >100 | |
| P. aeruginosa | >100 | >100 | >100 | >100 | >100 | |
| Mycobacteria | M. smegmatis | 6.25 | 3.12 | 100 | 6.25 | 25 |
| Fungi | A. flavus | >100 | >100 | >100 | >100 | >100 |
| A. niger | >100 | >100 | >100 | >100 | >100 | |
| A. ochraceus | >100 | >100 | >100 | >100 | >100 | |
| C. albicans | 1.5 | 1.5 | 6.25 | 3.12 | 6.25 | |
| F. graminearum | 25 | 50 | >100 | 25 | 25 | |
| F. oxysporum | >100 | >100 | >100 | >100 | >100 | |
| F. verticillioides | >100 | >100 | >100 | >100 | >100 | |
| P. verrucosum | >100 | >100 | >100 | >100 | >100 | |
| S. chartarum | >100 | >100 | >100 | >100 | >100 | |
| Cells | ENN A | ENN A1 | ENN B | ENN B1 | BEA |
|---|---|---|---|---|---|
| BEAS-2B | 5.7 ± 1.2 | 6.4 ± 1.5 | 43.7 ± 7.7 | 12.7 ± 2.8 | 6.3 ± 1.4 |
| Caco-2 | 1.1 ± 0.2 | 2.7 ± 0.5 | 4.6 ± 1.3 | 3.1 ± 0.5 | 3.9 ± 0.7 |
| HEK | 2.0 ±0.1 | 2.3 ± 0.1 | 54.2 ± 35.3 | 3.3 ± 0.5 | 5.4 ± 0.9 |
| HEPG2 | 3.0 ± 0.7 | 5.6 ± 1.1 | 3.4 ± 0.7 | 5.6 ± 1.4 | 3.4 ± 0.6 |
| HUVEC | 2.8 ± 0.6 | 4.6 ± 0.9 | 17.3 ± 3.3 | 7.0 ± 1.1 | 2.4 ± 0.5 |
| N87 | 0.01 ± 0.01 | 0.003 ± 0.002 | 1.7 ± 0.1 | 0.008 ± 0.008 | 27.5 ± 0.7 |
| Erythrocytes | 68.7 ± 9.6 | 162.7 ± 22.8 | 534.5 ± 86.5 | 356.4 ± 59.3 | 118.9 ± 15.8 |
| Peptides | POPC | POPE | POPG | Cardiolipin | LTA |
|---|---|---|---|---|---|
| ENN A | 40.28 | 38.12 | 44.75 | 49.72 | 56.1 |
| ENN A1 | 38.06 | 34.38 | 41.10 | 48.57 | 53.71 |
| ENN B | 32.92 | 30.15 | 34.67 | 33.29 | 38.9 |
| ENN B1 | 36.19 | 31.87 | 41.71 | 43.51 | 43.62 |
| BEA | 41.65 | 32.46 | -- | -- | -- |
| Condition | Known/possible target | Observed Morphology |
|---|---|---|
| Control | - |
|
| Amoxicillin | Cell wall biosynthesis |
|
| Gemifloxacin | DNA synthesis |
|
| Rifampicin | RNA biosynthesis |
|
| Tetracycline | Protein biosynthesis |
|
| ENN A | RNA biosynthesis |
|
| ENN A1 | RNA biosynthesis |
|
| ENN B | Protein biosynthesis |
|
| ENN B1 | Protein biosynthesis |
|
| BEA | Protein biosynthesis |
|
| Chemical Groups Present at R1, R2, R3 | Effects |
|---|---|
| iso-propyl | • Decrease in antimicrobial activity |
| • Decrease in hemolysis | |
| • Variable effect on cytotoxicity (either a decrease for BEAS-2B, Caco-2, HEK, and HUVEC cells or an increase for HEPG2 and N87 cells) | |
| • Decrease in insertion in bacterial and eukaryotic lipids | |
| • Inhibition of bacterial protein synthesis | |
| sec-butyl | • Increase in antimicrobial activity |
| • Increase in hemolysis | |
| • Increase in cytotoxicity | |
| • Increase in insertion in bacterial and eukaryotic lipids | |
| • Inhibition of bacterial RNA synthesis | |
| Phenylmethyl | • Variable effect on antimicrobial activity (either a decrease for C. albicans, C. perfringens, E. faecalis, and M. smegmatis or an increase for B. subtilis and S. aureus) |
| • Increase in hemolysis | |
| • Variable effect on cytotoxicity (either a decrease for BEAS-2B, Caco-2, HEK, HEPG2, and N87 cells or an increase for HUVEC cells) | |
| • Increase in insertion in eukaryotic lipid (POPC) | |
| • Inhibition of bacterial protein synthesis |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olleik, H.; Nicoletti, C.; Lafond, M.; Courvoisier-Dezord, E.; Xue, P.; Hijazi, A.; Baydoun, E.; Perrier, J.; Maresca, M. Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins 2019, 11, 514. https://doi.org/10.3390/toxins11090514
Olleik H, Nicoletti C, Lafond M, Courvoisier-Dezord E, Xue P, Hijazi A, Baydoun E, Perrier J, Maresca M. Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins. 2019; 11(9):514. https://doi.org/10.3390/toxins11090514
Chicago/Turabian StyleOlleik, Hamza, Cendrine Nicoletti, Mickael Lafond, Elise Courvoisier-Dezord, Peiwen Xue, Akram Hijazi, Elias Baydoun, Josette Perrier, and Marc Maresca. 2019. "Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin" Toxins 11, no. 9: 514. https://doi.org/10.3390/toxins11090514
APA StyleOlleik, H., Nicoletti, C., Lafond, M., Courvoisier-Dezord, E., Xue, P., Hijazi, A., Baydoun, E., Perrier, J., & Maresca, M. (2019). Comparative Structure–Activity Analysis of the Antimicrobial Activity, Cytotoxicity, and Mechanism of Action of the Fungal Cyclohexadepsipeptides Enniatins and Beauvericin. Toxins, 11(9), 514. https://doi.org/10.3390/toxins11090514








