Molecular Characterisation of Aflatoxigenic and Non-Aflatoxigenic Strains of Aspergillus Section Flavi Isolated from Imported Peanuts along the Supply Chain in Malaysia
Abstract
1. Introduction
2. Results
2.1. PCR Amplification and Basic Local Alignment Search Tool (BLAST) Search
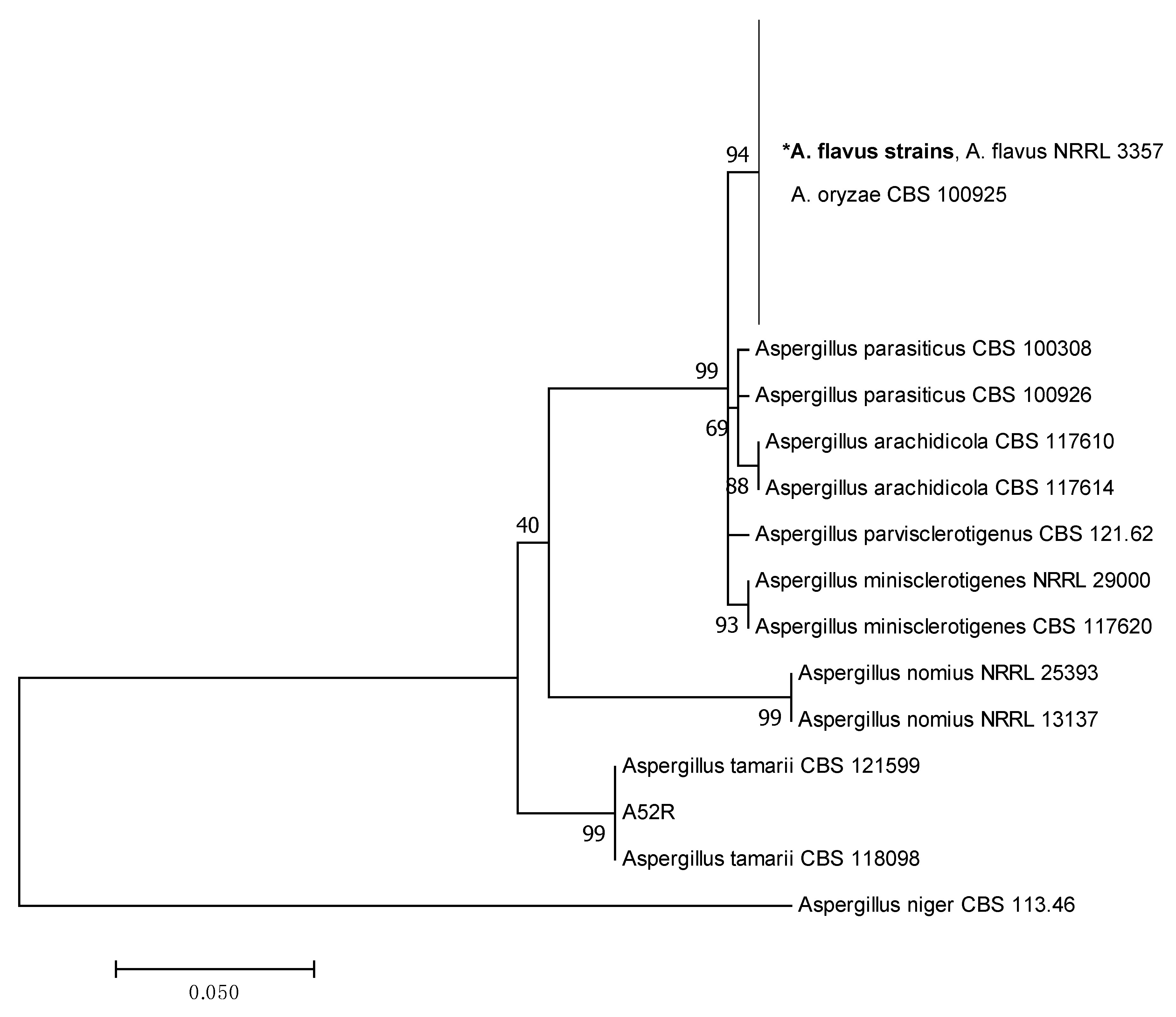
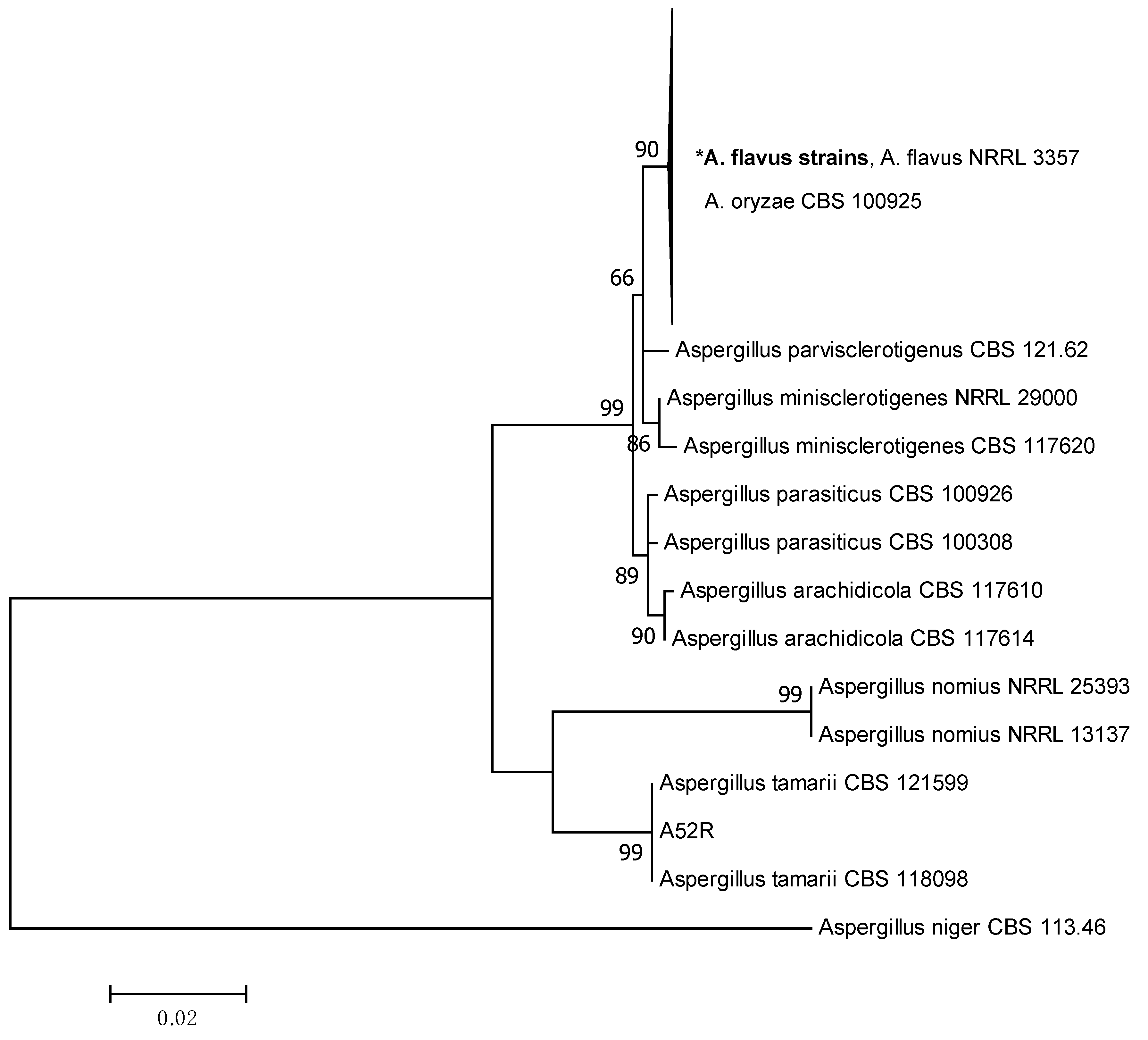
2.2. Phylogenetic Analysis
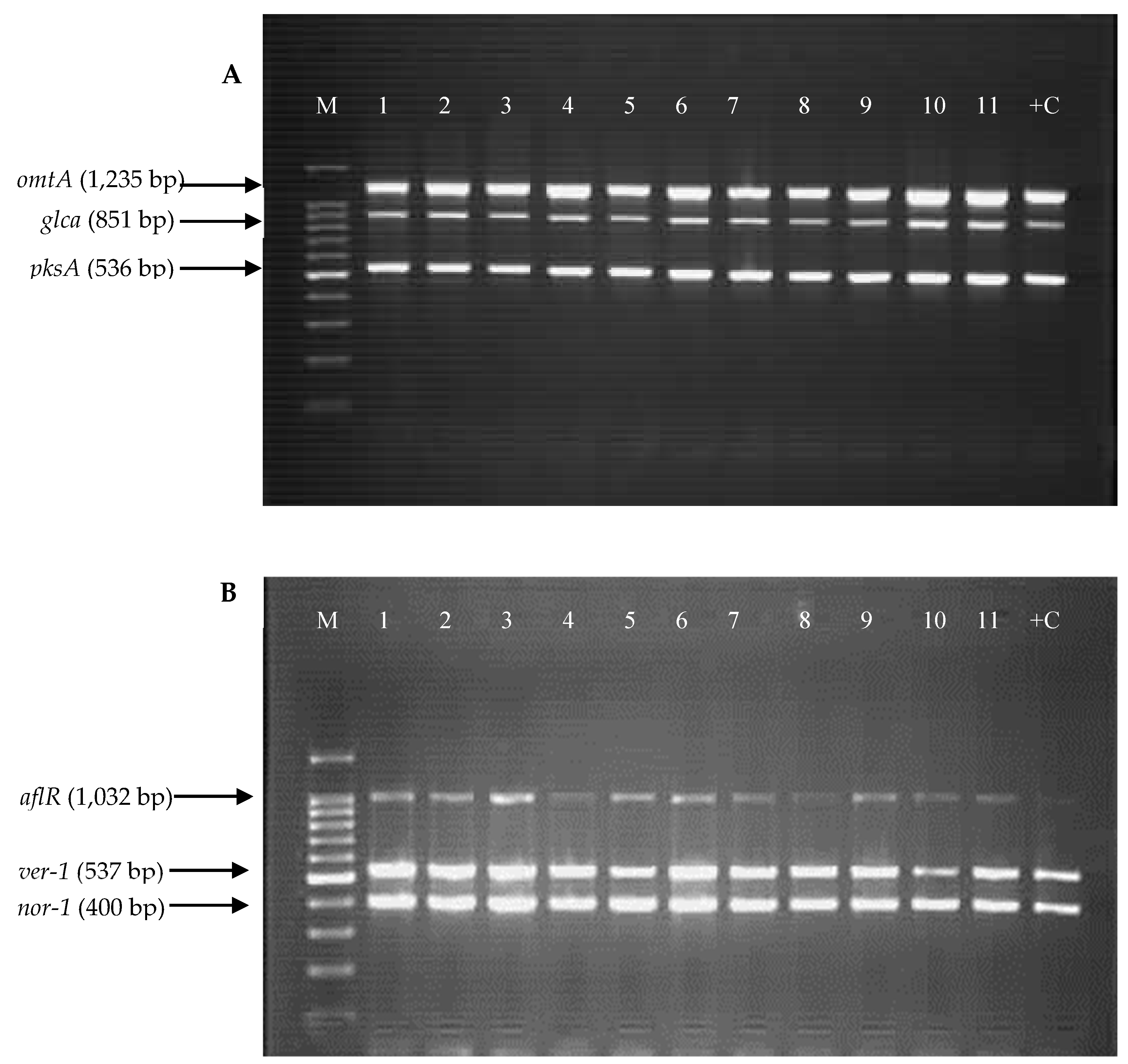
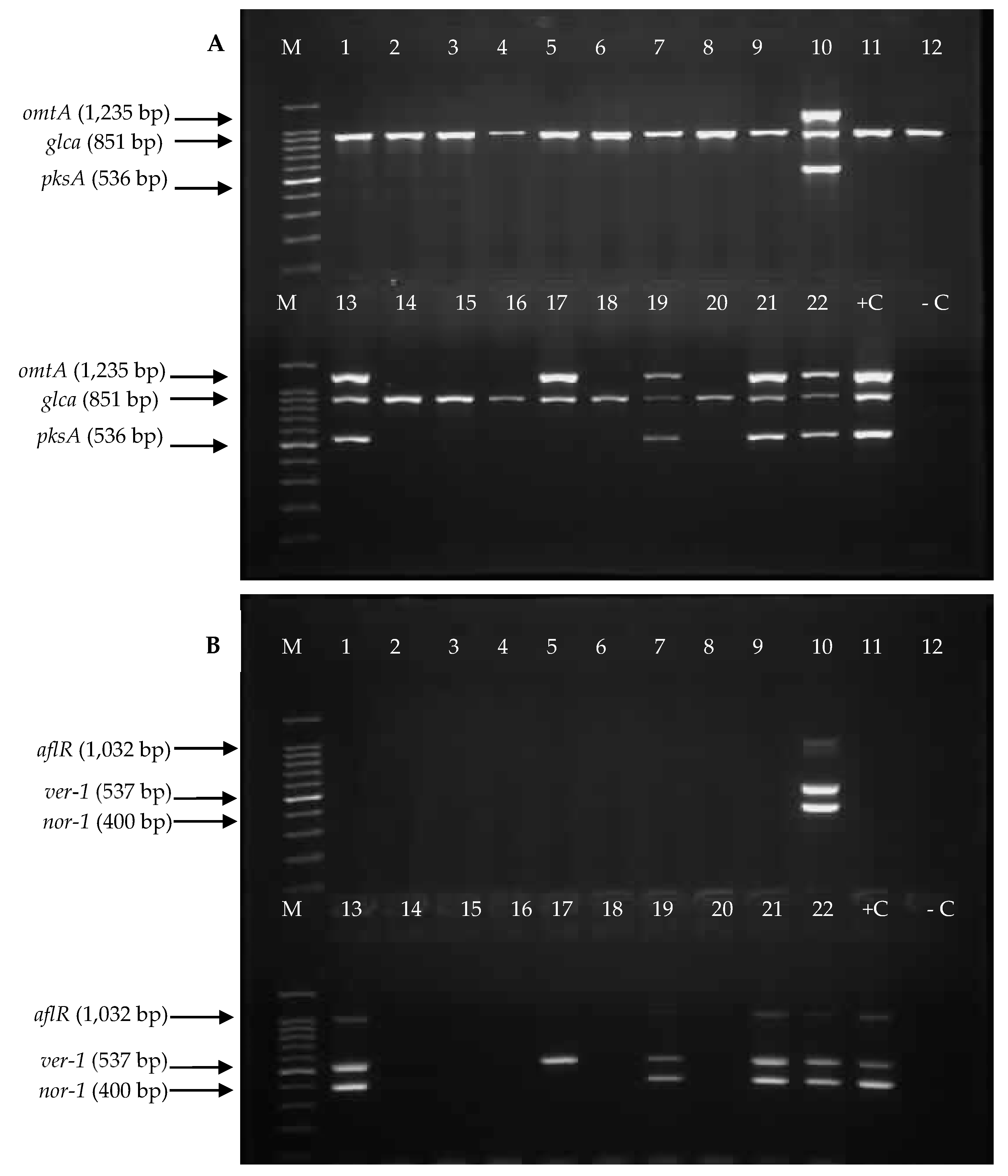
2.3. Detection of Aflatoxin Biosynthesis Genes in Aspergillus Section Flavi Strains
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Fungal Isolates
5.2. Molecular Identification of Aspergillus Section Flavi
5.2.1. Genomic DNA Extraction
5.2.2. PCR Amplification and Sequencing of ITS and β-Tubulin Genes
5.2.3. Sequence Alignment and Species Identification
5.3. Phylogenetic Analysis
5.4. PCR Amplification and Detection of Aflatoxin Biosynthesis Genes
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gnonlonfin, G.J.B.; Hell, K.; Adjovi, Y.; Fandohan, P.; Koudande, D.O.; Mensah, G.A.; Sanni, A.; Brimer, L. A review on aflatoxin contamination and its implications in the developing world: A sub-Saharan African perspective. Crit. Rev. Food Sci. Nutr. 2013, 53, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye, A.; Misore, A.; et al. Aflatoxin contamination of commercial maize products during an outbreak of acute aflatoxicoses in Eastern and Central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- IARC Aflatoxins. In Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Humans; International Agency for Research on Cancer: Lyon, France, 1993; Volume 56, pp. 245–395. ISBN 9283212568.
- Godet, M.; Munaut, F. Molecular strategy for identification in Aspergillus section Flavi. FEMS Microbiol. Lett. 2010, 304, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Nyirahakizimana, H.; Mwamburi, L.; Wakhisi, J.; Mutegi, C.K.; Christie, M.E.; Wagacha, J.M. Occurrence of Aspergillus species and aflatoxin contamination in raw and roasted peanuts from formal and informal markets in Eldoret and Kericho, Kenya. Adv. Microbiol. 2013, 03, 333–342. [Google Scholar] [CrossRef][Green Version]
- Ozbey, F.; Kabak, B. Natural co-occurrence of aflatoxins and ochratoxin A in spices. Food Control 2012, 28, 354–361. [Google Scholar] [CrossRef]
- Reddy, K.R.N.; Farhana, N.I.; Salleh, B. Occurrence of Aspergillus spp. and aflatoxin B1 in Malaysian foods used for human consumption. J. Food Sci. 2011, 76, 99–104. [Google Scholar] [CrossRef]
- Kim, D.M.; Chung, S.H.; Chun, H.S. Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in meju, a Korean fermented soybean food starter. Food Microbiol. 2011, 28, 1402–1408. [Google Scholar] [CrossRef]
- Kim, K.M.; Lim, J.; Lee, J.J.; Hurh, B.S.; Lee, I. Characterization of aspergillus sojae isolated from meju, korean traditional fermented soybean brick. J. Microbiol. Biotechnol. 2017, 27, 251–261. [Google Scholar] [CrossRef]
- Peterson, S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 2008, 100, 205–226. [Google Scholar] [CrossRef]
- Pildain, M.B.; Vaamonde, G.; Cabral, D. Analysis of population structure of Aspergillus flavus from peanut based on vegetative compatibility, geographic origin, mycotoxin and sclerotia production. Int. J. Food Microbiol. 2004, 93, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Reis, T.A.; Baquião, A.C.; Atayde, D.D.; Grabarz, F.; Corrêa, B. Characterization of Aspergillus section Flavi isolated from organic Brazil nuts using a polyphasic approach. Food Microbiol. 2014, 42, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Pildain, B.; Frisvad, J.C.; Vaamonde, G.; Cabral, D.; Varga, J.; Samson, R.A. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. Int. J. Syst. Evol. Microbiol. 2008, 58, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Frisvad, J.C.; Samson, R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 2011, 69, 57–80. [Google Scholar] [CrossRef]
- Afsah-Hejri, L.; Jinap, S.; Hajeb, P.; Radu, S.; Shakibazadeh, S. A review on mycotoxins in food and feed: Malaysia case study. Compr. Rev. Food Sci. Food Saf. 2013, 12, 629–651. [Google Scholar] [CrossRef]
- Norlia, M.; Nor-Khaizura, M.A.; Selamat, J.; Abu Bakar, F.; Radu, S.; Chin, C.K. Evaluation of aflatoxins and Aspergillus sp. contamination in raw peanuts and peanut-based products along the supply chain in Malaysia. Food Addit. Contam. Part A 2018, 23, 1–16. [Google Scholar] [CrossRef]
- Norlia, M.; Jinap, S.; Nor-khaizura, M.A.R.; Son, R.; Chin, C.K. Sardjono Polyphasic approach to the identification and characterization of aflatoxigenic strains of Aspergillus section Flavi isolated from peanuts and peanut-based products marketed in Malaysia. Int. J. Food Microbiol. 2018, 282, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Nováková, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenář, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Tam, E.W.T.; Chen, J.H.K.; Lau, E.C.L.; Ngan, A.H.Y.; Fung, K.S.C.; Lee, K.C.; Lam, C.W.; Yuen, K.Y.; Lau, S.K.P.; Woo, P.C.Y. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: Characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser des. J. Clin. Microbiol. 2014, 52, 1153–1160. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D.; Glenn, D.R. An improved medium for the detection fo Aspergillus flavus and A. parasiticus. J. Appl. Bacteriol. 1983, 54, 109–114. [Google Scholar] [CrossRef]
- Prencipe, S.; Siciliano, I.; Contessa, C.; Botta, R.; Garibaldi, A.; Gullino, M.L.; Spadaro, D. Characterization of Aspergillus section Flavi isolated from fresh chestnuts and along the chestnut flour process. Food Microbiol. 2018, 69, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Hong, S.B.; Frisvad, J.C. Old and new concepts of species differentiation in Aspergillus. Med. Mycol. 2006, 44, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.-B.; Hubka, V.; Klaassen, C.H.W.; Perrones, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chang, P.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, W.; et al. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.A.; Nierman, W.C.; Wortman, J.R.; Pritchard, B.L.; Brown, D.; Dean, R.A.; Bhatnagar, D.; Cleveland, T.E.; Machida, M.; Yu, J. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 2006, 44, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Z.; Liou, G.Y.; Yuan, G.F. Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology 2006, 152, 161–170. [Google Scholar] [CrossRef]
- Chang, P.K.; Horn, B.W.; Dorner, J.W. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genet. Biol. 2005, 42, 914–923. [Google Scholar] [CrossRef]
- Yin, Y.; Lou, T.; Yan, L.; Michailides, T.J.; Ma, Z. Molecular characterization of toxigenic and atoxigenic Aspergillus flavus isolates, collected from peanut fields in China. J. Appl. Microbiol. 2009, 107, 1857–1865. [Google Scholar] [CrossRef]
- Chang, P.K.; Ehrlich, K.C. What does genetic diversity of Aspergillus flavus tell us about Aspergillus oryzae? Int. J. Food Microbiol. 2010, 138, 189–199. [Google Scholar] [CrossRef]
- Geiser, D.M.; Pitt, J.I.; Taylor, J.W. Cryptic Speciation and Recombination in the Aflatoxin-producing Fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA 1998, 95, 388–393. [Google Scholar]
- Geiser, D.M.; Dorner, J.W.; Horn, B.W.; Taylor, J.W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 2000, 31, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Yamada, O.; Gomi, K. Genomics of Aspergillus oryzae: Learning from the history of koji mold and exploration of its future. DNA Res. 2008, 15, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.; Sant’Ana, A.S.; Fungaro, M.H.P.; Silva, J.J.; Nascimento, M. da S. do; Frisvad, J.C.; Taniwaki, M.H. The biodiversity of Aspergillus section Flavi and aflatoxins in the Brazilian peanut production chain. Food Res. Int. 2017, 94, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Passone, M.A.; Rosso, L.C.; Ciancio, A.; Etcheverry, M. Detection and quantification of Aspergillus section Flavi spp. in stored peanuts by real-time PCR of nor-1 gene, and effects of storage conditions on aflatoxin production. Int. J. Food Microbiol. 2010, 138, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Wagacha, J.M.; Mutegi, C.; Karanja, L.; Kimani, J.; Christie, M.E. Fungal species isolated from peanuts in major Kenyan markets: Emphasis on Aspergillus section Flavi. Crop Prot. 2013, 52, 1–9. [Google Scholar] [CrossRef]
- Rank, C.; Klejnstrup, M.L.; Petersen, L.M.; Kildgaard, S.; Frisvad, J.C.; Held Gotfredsen, C.; Ostenfeld Larsen, T. Comparative chemistry of Aspergillus oryzae (RIB40) and A. flavus (NRRL 3357). Metabolites 2012, 2, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Wicklow, D.T.; Wilson, D.M.; Nelsen, T.C. Survival of Aspergillus flavus sclerotia and conidia buried in soil in Illinois or Georgia. Phytopathology 1993, 83, 1141–1147. [Google Scholar] [CrossRef]
- Horn, B.; Sorensen, R.; Lamb, M.; Sobolev, V.; Olarte, R.; Worthington, C.; Carbone, I. Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 2014, 104, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Wicklow, D.T.; Ito, Y. Aflatoxin and cyclopiazonic acid production by a sclerotium-producing Aspergillus tamarii strain. Appl. Environ. Microbiol. 1996, 62, 4036–4038. [Google Scholar]
- Ito, Y.; Peterson, S.W.; Wicklow, D.T.; Goto, T. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 2001, 105, 233–239. [Google Scholar] [CrossRef]
- Gallo, A.; Stea, G.; Battilani, P.; Logrieco, A.F.; Perrone, G. Molecular characterization of an Aspergillus flavus population isolated from maize during the first outbreak of aflatoxin contamination in Italy. Phytopathol. Mediterr. 2012, 51, 198–206. [Google Scholar]
- Erami, M.; Hashemi, S.; Pourbakhsh, S.; Shahsavandi, S.; Mohammadi, S.; Shooshtari, A.; Jahanshiri, Z. Application of PCR on detection of aflatoxinogenic fungi. Arch. Razi Inst. 2007, 62, 95–100. [Google Scholar]
- Davari, E.; Mohsenzadeh, M.; Mohammadi, G.; Rezaeian-Doloei, R. Characterization of aflatoxigenic Aspergillus flavus and A. parasiticus strain isolates from animal feedstuffs in northeastern Iran. Iran. J. Vet. Res. 2015, 16, 150–155. [Google Scholar] [PubMed]
- Abdel-Hadi, A.; Carter, D.; Magan, N. Temporal monitoring of the nor-1 (aflD) gene of Aspergillus flavus in relation to aflatoxin B1 production during storage of peanuts under different water activity levels. J. Appl. Microbiol. 2010, 109, 1914–1922. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Venâncio, A.; Kozakiewicz, Z.; Lima, N. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus Section Flavi isolated from Portuguese almonds. Int. J. Food Microbiol. 2009, 129, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Criseo, G.; Bagnara, A.; Bisignano, G. Differentiation of aflatoxin-producing and non-producing strains of Aspergillus favus group. Lett. Appl. Microbiol. 2001, 33, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Heydt, M.; Abdel-Hadi, A.; Magan, N.; Geisen, R. Complex regulation of the aflatoxin biosynthesis gene cluster of Aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 2009, 135, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hadi, A.; Carter, D.; Magan, N. Discrimination between aflatoxigenic and non-aflatoxigenic strains of Aspergillus section Flavi group contaminationg Egyption peanuts using molecular and analystical techniques. World Mycotoxin J. 2011, 4, 69–77. [Google Scholar] [CrossRef]
- Mutegi, C.K.; Ngugi, H.K.; Hendriks, S.L.; Jones, R.B. Factors associated with the incidence of Aspergillus section Flavi and aflatoxin contamination of peanuts in the Busia and Homa bay districts of western Kenya. Plant Pathol. 2012, 61, 1143–1153. [Google Scholar] [CrossRef]
- Zorzete, P.; Baquião, A.C.; Atayde, D.D.; Reis, T.A.; Gonçalez, E.; Corrêa, B. Mycobiota, aflatoxins and cyclopiazonic acid in stored peanut cultivars. Food Res. Int. 2013, 52, 380–386. [Google Scholar] [CrossRef]
- Kachapulula, P.W.; Akello, J.; Bandyopadhyay, R.; Cotty, P.J. Aspergillus section Flavi community structure in Zambia influences aflatoxin contamination of maize and groundnut. Int. J. Food Microbiol. 2017, 261, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.M.; Barros, G.G.; Palacios, S.A.; Chulze, S.N.; Battilani, P. Review on pre- and post-harvest management of peanuts to minimize aflatoxin contamination. Food Res. Int. 2014, 62, 11–19. [Google Scholar] [CrossRef]
- Wagacha, J.M.; Mutegi, C.K.; Christie, M.E.; Karanja, L.W.; Kimani, J. Changes in fungal population and aflatoxin levels and assessment of major aflatoxin types in stored peanuts (Arachis hypogaea Linnaeus). J. Food Res. 2013, 2, 10–23. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donalson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols. A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. ISBN 008088671X. [Google Scholar]
- Shapira, R.; Paster, N.; Eyal, O.; Menasherov, M.; Mett, A.; Salomon, R. Detection of aflatoxigenic molds in grains by PCR. Appl. Environ. Microbiol. 1996, 62, 3270–3273. [Google Scholar]
- Siti-Aminah, I. Screening of Toxigenic and Atoxigenic Aspergillus flavus Isolates Collected from Corn Fields in UPM, Serdang. Master’s Thesis, Universiti Putra Malaysia, Selangor, Malaysia, 2017. [Google Scholar]

| Isolate No. | Species | Stakeholder | * Source | ** Chemotype | GenBank Accession Number | |
|---|---|---|---|---|---|---|
| ITS | β-Tubulin | |||||
| A8R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095114 | MN148806 |
| A34R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095115 | MN148807 |
| A35R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095116 | MN148808 |
| A42R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095117 | MN148809 |
| A45R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095118 | MN148810 |
| A46R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095119 | MN148811 |
| A47R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095120 | MN148812 |
| A50R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095121 | MN148813 |
| A53R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095122 | MN148814 |
| A54R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095123 | MN148815 |
| A55R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095124 | MN148816 |
| A57R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095125 | MN148817 |
| A58R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095126 | MN148818 |
| A59R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095127 | MN148819 |
| A60R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095128 | MN148820 |
| A61R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095129 | MN148821 |
| A63R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095130 | MN148822 |
| A68R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095131 | MN148823 |
| A74R | A. flavus | Manufacturer | Raw peanut kernel (India) | I | MN095132 | MN148824 |
| A81R | A. flavus | Manufacturer | Raw peanut kernel (China) | I | MN095133 | MN148825 |
| A90R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095134 | MN148826 |
| A91R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095135 | MN148827 |
| A92R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095136 | MN148828 |
| A95R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095137 | MN148829 |
| A96R | A. flavus | Retailer | Raw peanut kernel (India) | I | MN095138 | MN148830 |
| A116P | A. flavus | Retailer | Peanut-based product (roasted peanut) | I | MN095139 | MN148831 |
| A87R | A. flavus | Manufacturer | Raw peanut kernel (China) | II | MN095140 | MN148832 |
| A88R | A. flavus | Manufacturer | Raw peanut kernel (China) | II | MN095141 | MN148833 |
| A5R | A. flavus | Importer | Raw peanut kernel (China) | III | MN095142 | MN148834 |
| A24R | A. flavus | Importer | Raw peanut kernel (China) | III | MN095143 | MN148835 |
| A40R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095144 | MN148836 |
| A43R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095145 | MN148837 |
| A48R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095146 | MN148838 |
| A49R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095147 | MN148839 |
| A71R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095148 | MN148840 |
| A73R | A. flavus | Manufacturer | Raw peanut kernel (India) | III | MN095149 | MN148841 |
| A77R | A. flavus | Importer | Raw peanut in shell (Indonesia) | III | MN095150 | MN148842 |
| A78R | A. flavus | Manufacturer | Raw peanut kernel (China) | III | MN095151 | MN148843 |
| A94R | A. flavus | Retailer | Raw peanut kernel (India) | III | MN095152 | MN148844 |
| A97R | A. flavus | Retailer | Raw peanut kernel (India) | III | MN095153 | MN148845 |
| A108P | A. flavus | Manufacturer | Peanut-based product (roasted peanut) | III | MN095154 | MN148846 |
| A109P | A. flavus | Retailer | Peanut-based product (peanut candy) | III | MN095155 | MN148847 |
| A118P | A. flavus | Retailer | Peanut-based product (roasted peanut) | III | MN095156 | MN148848 |
| A9R | A. flavus | Retailer | Raw peanut kernel (China) | IV | MN095157 | MN148849 |
| A12R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095158 | MN148850 |
| A13R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095159 | MN148851 |
| A14R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095160 | MN148852 |
| A16R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095161 | MN148853 |
| A19R | A. flavus | Importer | Raw peanut in shell (China) | IV | MN095162 | MN148854 |
| A20R | A. flavus | Importer | Raw peanut in shell (China) | IV | MN095163 | MN148855 |
| A21R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095164 | MN148856 |
| A22R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095165 | MN148857 |
| A23R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095166 | MN148858 |
| A26R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095167 | MN148859 |
| A27R | A. flavus | Importer | Raw peanut kernel (China) | IV | MN095168 | MN148860 |
| A67R | A. flavus | Manufacturer | Raw peanut kernel (India) | IV | MN095169 | MN148861 |
| A75R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095170 | MN148862 |
| A76R | A. flavus | Importer | Raw peanut in shell (Vietnam) | IV | MN095171 | MN148863 |
| A98P | A. flavus | Manufacturer | Peanut-based product (peanut snack) | IV | MN095172 | MN148864 |
| A104P | A. flavus | Manufacturer | Peanut-based product (peanut sauce) | IV | MN095173 | MN148865 |
| A111P | A. flavus | Retailer | Peanut-based product (peanut snack) | IV | MN095174 | MN148866 |
| A114P | A. flavus | Retailer | Peanut-based product (roasted peanut) | IV | MN095175 | MN148867 |
| A115P | A. flavus | Retailer | Peanut-based product (roasted peanut) | IV | MN095176 | MN148868 |
| A122R | A. flavus | Retailer | Raw peanut kernel (India) | IV | MN095177 | MN148869 |
| A123R | A. flavus | Retailer | Raw peanut kernel (India) | IV | MN095178 | MN148870 |
| A1R | A. flavus | Importer | Raw peanut kernel (India) | V | MN095179 | MN148871 |
| A15R | A. flavus | Importer | Raw peanut kernel (China) | V | MN095180 | MN148872 |
| A25R | A. flavus | Importer | Raw peanut kernel (China) | V | MN095181 | MN148873 |
| A29R | A. flavus | Importer | Raw peanut in shell (China) | V | MN095182 | MN148874 |
| A41R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095183 | MN148875 |
| A44R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095184 | MN148876 |
| A69R | A. flavus | Manufacturer | Raw peanut kernel (India) | V | MN095185 | MN148877 |
| A80R | A. flavus | Manufacturer | Raw peanut kernel (China) | V | MN095186 | MN148878 |
| A82R | A. flavus | Manufacturer | Raw peanut kernel (China) | V | MN095187 | MN148879 |
| A102P | A. flavus | Manufacturer | Peanut-based product (peanut cookies) | V | MN095188 | MN148880 |
| A107P | A. flavus | Manufacturer | Peanut-based product (peanut cookies) | V | MN095189 | MN148881 |
| A52R | A. tamarii | Manufacturer | Raw peanut in shell (China) | VI | MN095190 | MN148882 |
| A. flavus NRRL 3357 | n.a | n.a | MF966967 | M38265 | ||
| A. oryzae CBS 100925 | n.a | n.a | KJ175432 | EF203138 | ||
| A. minisclerotigenes NRRL 29000 | n.a | n.a | KY937929 | KY924668 | ||
| A. minisclerotigenes CBS 117620 | n.a | n.a | JF422073 | EF203150 | ||
| A. parvisclerotigenus CBS 121.62 | n.a | n.a | EF409240 | EF203130 | ||
| A. parasiticus CBS 100926 | n.a | n.a | KJ175437 | KJ175497 | ||
| A. parasiticus CBS 100308 | n.a | n.a | KJ175436 | KJ175496 | ||
| A. nomius NRRL 25393 | n.a | n.a | AF027864 | AF255068 | ||
| A. nomius NRRL 13137 | n.a | n.a | AF027860 | AF255067 | ||
| A. tamarii CBS 121599 | n.a | n.a | KJ175443 | KJ175501 | ||
| A. tamarii CBS 118098 | n.a | n.a | KJ175442 | KJ175500 | ||
| A. arachidicola CBS 117610 | n.a | n.a | EF409241 | EF203158 | ||
| A. arachidicola CBS 117614 | n.a | n.a | KY937923 | KY924665 | ||
| A. niger CBS 113.46 | n.a | n.a | FJ629351 | FJ629302 | ||
| No. | Strain | * Chemotype | Aflatoxin Biosynthesis Gene | |||||
|---|---|---|---|---|---|---|---|---|
| aflR | aflP (omtA) | aflD (nor-1) | aflM (ver-1) | pksA | glcA | |||
| 1 | A8R | I | + | + | + | + | + | + |
| 2 | A34R | I | + | + | + | + | + | + |
| 3 | A35R | I | + | + | + | + | + | + |
| 4 | A42R | I | + | + | + | + | + | + |
| 5 | A45R | I | + | + | + | + | + | + |
| 6 | A46R | I | + | + | + | + | + | + |
| 7 | A47R | I | + | + | + | + | + | + |
| 8 | A50R | I | + | + | + | + | + | + |
| 9 | A53R | I | + | + | + | + | + | + |
| 10 | A54R | I | + | + | + | + | + | + |
| 11 | A55R | I | + | + | + | + | + | + |
| 12 | A57R | I | + | + | + | + | + | + |
| 13 | A58R | I | + | + | + | + | + | + |
| 14 | A59R | I | + | + | + | + | + | + |
| 15 | A60R | I | + | + | + | + | + | + |
| 16 | A61R | I | + | + | + | + | + | + |
| 17 | A63R | I | + | + | + | + | + | + |
| 18 | A68R | I | + | + | + | + | + | + |
| 19 | A74R | I | + | + | + | + | + | + |
| 20 | A81R | I | + | + | + | + | + | + |
| 21 | A90R | I | + | + | + | + | + | + |
| 22 | A91R | I | + | + | + | + | + | + |
| 23 | A92R | I | + | + | + | + | + | + |
| 24 | A95R | I | + | + | + | + | + | + |
| 25 | A96R | I | + | + | + | + | + | + |
| 26 | A116P | I | + | + | + | + | + | + |
| 27 | A87R | II | + | + | + | + | + | + |
| 28 | A88R | II | + | + | + | + | + | + |
| 29 | A5R | III | + | + | + | + | + | + |
| 30 | A24R | III | + | + | + | + | + | + |
| 31 | A40R | III | + | + | + | − | + | + |
| 32 | A43R | III | + | + | + | − | + | + |
| 33 | A48R | III | + | + | + | − | + | + |
| 34 | A49R | III | + | + | + | + | + | + |
| 35 | A71R | III | + | + | + | + | + | + |
| 36 | A73R | III | + | + | + | + | + | + |
| 37 | A77R | III | + | + | + | + | + | + |
| 38 | A78R | III | + | + | + | + | + | + |
| 39 | A94R | III | + | + | + | + | + | + |
| 40 | A97R | III | + | + | + | + | + | + |
| 41 | A108P | III | + | − | + | + | + | + |
| 42 | A109P | III | + | + | + | + | + | + |
| 43 | A118P | III | + | + | + | + | + | + |
| 44 | A9R | IV | − | − | − | − | − | + |
| 45 | A12R | IV | − | − | − | − | − | + |
| 46 | A13R | IV | − | − | − | − | − | + |
| 47 | A14R | IV | − | − | − | − | − | + |
| 48 | A16R | IV | − | − | − | − | − | + |
| 49 | A19R | IV | − | − | − | − | − | + |
| 50 | A20R | IV | − | − | − | − | − | + |
| 51 | A21R | IV | − | − | − | − | − | + |
| 52 | A22R | IV | − | − | − | − | − | + |
| 53 | A23R | IV | + | + | + | + | + | + |
| 54 | A26R | IV | − | − | − | − | − | + |
| 55 | A27R | IV | − | − | − | − | − | + |
| 56 | A67R | IV | + | + | + | + | + | + |
| 57 | A75R | IV | − | − | − | − | − | + |
| 58 | A76R | IV | − | − | − | − | − | + |
| 59 | A98P | IV | − | − | − | − | − | + |
| 60 | A104P | IV | − | + | − | + | − | + |
| 61 | A111P | IV | − | − | − | − | − | + |
| 62 | A114P | IV | − | + | + | + | + | + |
| 63 | A115P | IV | − | − | − | − | − | + |
| 64 | A122R | IV | + | + | + | + | + | + |
| 65 | A123R | IV | + | + | + | + | + | + |
| 66 | A1R | V | + | + | + | + | + | + |
| 67 | A15R | V | + | + | + | + | + | + |
| 68 | A25R | V | + | + | + | + | + | + |
| 69 | A29R | V | + | + | + | + | + | + |
| 70 | A41R | V | + | + | + | + | + | + |
| 71 | A44R | V | + | + | + | + | + | + |
| 72 | A69R | V | + | + | + | + | + | + |
| 73 | A80R | V | + | + | + | + | + | + |
| 74 | A82R | V | + | + | + | + | + | + |
| 75 | A102P | V | + | + | + | + | + | + |
| 76 | A107P | V | + | + | + | + | + | + |
| 77 | A52R | VI | + | + | + | + | + | + |
| Target Gene | Primer | Primer Sequences | Size (bp) | References |
|---|---|---|---|---|
| ITS region | ITS 1 | 5′-TCC GTA GGT GAA CCT GCG G-3′ | 600 | [57,58] |
| ITS 4 | 5′-TCC TCC GCT TAT TGA TAT GC-3′ | |||
| β-tubulin | Bt2a | 5′- GGT AAC CAA ATC GGT GCT GCT TTC-3′ | 495 | [57] |
| Bt2b | 5′-ACC CTC AGT GTA GTG ACC CTT GGC-3′ | |||
| aflR | aflr1 | 5′-TAT CTC CCC CCG GGC ATC TCC CGG-3′ | 1032 | [48,59] |
| aflr2 | 5′-CCG TCA GAC AGC CAC TGG ACA CGG-3′ | |||
| aflP (omtA) | omt1 | 5′-GGC CCG GTT CCT TGG CTC CTA AGC-3′ | 1024 | [59] |
| omt2 | 5′-CGC CCC AGT GAG ACC CTT CCT CG-3′ | |||
| aflD (nor-1) | nor1 | 5′-ACC GCT ACG CCG GCA CTC TCG GCA C-3′ | 400 | [48] |
| nor2 | 5′-GTT GGC CGC CAG CTT CGA CAC TCC G-3′ | |||
| aflM (ver-1) | ver1 | 5′-GCC GCA GGC CGC GGA GAA AGT GGT-3′ | 537 | [48] |
| ver2 | 5′-GGG GAT ATA CTC CCG CGA CAC AGC C-3′ | |||
| pksA | pksa1 | 5′-GCT GGG ATT CTG CAT GGG TT-3′ | 536 | [30] |
| pksa2 | 5′-CAG TTG CTC CCA AGG AGT GGT-3′ | |||
| glcA | glca1 | 5′-GTA CGA TGC AAA TGG CGT CC-3′ | 851 | [60] |
| glca2 | 5′-GAA GCT CTG TGT CGT TGG GA-3′ |
| Set | Primers | PCR Reaction Condition | Cycle |
|---|---|---|---|
| 1 | omt1/omt2, pksa1/pksa2, glca1/glca2 | Initial denaturation: 95 °C, 1 min Denaturation: 95 °C, 1 min Annealing: 61 °C, 1 min Extension: 72 °C, 1 min Final extension: 72 °C, 5 min | 1 30 30 30 1 |
| 2 | aflr1/aflr2, ver1/ver2, nor1/nor2 | Initial denaturation: 95 °C, 1 min Denaturation: 95 °C, 1 min Annealing: 67 °C, 1 min Extension: 72 °C, 1 min Final extension: 72 °C, 5 min | 1 30 30 30 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norlia, M.; Jinap, S.; Nor-Khaizura, M.A.R.; Radu, S.; Chin, C.K.; Samsudin, N.I.P.; Farawahida, A.H. Molecular Characterisation of Aflatoxigenic and Non-Aflatoxigenic Strains of Aspergillus Section Flavi Isolated from Imported Peanuts along the Supply Chain in Malaysia. Toxins 2019, 11, 501. https://doi.org/10.3390/toxins11090501
Norlia M, Jinap S, Nor-Khaizura MAR, Radu S, Chin CK, Samsudin NIP, Farawahida AH. Molecular Characterisation of Aflatoxigenic and Non-Aflatoxigenic Strains of Aspergillus Section Flavi Isolated from Imported Peanuts along the Supply Chain in Malaysia. Toxins. 2019; 11(9):501. https://doi.org/10.3390/toxins11090501
Chicago/Turabian StyleNorlia, Mahror, Selamat Jinap, Mahmud Ab Rashid Nor-Khaizura, Son Radu, Cheow Keat Chin, Nik Iskandar Putra Samsudin, and Abdul Halim Farawahida. 2019. "Molecular Characterisation of Aflatoxigenic and Non-Aflatoxigenic Strains of Aspergillus Section Flavi Isolated from Imported Peanuts along the Supply Chain in Malaysia" Toxins 11, no. 9: 501. https://doi.org/10.3390/toxins11090501
APA StyleNorlia, M., Jinap, S., Nor-Khaizura, M. A. R., Radu, S., Chin, C. K., Samsudin, N. I. P., & Farawahida, A. H. (2019). Molecular Characterisation of Aflatoxigenic and Non-Aflatoxigenic Strains of Aspergillus Section Flavi Isolated from Imported Peanuts along the Supply Chain in Malaysia. Toxins, 11(9), 501. https://doi.org/10.3390/toxins11090501





