Bacteriocin Occurrence and Activity in Escherichia coli Isolated from Bovines and Wastewater
Abstract
1. Introduction
2. Results
2.1. Functional Screening and Genomic Identification of E. coli Bacteriocins
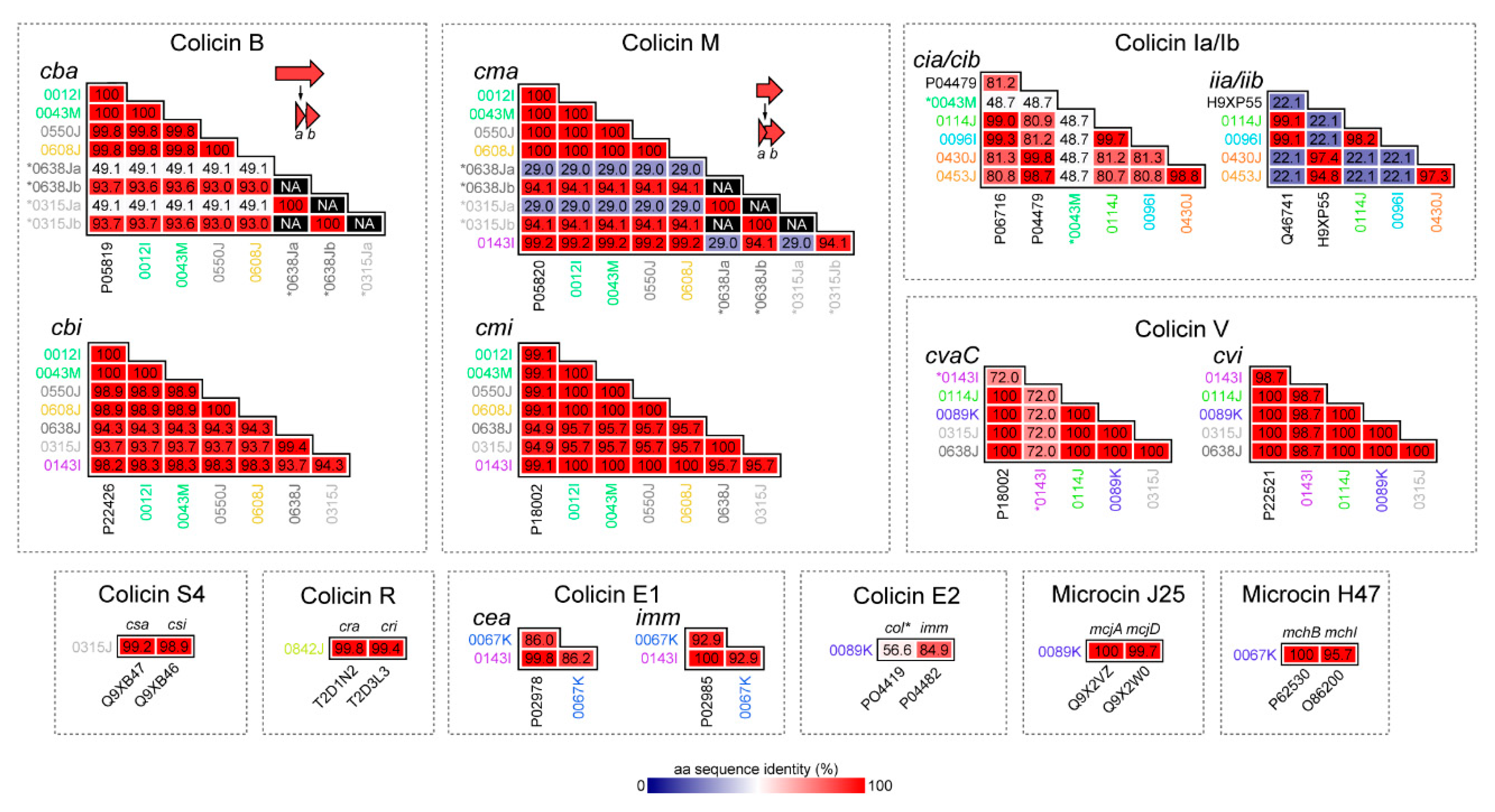
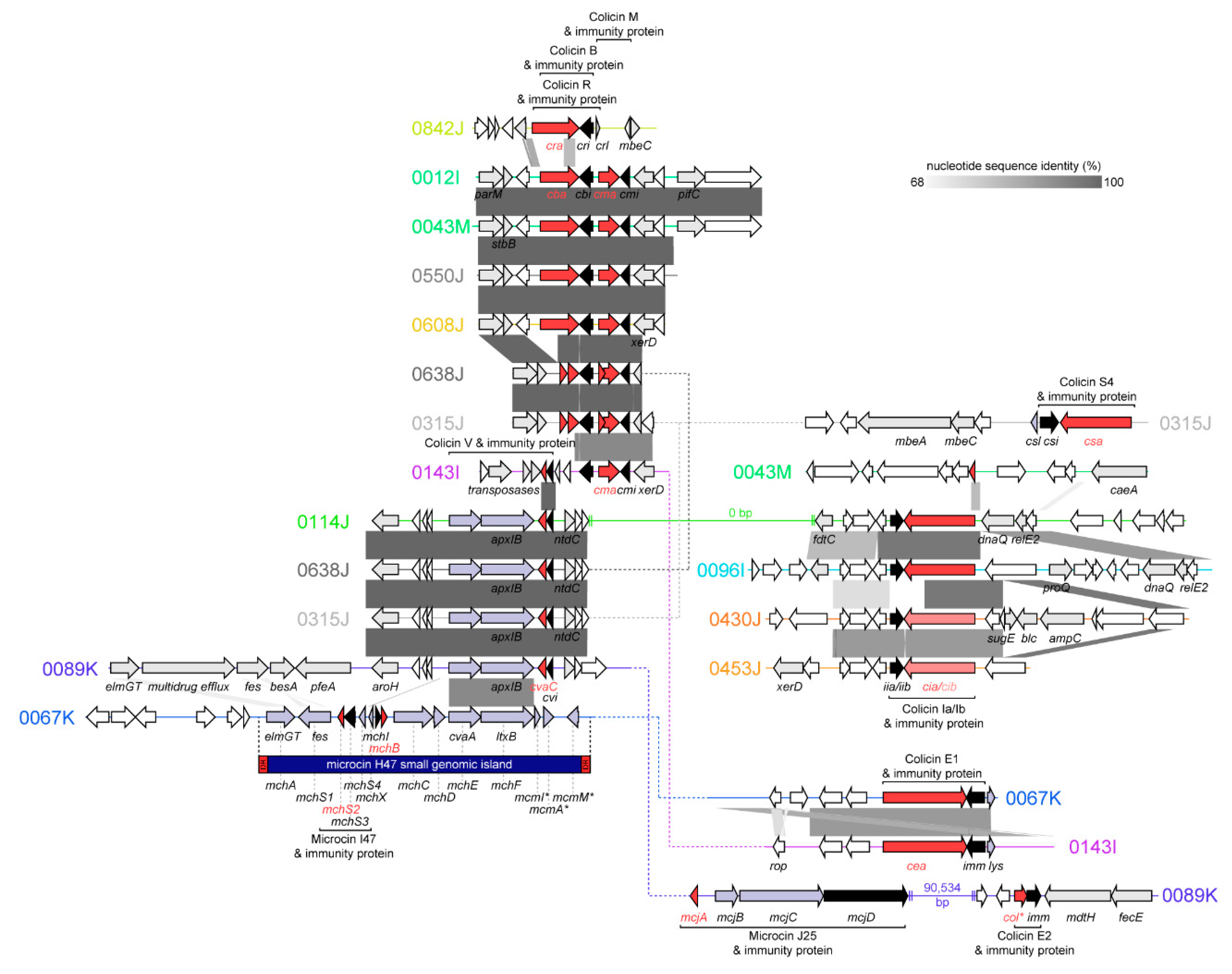
2.2. Bacteriocins Identified, Genomic Context, and Similarity to Known Colicins and Microcins
2.2.1. Colicins B and M
2.2.2. Colicin S4
2.2.3. Colicin R
2.2.4. Colicins Ia and Ib
2.2.5. Colicins E1 and E2
2.2.6. Colicin V (Microcin)
2.2.7. Microcin J25
2.2.8. Microcin H47
2.3. Activity against Shiga Toxin-Producing E. coli and Other Enteric Pathogens
2.4. Activity of Bacteriocin-Producing E. coli against Antimicrobial Resistant Isolates from Bovines and Wastewater
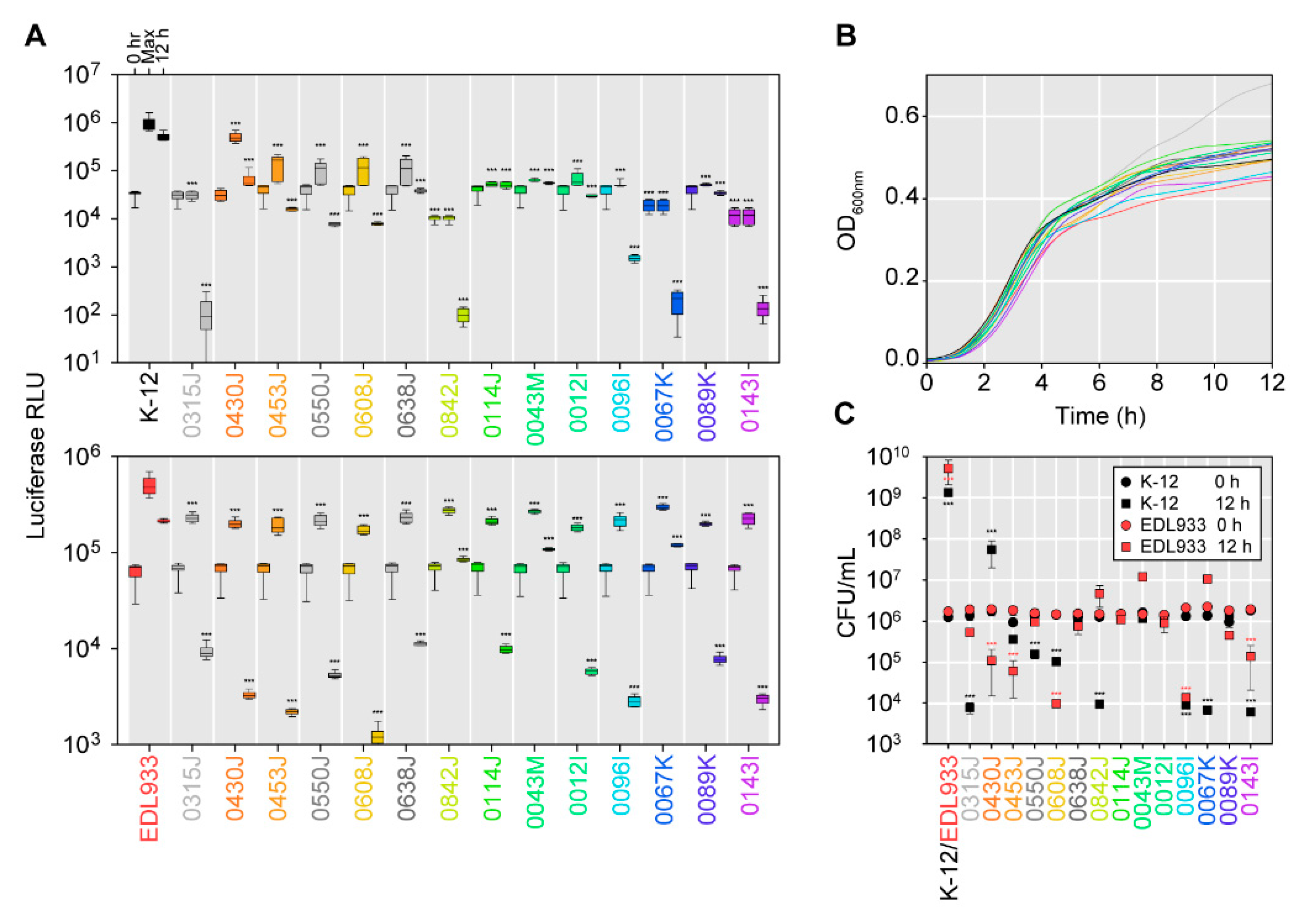
2.5. Effectiveness of Bacteriocin-Producers in Broth-Based Competitions with E. coli K-12 Strain MG1655 or the Shiga Toxin-Producing E. coli Strain EDL933
3. Discussion
4. Materials and Methods
4.1. Enrichment and Isolation of E. coli
4.2. Screening for Bacteriocin Production and Activity against Enteric Bacteria
4.3. Antimicrobial Susceptibility Assays
4.4. Broth-Based Co-culture and Activity Assay with Luciferase-Expressing E. coli
4.5. Whole-Genome Sequencing and Bioinformatic Identification of Bacteriocin Genes
4.6. Data Visualization and Statistical Analyses
4.7. Nucleotide Sequence Accession Numbers
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flaherty, R.A.; Freed, S.D.; Lee, S.W. The wide world of ribosomally encoded bacterial peptides. PLoS Pathog. 2014, 10, e1004221. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubes, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Focus on modified microcins: Structural features and mechanisms of action. Biochimie 2002, 84, 511–519. [Google Scholar] [CrossRef]
- Gordon, D.M.; O’Brien, C.L. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 2006, 152, 3239–3244. [Google Scholar] [CrossRef] [PubMed]
- Paquette, S.-J.; Zaheer, R.; Stanford, K.; Thomas, J.; Reuter, T. Competition among Escherichia coli strains for space and resources. Veter. Sci. 2018, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Nolan, E.M.; Walsh, C.T. Investigations of the MceIJ-catalyzed posttranslational modification of the microcin E492 C-terminus: Linkage of ribosomal and nonribosomal peptides to form “trojan horse” antibiotics. Biochemistry 2008, 47, 9289–9299. [Google Scholar] [CrossRef] [PubMed]
- Lazzaroni, J.-C.; Dubuisson, J.-F.; Vianney, A. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 2002, 84, 391–397. [Google Scholar] [CrossRef]
- Suit, J.L.; Fan, M.-L.J.; Sabik, J.F.; Labarre, R.; Luria, S. Alternative forms of lethality in mitomycin C-induced bacteria carrying ColE1 plasmids. Proc. Natl. Acad. Sci. USA 1983, 80, 579–583. [Google Scholar] [CrossRef]
- Azpiroz, M.F.; Laviña, M. Modular structure of microcin H47 and colicin V. Antimicrob. Agents Chemother. 2007, 51, 2412–2419. [Google Scholar] [CrossRef]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Schamberger, G.P.; Diez-Gonzalez, F. Characterization of colicinogenic Escherichia coli strains inhibitory to enterohemorrhagic Escherichia coli. J. Food Prot. 2004, 67, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Wisener, L.; Sargeant, J.; O’connor, A.; Faires, M.; Glass-Kaastra, S. The use of direct-fed microbials to reduce shedding of Escherichia coli O157 in beef cattle: A systematic review and meta-analysis. Zoonoses Public Health 2015, 62, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Lagha, A.B.; Haas, B.; Gottschalk, M.; Grenier, D. Antimicrobial potential of bacteriocins in poultry and swine production. Veter. Res. 2017, 48, 22. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P.; Ross, R. Bacteriocins: Developing innate immunity for food. Nat. Rev. Genet. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Šmajs, D.; Micenková, L.; Šmarda, J.; Vrba, M.; Ševčíková, A.; Vališová, Z.; Woznicová, V. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: Colicin E1 is a potential virulence factor. BMC Microbiol. 2010, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Gillor, O.; Etzion, A.; Riley, M. The dual role of bacteriocins as anti-and probiotics. Appl. Microbiol. Biotechnol. 2008, 81, 591–606. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, J.W.; Guinane, C.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocin production: A relatively unharnessed probiotic trait? F1000Research 2016, 5, 2587. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bano, J.; Gutierrez-Gutierrez, B.; Machuca, I.; Pascual, A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018, 31, e00079-17. [Google Scholar] [CrossRef] [PubMed]
- Oteo, J.; Pérez-Vázquez, M.; Campos, J. Extended-spectrum β-lactamase producing Escherichia coli: Changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 2010, 23, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Hovde, C.J. Enterohemorrhagic Escherichia coli and Other Shiga Toxin-Producing E. coli; American Society for Microbiology (ASM): Washington, DC, USA, 2015. [Google Scholar]
- Hussein, H. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 2007, 85, E63–E72. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.M.; Oliver, E.; Littlefield-Wyer, J. The diversity of bacteriocins in Gram-negative bacteria. In Bacteriocins; Springer: Berlin, Germany, 2007; pp. 5–18. [Google Scholar]
- El Ghachi, M.; Bouhss, A.; Barreteau, H.; Touzé, T.; Auger, G.; Blanot, D.; Mengin-Lecreulx, D. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 2006, 281, 22761–22772. [Google Scholar] [CrossRef] [PubMed]
- Zschüttig, A.; Auerbach, C.; Meltke, S.; Eichhorn, C.; Brandt, M.; Blom, J.; Goesmann, A.; Jarek, M.; Scharfe, M.; Zimmermann, K. Complete sequence of probiotic symbioflor 2 Escherichia coli strain G3/10 and draft sequences of symbioflor 2 E. coli strains G1/2, G4/9, G5, G6/7, and G8. Genome Announc. 2015, 3, e01330-14. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, O.; Beloin, C.; Latour-Lambert, P.; Ghigo, J.-M. A new biofilm-associated colicin with increased efficiency against biofilm bacteria. ISME J. 2014, 8, 1275. [Google Scholar] [CrossRef] [PubMed]
- Cursino, L.; Šmajs, D.; Šmarda, J.; Nardi, R.; Nicoli, J.; Chartone-Souza, E.; Nascimento, A. Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. J. Appl. Microbiol. 2006, 100, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.E.; Walsh, T.R.; Liu, B.T.; Zou, M.T.; Deng, H.; Fang, L.X.; Liao, X.P.; Sun, J.; Liu, Y.H. Complete sequence of the FII plasmid p42-2, carrying blaCTX-M-55, oqxAB, fosA3, and floR from Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 4336–4338. [Google Scholar] [CrossRef] [PubMed]
- Gilson, L.; Mahanty, H.K.; Kolter, R. Genetic analysis of an MDR-like export system: The secretion of colicin V. EMBO J. 1990, 9, 3875–3884. [Google Scholar] [CrossRef] [PubMed]
- Touzain, F.; Le Devendec, L.; De Boisseson, C.; Baron, S.; Jouy, E.; Perrin-Guyomard, A.; Blanchard, Y.; Kempf, I. Characterization of plasmids harboring blaCTX-M and blaCMY genes in E. coli from French broilers. PLoS ONE 2018, 13, e0188768. [Google Scholar] [CrossRef] [PubMed]
- Fricke, W.F.; McDermott, P.F.; Mammel, M.K.; Zhao, S.; Johnson, T.J.; Rasko, D.A.; Fedorka-Cray, P.J.; Pedroso, A.; Whichard, J.M.; LeClerc, J.E. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 2009, 75, 5963–5971. [Google Scholar] [CrossRef]
- Mukhopadhyay, J.; Sineva, E.; Knight, J.; Levy, R.M.; Ebright, R.H. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 2004, 14, 739–751. [Google Scholar] [CrossRef]
- Bountra, K.; Hagelueken, G.; Choudhury, H.G.; Corradi, V.; El Omari, K.; Wagner, A.; Mathavan, I.; Zirah, S.; Wahlgren, W.Y.; Tieleman, D.P. Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 2017, 36, 3062–3079. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, M.F.; Bascuas, T.; Laviña, M. Microcin H47 system: An Escherichia coli small genomic island with novel features. PLoS ONE 2011, 6, e26179. [Google Scholar] [CrossRef] [PubMed]
- Czárán, T.L.; Hoekstra, R.F.; Pagie, L. Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. USA 2002, 99, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Lopez, F.E.; Vincent, P.A.; Zenoff, A.M.; Salomon, R.A.; Farías, R.N. Efficacy of microcin J25 in biomatrices and in a mouse model of Salmonella infection. J. Antimicrob. Chemother. 2007, 59, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ding, X.; Li, N.; Zhang, X.; Zeng, X.; Wang, S.; Liu, H.; Wang, Y.; Jia, H.; Qiao, S. Dietary supplemented antimicrobial peptide microcin J25 improves the growth performance, apparent total tract digestibility, fecal microbiota, and intestinal barrier function of weaned pigs. J. Anim. Sci. 2017, 95, 5064–5076. [Google Scholar] [CrossRef] [PubMed]
- El Kheir, S.M.; Cherrat, L.; Awussi, A.A.; Ramia, N.E.; Taha, S.; Rahman, A.; Passerini, D.; Leroi, F.; Petit, J.; Mangavel, C. High-throughput identification of candidate strains for biopreservation by using bioluminescent Listeria monocytogenes. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Mollenkopf, D.F.; Weeman, M.F.; Daniels, J.B.; Abley, M.J.; Mathews, J.L.; Gebreyes, W.A.; Wittum, T.E. Variable within-and between-herd diversity of CTX-M cephalosporinase-bearing Escherichia coli isolates from dairy cattle. Appl. Environ. Microbiol. 2012, 78, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.; Johnson, R.P.; Alexander, T.W.; McAllister, T.A.; Reuter, T. Influence of season and feedlot location on prevalence and virulence factors of seven serogroups of Escherichia coli in feces of western-Canadian slaughter cattle. PLoS ONE 2016, 11, e0159866. [Google Scholar] [CrossRef] [PubMed]
- Hockett, K.L.; Baltrus, D.A. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J. Vis. Exp. 2017, 119, e55064. [Google Scholar] [CrossRef]
- CLSI. Performance standards for antimicrobial disk susceptibility tests; approved standard twelfth edition. In CLSI Document M02-A12; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume CLSI document M02-A12. [Google Scholar]
- CLSI. Performance standards for antimicrobial susceptibility testing. In CLSI supplement M100S, 26th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Karsi, A.; Lawrence, M.L. Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid 2007, 57, 286–295. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Boil. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE second release: A database and tool platform for bacteriocin characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Montalban-Lopez, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
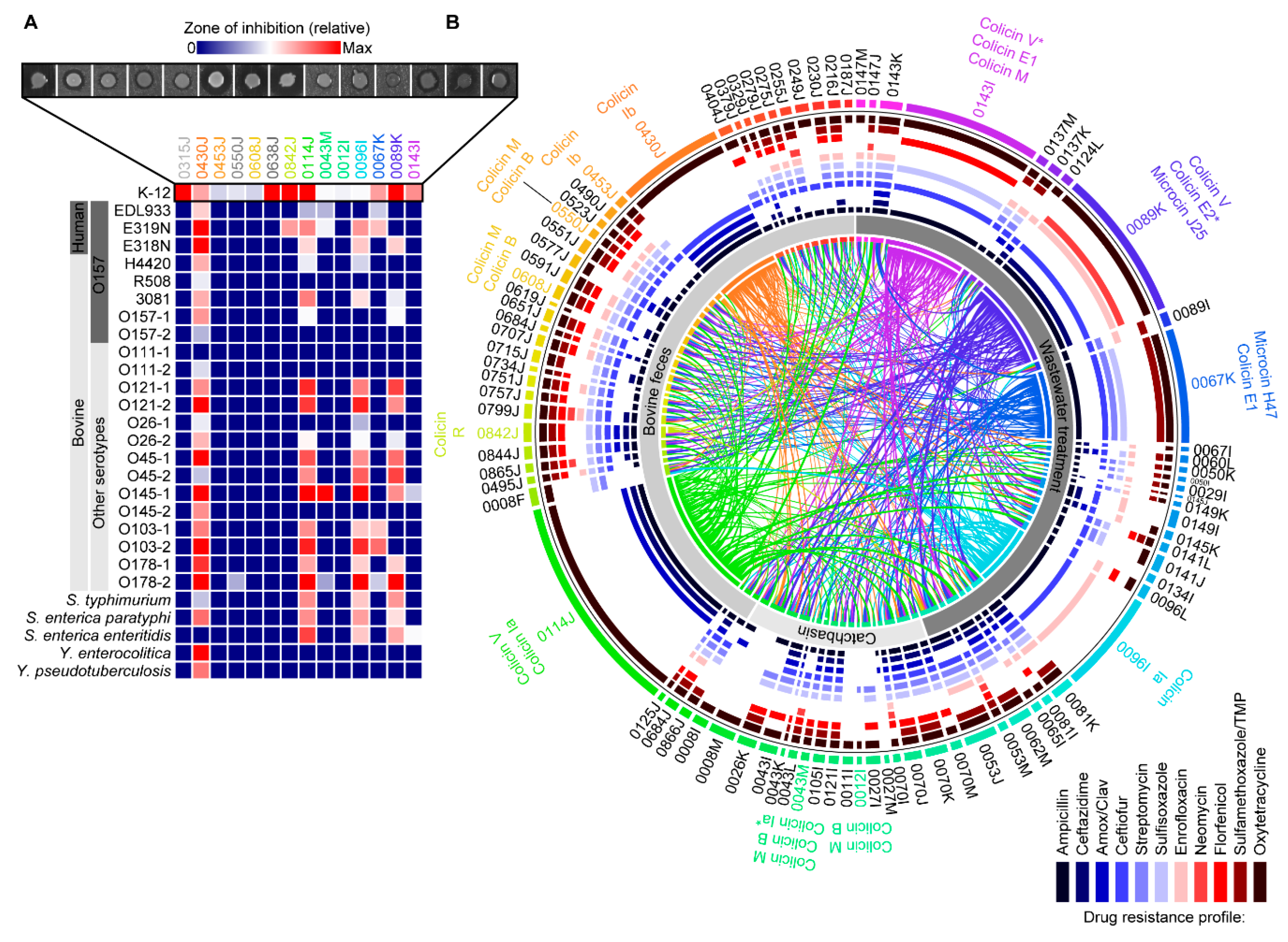
| E. coli Isolate | Isolation Source | AMR Profile1 | AMR Genes2 | Bacteriocins Identified | Contig BLAST Hit3 | Accession Numbers |
|---|---|---|---|---|---|---|
| 0315J | Bovine feces | AMP, AMXC, STR, OXYT | VmacAB | colicin M * colicin B * colicin S4 colicin V | M,BS. enterica Saintpaul pCFSAN004174 S4pSYM12 VS. enterica Kentucky pCVM29188_146 | M,BCP019207 S4KM107848 VCP001122 |
| 0430J | Bovine feces | AMP, AMXC, CTZD, CTIO, OXYT | IbblaCMY-2IbsugE | colicin Ib | IbS. enterica pFDAARGOS-312-4 | IbCP022064 |
| 0453J | Bovine feces | AMP, CTIO, SULF, TMSZ, OXYT | colicin Ib | Ibp2HS-C-2 | 1ACP038182 | |
| 0550J | Bovine feces | AMP, CTIO, STR, SULF, ENRO, FLOR, OXYT | colicin M colicin B | M,BFHI82 plasmid contig | M,BLM996773 | |
| 0608J | Bovine feces | AMP, CTZD, CTIO, STR, SULF, ENRO, FLOR, TMSZ, OXYT | colicin M colicin B | M,Bp2013C-4404 | M,BCP027378 | |
| 0638J | Bovine feces | AMP, AMXC, STR, SULF, OXYT | colicin M * colicin B * colicin V | M,Bp2013C-4404-2 VpDSM30083 | M,BCP027378 VCP033091 | |
| 0842J | Bovine feces | AMP, AMXC, CTZD, CTIO, STR, SULF, FLOR, TMSZ, OXYT | colicin R | Rp14408-3 | RLT599828 | |
| 0114J | Bovine feces | AMP, AMXC, CTZD, FLOR, TMSZ, OXYT | colicin V colicin Ia | V,IapCOV8 | V,1AMG648896 | |
| 0043M | Feedlot catchbasin | AMP, AMXC, CTZD, CTIO, STR, SULF, FLOR, TMSZ, OXYT | M,Baph(6)-Id | colicin M, colicin B colicin Ia * | M,BS. enterica Heidelberg p12-4373-62 Iap2014C-3075 | M,BCP012928 1ACP027448 |
| 0012I | Feedlot catchbasin | STR, OXYT | aph(6)-Id tet(C) | colicin M colicin B | M,BpExPEC-XM | M,BCP025329 |
| 0096I | Wastewater influent | AMP, CTIO, ENRO | colicin Ia | IapLKSZ01 | 1ACP030282 | |
| 0067K | Wastewater influent | AMP, CTIO, STR, SULF, TMSZ, OXYT | colicin E1 microcin H47 | E1pCOLE1-H22 H47NCTC10444 | E1AY913943 H47LR134092 | |
| 0089K | Wastewater Influent | AMP, CTZD, CTIO, ENRO, NMYN, OXYT | E2,J25bmrA | colicin V colicin E2 * microcin J25 | VpAMSC2 E2,J25pH17-5 | VCP031107 E2,J25CP021198 |
| 0143I | Wastewater influent | AMPI, CTIO, SULF, FLOR, OXYT | M,VblaCTX-M-55 | colicin E1 colicin M colicin V * | E1pEC276_KPC M,VBR02-DEC | E1CP018949 M,VCP035320 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cameron, A.; Zaheer, R.; Adator, E.H.; Barbieri, R.; Reuter, T.; McAllister, T.A. Bacteriocin Occurrence and Activity in Escherichia coli Isolated from Bovines and Wastewater. Toxins 2019, 11, 475. https://doi.org/10.3390/toxins11080475
Cameron A, Zaheer R, Adator EH, Barbieri R, Reuter T, McAllister TA. Bacteriocin Occurrence and Activity in Escherichia coli Isolated from Bovines and Wastewater. Toxins. 2019; 11(8):475. https://doi.org/10.3390/toxins11080475
Chicago/Turabian StyleCameron, Andrew, Rahat Zaheer, Emelia H. Adator, Ruth Barbieri, Tim Reuter, and Tim A. McAllister. 2019. "Bacteriocin Occurrence and Activity in Escherichia coli Isolated from Bovines and Wastewater" Toxins 11, no. 8: 475. https://doi.org/10.3390/toxins11080475
APA StyleCameron, A., Zaheer, R., Adator, E. H., Barbieri, R., Reuter, T., & McAllister, T. A. (2019). Bacteriocin Occurrence and Activity in Escherichia coli Isolated from Bovines and Wastewater. Toxins, 11(8), 475. https://doi.org/10.3390/toxins11080475