Rapid and Differential Evolution of the Venom Composition of a Parasitoid Wasp Depending on the Host Strain
Abstract
1. Introduction
2. Results
2.1. Experimental Evolution Protocol
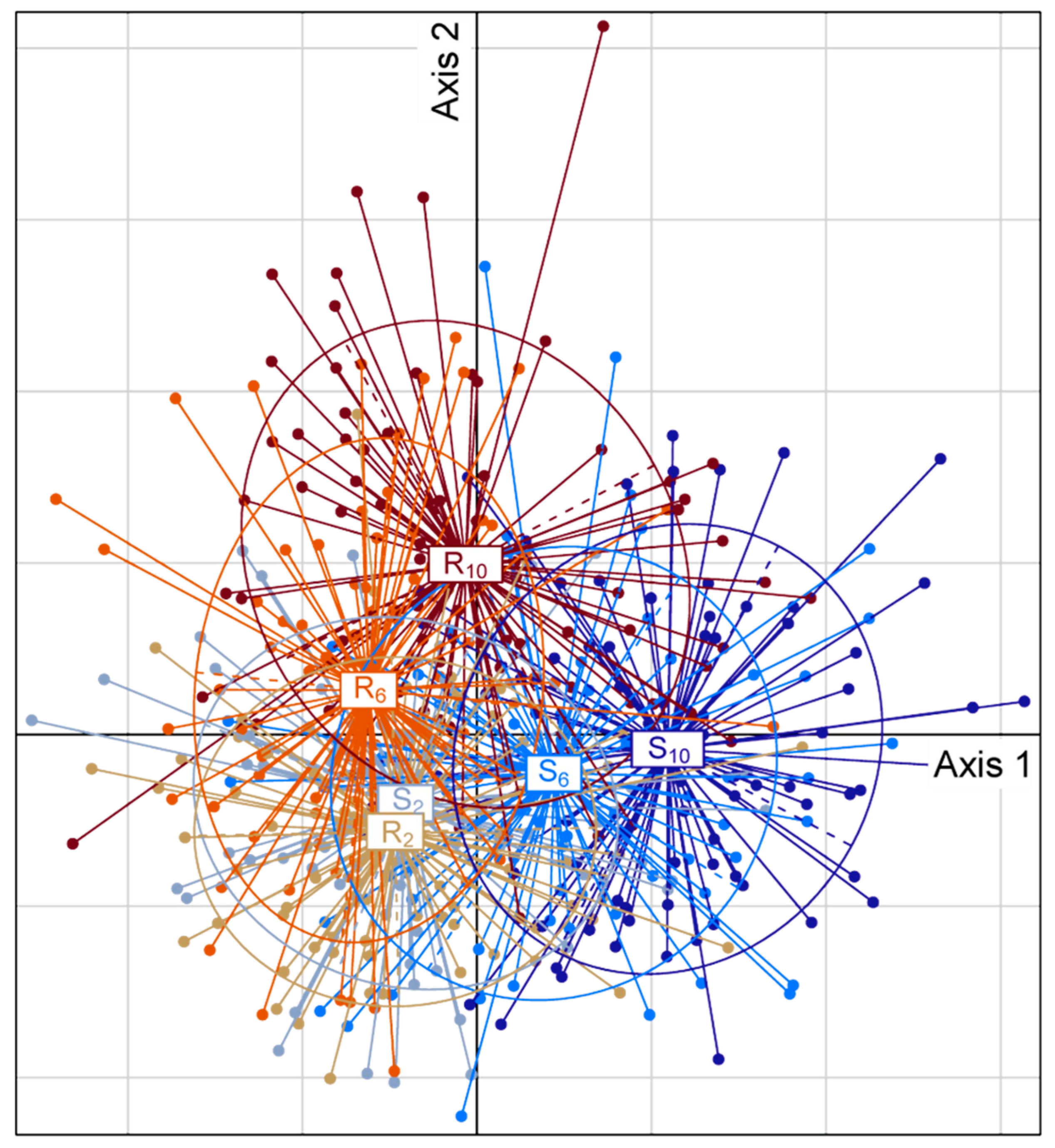
2.2. Host Specific Evolution of Venom Composition Evidenced by a Global Approach
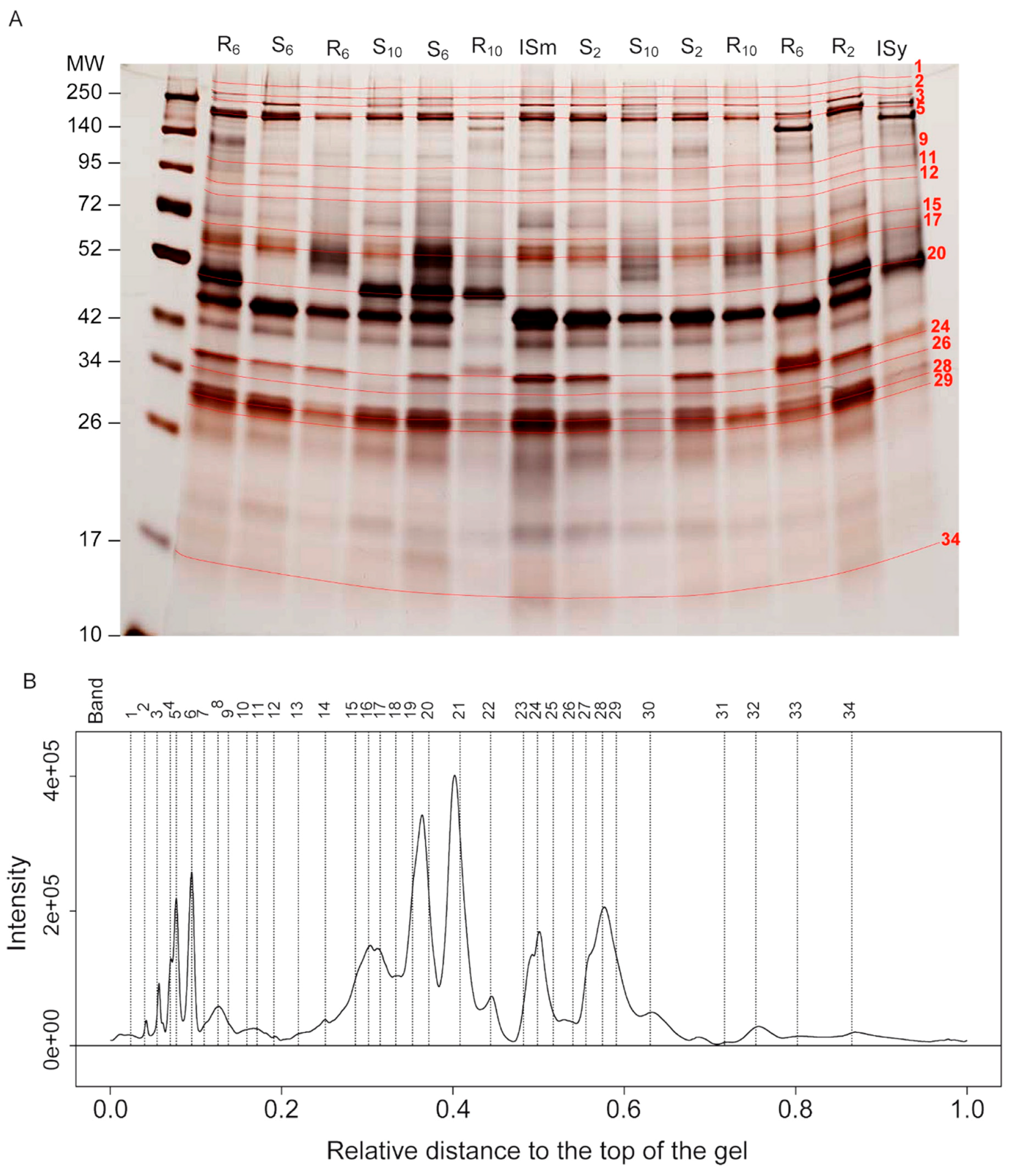
2.3. Identification of Protein Bands Whose Intensity Has Changed by Selection on Different Host Strains
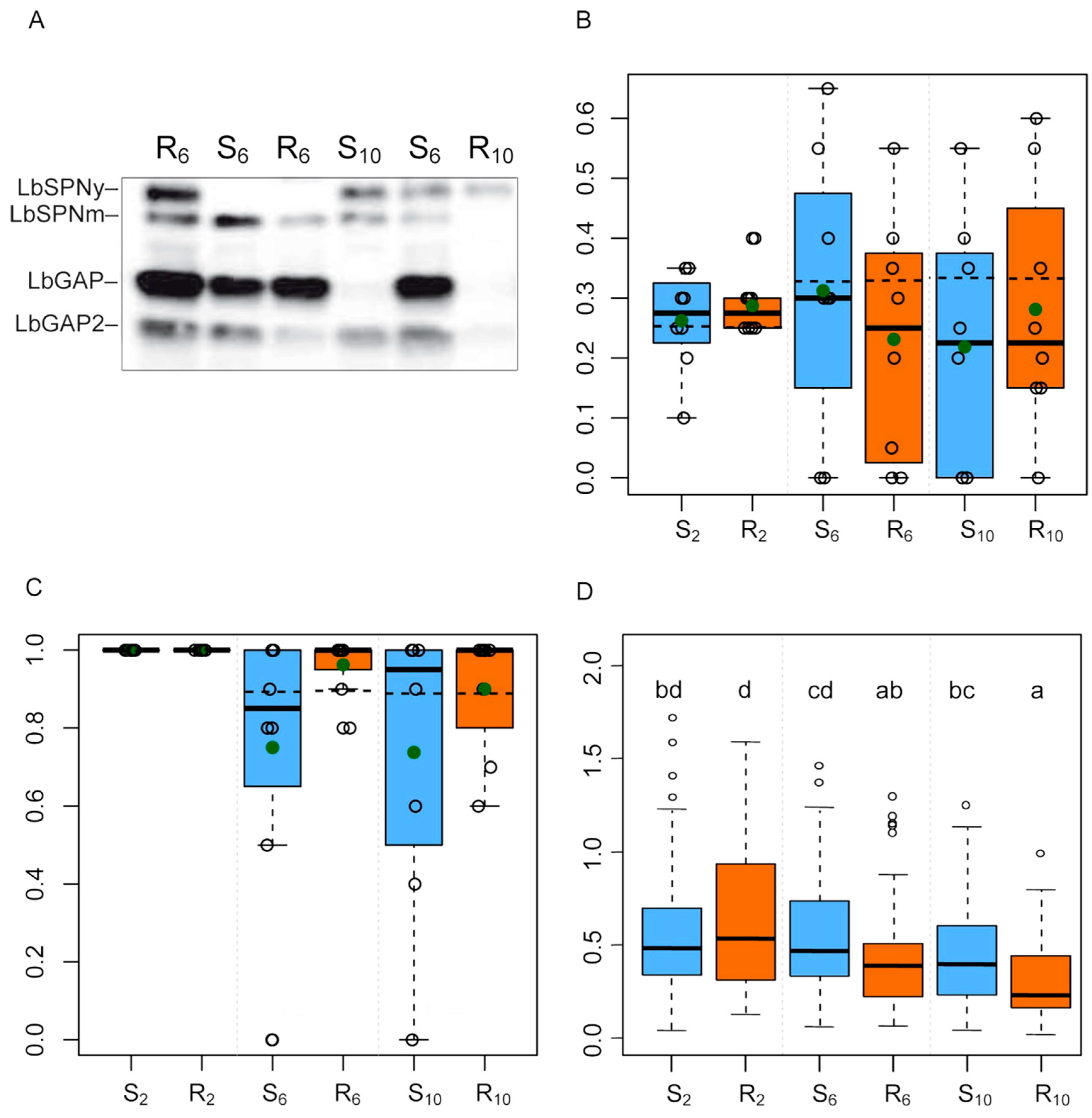
2.4. Putative Identification of Venom Proteins Whose Amount Evolved in Response to the Host Strain
2.5. Trends of Venom Band Evolution
2.6. Host Specific Evolution of Venom Composition Evidenced by a Specific Approach
3. Discussion
3.1. The Composition of Venom Evolves Rapidly Under Selection on Host Strains
3.2. A Majority of Bands Whose Intensity Changed Are of ISm Origin
3.3. Venom Components Can Be Costly
3.4. Proteins Whose Quantity Potentially Evolved
4. Materials and Methods
4.1. Biological Material
4.2. Experimental Evolution Protocol
4.3. Venom Analysis and Data Acquisition
4.4. Statistical Analysis for the Global Analysis of Venom Composition
4.4.1. Evolution of the Global Venom Composition
4.4.2. Evolution of Venom Protein Bands
4.5. Statistical Analysis for the Specific Study of Venom Evolution
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Daltry, J.C.; Wüster, W.; Thorpe, R.S. Diet and snake venom evolution. Nature 1996, 379, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wüster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. B Biol. Sci. 2009, 276, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Olenzek, A.M.; Duda, T.F. Effects of geographical heterogeneity in species interactions on the evolution of venom genes. Proc. R. Soc. B Biol. Sci. 2015, 282. [Google Scholar] [CrossRef] [PubMed]
- Zancolli, G.; Calvete, J.J.; Cardwell, M.D.; Greene, H.W.; Hayes, W.K.; Hegarty, M.J.; Herrmann, H.W.; Holycross, A.T.; Lannutti, D.I.; Mulley, J.F.; et al. When one phenotype is not enough: Divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. R. Soc. B Biol. Sci. 2019, 286. [Google Scholar] [CrossRef] [PubMed]
- Poirié, M.; Colinet, D.; Gatti, J.L. Insights into function and evolution of parasitoid wasp venoms. Curr. Opin. Insect Sci. 2014, 6, 52–60. [Google Scholar] [CrossRef]
- Asgari, S.; Rivers, D.B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef]
- Moreau, S.J.M.; Asgari, S. Venom proteins from parasitoid wasps and their biological functions. Toxins (Basel) 2015, 7, 2385–2412. [Google Scholar] [CrossRef]
- Martinson, E.O.; Werren, J.H. Venom is beneficial but not essential for development and survival of Nasonia. Ecol. Entomol. 2018, 43, 146–153. [Google Scholar] [CrossRef]
- Mrinalini; Siebert, A.L.; Wright, J.; Martinson, E.; Wheeler, D.; Werren, J.H. Parasitoid venom induces metabolic cascades in fly hosts. Metabolomics 2015, 11, 350–366. [Google Scholar] [CrossRef]
- Colinet, D.; Deleury, E.; Anselme, C.; Cazes, D.; Poulain, J.; Azema-Dossat, C.; Belghazi, M.; Gatti, J.L.; Poirié, M. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: The case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 2013, 43, 601–611. [Google Scholar] [CrossRef]
- Colinet, D.; Mathé-Hubert, H.; Allemand, R.; Gatti, J.L.; Poirié, M. Variability of venom components in immune suppressive parasitoid wasps: From a phylogenetic to a population approach. J. Insect Physiol. 2013, 59, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Mathé-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirié, M.; Gatti, J.L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 β-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar]
- Colinet, D.; Schmitz, A.; Cazes, D.; Gatti, J.L.; Poirié, M. The origin of intraspecific variation of virulence in an eukaryotic immune suppressive parasite. PLoS Pathog. 2010, 6, e1001206. [Google Scholar] [CrossRef] [PubMed]
- Labrosse, C.; Stasiak, K.; Lesobre, J.; Grangeia, A.; Huguet, E.; Drezen, J.M.; Poirie, M. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulardi (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect Biochem. Mol. Biol. 2005, 35, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Labrosse, C.; Eslin, P.; Doury, G.; Drezen, J.M.; Poirié, M. Haemocyte changes in D. melanogaster in response to long gland components of the parasitoid wasp Leptopilina boulardi: A Rho-GAP protein as an important factor. J. Insect Physiol. 2005, 51, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Schmitz, A.; Depoix, D.; Crochard, D.; Poirié, M. Convergent use of RhoGAP toxins by eukaryotic parasites and bacterial pathogens. PLoS Pathog. 2007, 3, e203. [Google Scholar] [CrossRef]
- Wan, B.; Goguet, E.; Ravallec, M.; Pierre, O.; Lemauf, S.; Volkoff, A.-N.; Gatti, J.-L.; Poirié, M. Venom atypical extracellular vesicles as interspecies vehicles of virulence factors involved in host specificity: The case of a Drosophila parasitoid wasp. Front. Immunol. 2019, 10, 1688. [Google Scholar] [CrossRef]
- Colinet, D.; Dubuffet, A.; Cazes, D.; Moreau, S.; Drezen, J.M.; Poirié, M. A serpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 2009, 33, 681–689. [Google Scholar] [CrossRef]
- Mathé-Hubert, H.; Kremmer, L.; Colinet, D.; Gatti, J.L.; Van Baaren, J.; Delava, É.; Poirié, M. Variation in the venom of parasitic wasps, drift, or selection? Insights from a multivariate QST analysis. Front. Ecol. Evol. 2019, 7, 156. [Google Scholar] [CrossRef]
- Dupas, S.; Carton, Y.; Poirié, M. Genetic dimension of the coevolution of virulence-resistance in Drosophila—Parasitoid wasp relationships. Heredity (Edinb.) 2003, 90, 84–89. [Google Scholar] [CrossRef]
- Dubuffet, A.; Doury, G.; Labrousse, C.; Drezen, J.-M.; Carton, Y.; Poirié, M. Variation of success of Leptopilina boulardi in Drosophila yakuba: The mechanisms explored. Dev. Comp. Immunol. 2008, 32, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Carton, Y.; Frey, F.; Nappi, A. Genetic determinism of the cellular immune reaction in Drosophila melanogaster. Heredity (Edinb.) 1992, 69, 393–399. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benassi, V.; Frey, F.; Carton, Y. A new specific gene for wasp cellular immune resistance in Drosophila. Heredity (Edinb.) 1998, 80, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Poirié, M.; Frey, F.; Hita, M.; Huguet, E.; Lemeunier, F.; Periquet, G.; Carton, Y. Drosophila resistance genes to parasitoids: Chromosomal location and linkage analysis. Proc. R. Soc. B Biol. Sci. 2000, 267, 1417–1421. [Google Scholar] [CrossRef] [PubMed]
- Hita, M.T.; Poirié, M.; Leblanc, N.; Lemeunier, F.; Lutcher, F.; Frey, F.; Periquet, G.; Carton, Y. Genetic localization of a Drosophila melanogaster resistance gene to a parasitoid wasp and physical mapping of the region. Genome Res. 1999, 9, 471–481. [Google Scholar]
- Hita, M.; Espagne, E.; Lemeunier, F.; Pascual, L.; Carton, Y.; Periquet, G.; Poirié, M. Mapping candidate genes for Drosophila melanogaster resistance to the parasitoid wasp Leptopilina boulardi. Genet. Res. 2006, 88, 81–91. [Google Scholar] [CrossRef]
- Mathé-Hubert, H.; Gatti, J.-L.; Colinet, D.; Poirié, M.; Malausa, T. Statistical analysis of the individual variability of 1D protein profiles as a tool in ecology: An application to parasitoid venom. Mol. Ecol. Resour. 2015, 15, 1120–1132. [Google Scholar] [CrossRef]
- Board, J.E.; Kang, M.S.; Harville, B.G. Path analyses identify indirect selection criteria for yield of late-planted soybean. Crop Sci. 1997, 37, 879–884. [Google Scholar] [CrossRef]
- Russo, J.; Brehélin, M.; Carton, Y. Haemocyte changes in resistant and susceptible strains of D. melanogaster caused by virulent and avirulent strains of the parasitic wasp Leptopilina boulardi. J. Insect Physiol. 2001, 47, 167–172. [Google Scholar] [CrossRef]
- Dubuffet, A.; Dupas, S.; Frey, F.; Drezen, J.M.; Poirié, M.; Carton, Y. Genetic interactions between the parasitoid wasp Leptopilina boulardi and its Drosophila hosts. Heredity (Edinb.) 2007, 98, 21–27. [Google Scholar] [CrossRef]
- Dupas, S.; Boscaro, M. Geographic variation and evolution of immunosuppressive genes in a Drosophila parasitoid. Ecography (Cop.) 1999, 22, 284–291. [Google Scholar] [CrossRef]
- Dupas, S.; Poirié, M.; Frey, F.; Carton, Y. Is parasitoid virulence against multiple hosts adaptive or constrained by phylogeny? A study of Leptopilina spp.(Hymenoptera: Figitidae)/Drosophila (Diptera: Drosophilidae) interactions. Ann. Soc. Entomol. Fr. 2013, 49, 222–231. [Google Scholar] [CrossRef]
- .Morgenstern, D.; King, G.F. The venom optimization hypothesis revisited. Toxicon 2013, 63, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.J.; Wiklund, M.-L.; Wikman, S.; Hultmark, D. Rac1 signalling in the Drosophila larval cellular immune response. J. Cell Sci. 2006, 119, 2015–2024. [Google Scholar] [CrossRef]
- Williams, M.J.; Ando, I.; Hultmark, D. Drosophila melanogaster Rac2 is necessary for a proper cellular immune response. Genes to Cells 2005, 10, 813–823. [Google Scholar] [CrossRef]
- Martinson, E.O.; Mrinalini; Kelkar, Y.D.; Chang, C.H.; Werren, J.H. The evolution of venom by co-option of single-copy genes. Curr. Biol. 2017, 27, 2007–2013.e8. [Google Scholar] [CrossRef]
- Dennis, A.B.; Patel, V.; Oliver, K.M.; Vorburger, C. Parasitoid gene expression changes after adaptation to symbiont-protected hosts. Evolution 2017, 71, 2599–2617. [Google Scholar] [CrossRef]
- Cavigliasso, F.; Mathé-hubert, H.; Gatti, J.; Colinet, D.; Poirié, M. Experimental evolution of parasitism success and venom composition of the parasitoid wasp Leptopilina boulardi on different Drosophila host species. Manuscript in preparation.
- Dupas, S.; Frey, F.; Carton, Y. A single parasitoid segregating factor controls immune suppression in Drosophila. J. Hered. 1998, 89, 306–311. [Google Scholar] [CrossRef]
- Carton, Y.; Nappi, A.J. Drosophila cellular immunity against parasitoids. Parasitol. Today 1997, 13, 218–227. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Bennett, A.F. The importance of experimental studies in evolutionary biology. In Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments; University of California Press: Oakland, CA, USA, 2009; pp. 15–30. [Google Scholar]
- Morrissey, J.H. Silver stain for proteins in polyacrylamide gels: A modified procedure with enhanced uniform sensitivity. Anal. Biochem. 1981, 117, 307–310. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Kremmer, L.; Lemauf, S.; Rebuf, C.; Gatti, J.L.; Poirié, M. Development of RNAi in a Drosophila endoparasitoid wasp and demonstration of its efficiency in impairing venom protein production. J. Insect Physiol. 2014, 63, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Peng, B.; Amos, C.I. Forward-time simulations of non-random mating populations using simuPOP. Bioinformatics 2008, 24, 1408–1409. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; The R Core Team. nlme: Linear and nonlinear mixed effects models. R Packag. Vers. 2011. [Google Scholar]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biometrical J. 2008, 50, 346–363. [Google Scholar] [CrossRef]

| Model | Variance Partition | Df | Sums of Sqs | F | R² | Pr (>F) |
|---|---|---|---|---|---|---|
| Evolution on the two host strains | Generation | 1 | 3 × 109 | 3.47 | 0.01 | 0.02 * |
| Host strain | 1 | 9 × 109 | 10.36 | 0.02 | 2 × 10−4 *** | |
| Generation × Host | 1 | 5 × 109 | 6.19 | 0.01 | 1 × 10−3 ** | |
| Population | 14 | 8 × 1010 | 6.87 | 0.17 | 2 × 10−4 *** | |
| Residuals | 456 | 4 × 1011 | 0.80 | |||
| Total | 473 † | 4 × 1011 | 1.00 | |||
| Evolution on the (R) host strain | Generation | 1 | 4 × 109 | 4.28 | 0.01 | 1 × 10−3 ** |
| Population | 7 | 3 × 1010 | 5.28 | 0.14 | 2 × 10−4 *** | |
| Residuals | 231 | 2 × 1011 | 0.85 | |||
| Total | 239 † | 2 × 1011 | 1.00 | |||
| Evolution on the (S) host strain | Generation | 1 | 4 × 109 | 5.46 | 0.02 | 2 × 10−3 ** |
| Population | 7 | 5 × 1010 | 8.57 | 0.21 | 2 × 10−4 *** | |
| Residuals | 225 | 2 × 1011 | 0.77 | |||
| Total | 233 † | 2 × 1011 | 1.00 |
| Band Number | Cluster Number | Correlation with Axis 1 (Evolution on S Strain) | Correlation with Axis 2 (Evolution on R Strain) | Band Origin | Band Evolution According to the Origin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Before Partial Correlations | After Partial Correlations | Before Partial Correlations | After Partial Correlations | S Strain | R Strain | |||||||||
| ISm | ISy | ISm | ISy | |||||||||||
| 1 | None | + | * | - | n.s. | 0.84 | ↗ | |||||||
| 2 | None | + | *** | + | n.s. | 0.70 | ↗ | |||||||
| 3 | None | + | *** | - | * | 0.86 | ↗ | ↘ | ||||||
| 11 | None | + | *** | + | n.s. | 0.24 | ↗ | |||||||
| 12 | None | + | * | + | ** | 0.38 | ↗ | ↗ | ||||||
| 26 | 1 | - | * | - | * | + | *** | + | *** | 0.61 | ↘ | ↗ | ||
| 6 | 2 | - | *** | - | n.s. | + | *** | + | n.s. | 0.12 | ||||
| 22 | 2 | + | *** | + | n.s. | - | *** | - | n.s. | 0.88 | ||||
| 23 | 2 | - | *** | + | n.s. | + | *** | + | n.s. | 0.03 | ||||
| 24 | 2 | - | *** | - | *** | + | *** | + | n.s. | 0.62 | ↘ | |||
| 4 | 3 | + | n.s. | + | n.s. | - | *** | - | n.s. | 0.46 | ||||
| 5 | 3 | + | *** | + | *** | - | *** | - | * | 0.60 | ↗ | ↘ | ||
| 20 | 4 | - | n.s. | - | n.s. | - | * | - | * | 0.06 | ↘ | |||
| 27 | 5 | + | *** | + | n.s. | + | n.s. | + | *** | 0.65 | ||||
| 28 | 5 | + | *** | - | ** | - | *** | - | *** | 0.79 | ↘ | |||
| 29 | 5 | + | *** | + | *** | - | *** | - | *** | 0.82 | ↗ | ↘ | ||
| 30 | 5 | + | *** | - | n.s. | - | * | + | n.s. | 0.85 | ||||
| 34 | 5 | + | *** | + | ** | - | *** | - | n.s. | 0.93 | ↗ | |||
| 8 | 6 | + | *** | + | n.s. | - | n.s. | - | n.s. | 0.10 | ||||
| 9 | 6 | + | *** | + | n.s. | - | ** | - | *** | 0.43 | ↘ | |||
| 10 | 6 | + | *** | + | n.s. | + | n.s. | + | n.s. | 0.07 | ||||
| 13 | 7 | - | n.s. | - | n.s. | + | *** | - | n.s. | 0.77 | ||||
| 14 | 7 | - | n.s. | - | n.s. | + | *** | + | n.s. | 0.69 | ||||
| 15 | 7 | - | *** | - | ** | + | *** | + | n.s. | 0.48 | ↘ | |||
| 16 | 7 | - | n.s. | - | n.s. | + | *** | + | n.s. | 0.43 | ||||
| 17 | 7 | - | *** | - | *** | + | *** | - | n.s. | 0.24 | ↘ | |||
| 18 | 7 | - | *** | + | n.s. | + | ** | + | n.s. | 0.07 | ||||
| Reference Band | Number of Proteins in the Band | Putative Function | Number of Peptides Matches | Band Origin | Evolution on the R Strain | Evolution on the S Strain |
|---|---|---|---|---|---|---|
| 3 | 1 | Unknown | 12 | ISm | ↘ | ↗ |
| 5 | 6 | Unknown | 49 | ISm | ↘ | ↗ |
| Unknown | 46 | |||||
| Unknown | 34 | |||||
| Sushi/SCR/CCP domain containing protein | 18 | |||||
| Unknown | 14 | |||||
| Unknown | 11 | |||||
| 11 | 2 | Sushi/SCR/CCP domain containing protein | 10 | ISy | ↗ | |
| Unknown | 10 | |||||
| 15 | 3 | Unknown a | 39 | ISy | ↘ | |
| Unknown b | 36 | |||||
| Unknown | 28 | |||||
| 17 | 2 | Unknown a | 58 | ISy | ↘ | |
| Unknown b | 36 | |||||
| 20 | 1 | Serpin (LbSPNy) | 81 | ISy | ↘ | |
| 24 | 5 | RhoGAP (LbGAP) | 52 | ISm | ↘ | |
| Unknown | 21 | |||||
| Serpin (LbSPNm) | 17 | |||||
| Unknown | 12 | |||||
| Unknown | 11 | |||||
| 26 | 1 | RhoGAP (LbGAPy4) | 24 | ISm | ↗ | ↘ |
| 28 | 3 | RhoGAP (LbGAP2) | 43 | ISm | ↘ | |
| RhoGAP (LbGAP1) | 23 | |||||
| Serpine (LbSPNm) | 15 | |||||
| 29 | 4 | RhoGAP (LbGAP2) | 68 | ISm | ↘ | ↗ |
| Unknown | 21 | |||||
| RhoGAP (LbGAPy2) | 19 | |||||
| Unknown | 11 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavigliasso, F.; Mathé-Hubert, H.; Kremmer, L.; Rebuf, C.; Gatti, J.-L.; Malausa, T.; Colinet, D.; Poirié, M. Rapid and Differential Evolution of the Venom Composition of a Parasitoid Wasp Depending on the Host Strain. Toxins 2019, 11, 629. https://doi.org/10.3390/toxins11110629
Cavigliasso F, Mathé-Hubert H, Kremmer L, Rebuf C, Gatti J-L, Malausa T, Colinet D, Poirié M. Rapid and Differential Evolution of the Venom Composition of a Parasitoid Wasp Depending on the Host Strain. Toxins. 2019; 11(11):629. https://doi.org/10.3390/toxins11110629
Chicago/Turabian StyleCavigliasso, Fanny, Hugo Mathé-Hubert, Laurent Kremmer, Christian Rebuf, Jean-Luc Gatti, Thibaut Malausa, Dominique Colinet, and Marylène Poirié. 2019. "Rapid and Differential Evolution of the Venom Composition of a Parasitoid Wasp Depending on the Host Strain" Toxins 11, no. 11: 629. https://doi.org/10.3390/toxins11110629
APA StyleCavigliasso, F., Mathé-Hubert, H., Kremmer, L., Rebuf, C., Gatti, J.-L., Malausa, T., Colinet, D., & Poirié, M. (2019). Rapid and Differential Evolution of the Venom Composition of a Parasitoid Wasp Depending on the Host Strain. Toxins, 11(11), 629. https://doi.org/10.3390/toxins11110629






