Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05
Abstract
:1. Introduction
2. Results
2.1. Enzymatic Activity Detection of Recombinant CEs
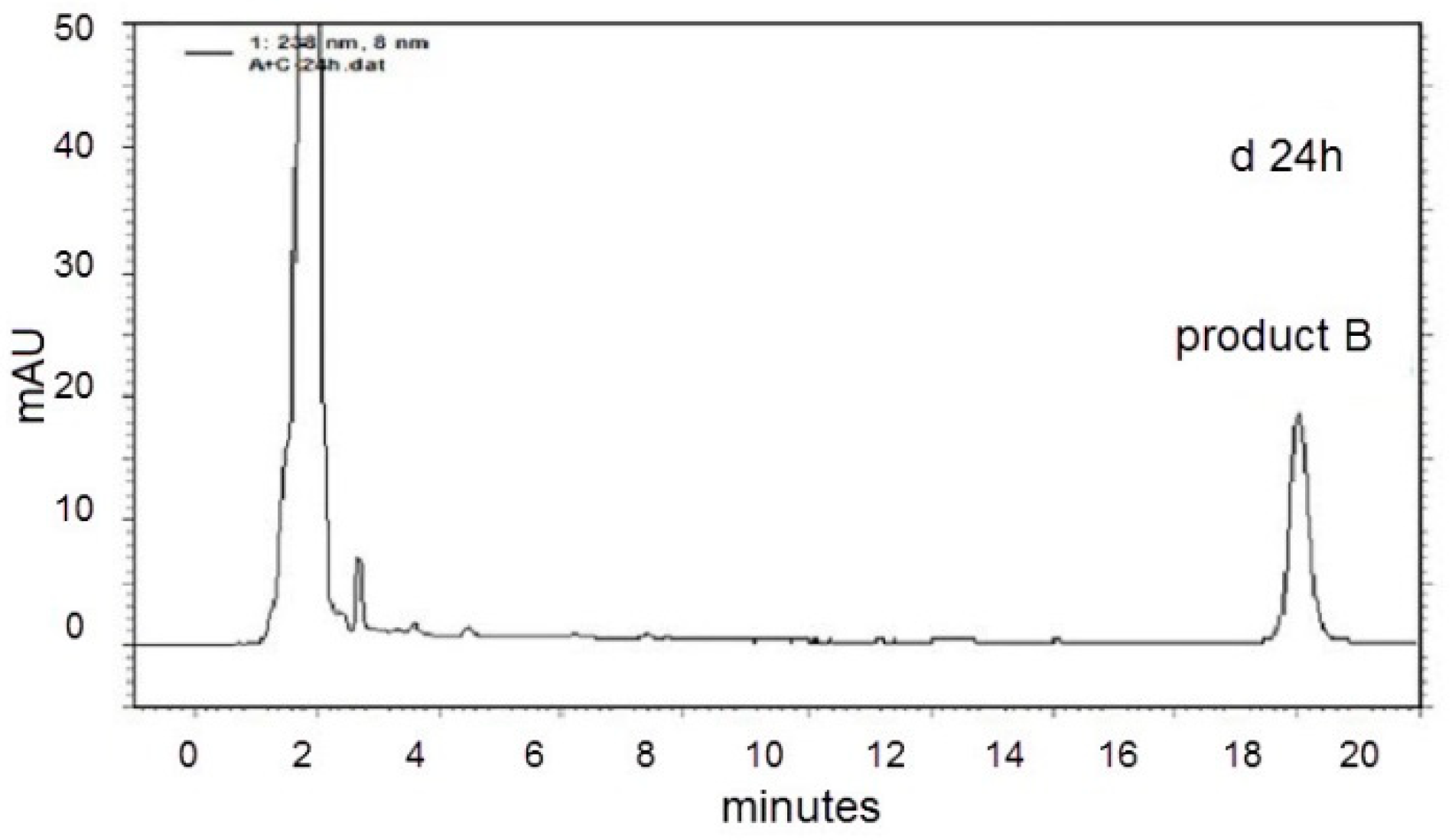
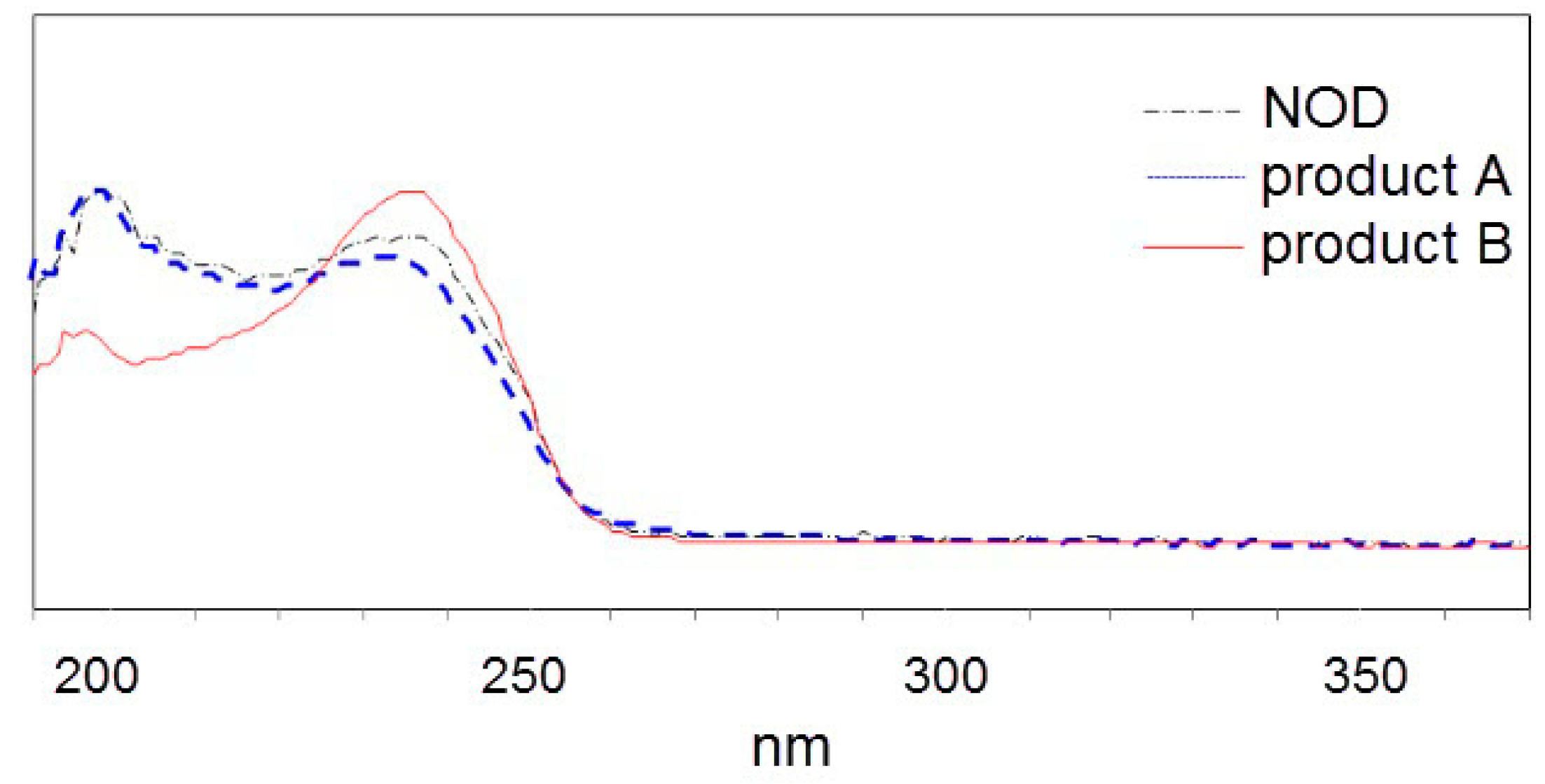
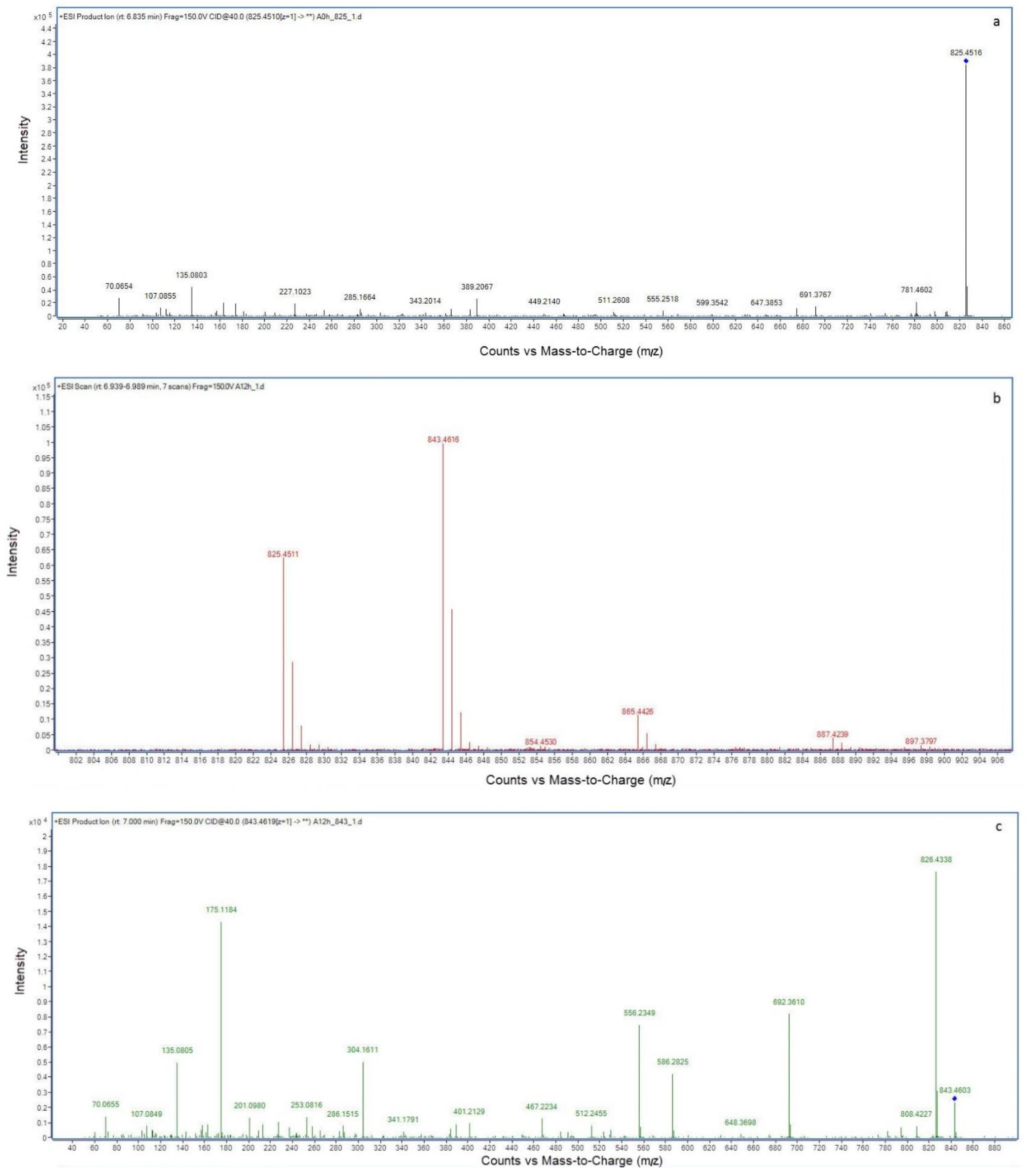
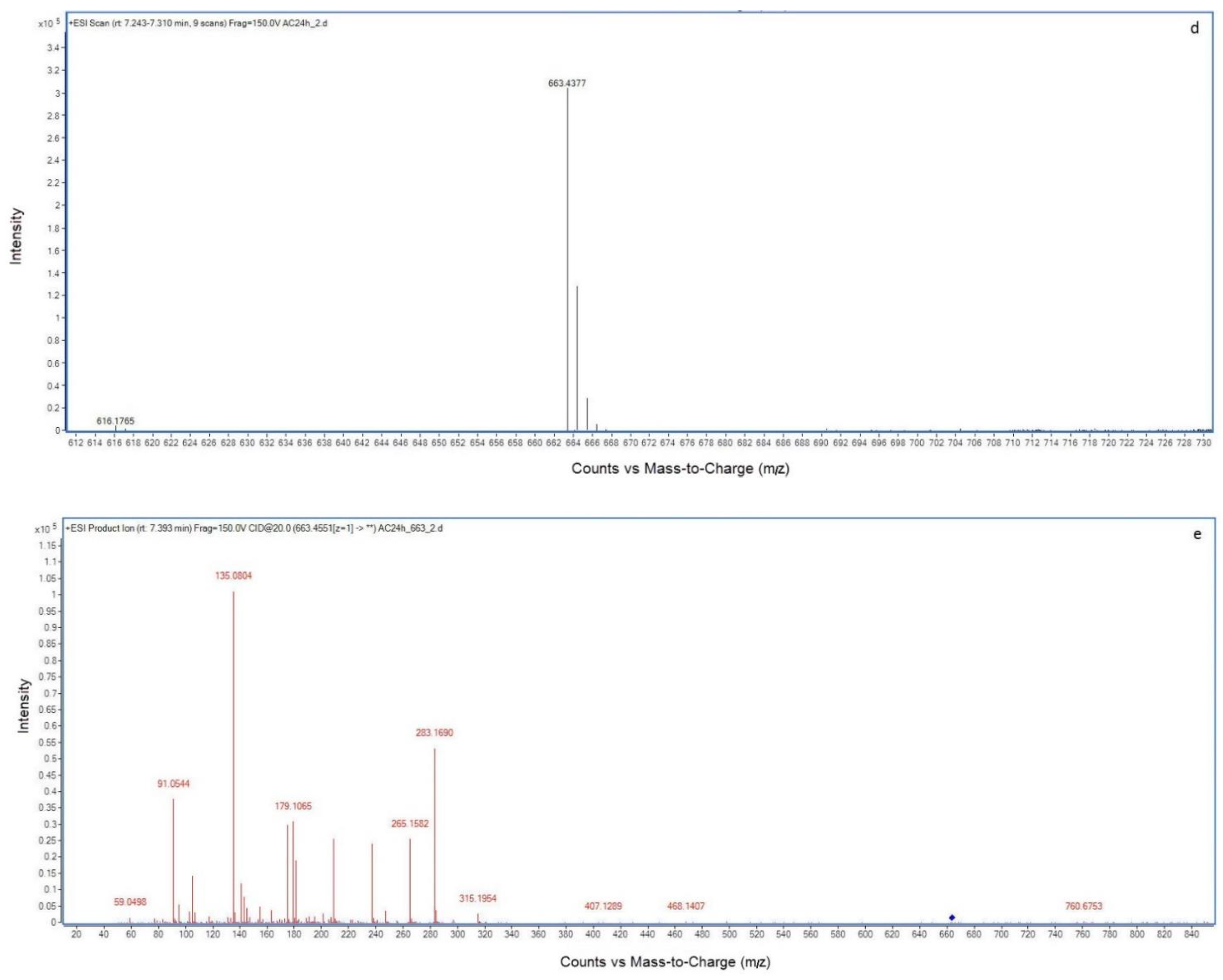
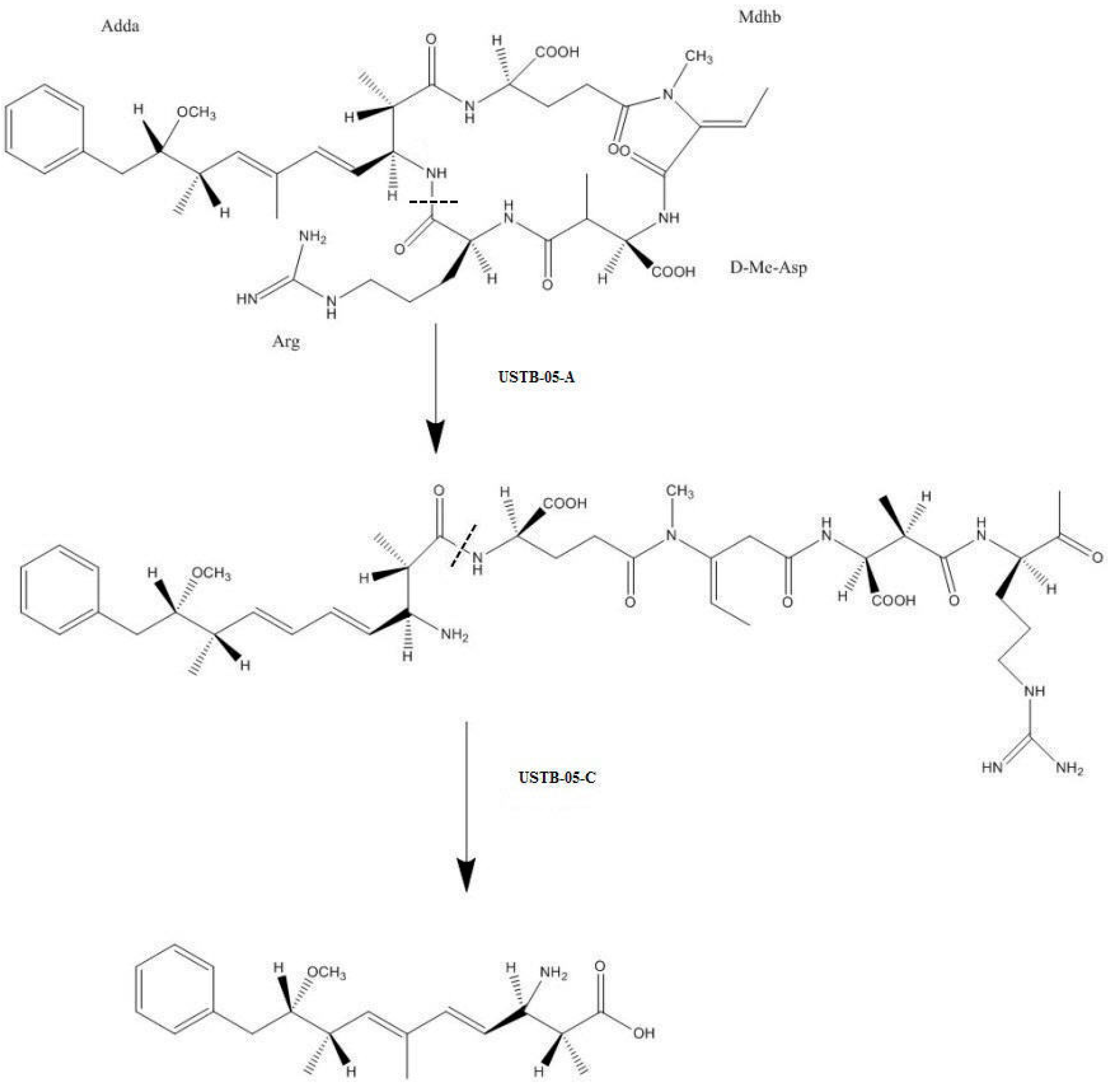
2.2. LC-MS Analysis of NOD and Its Biodegradation Products
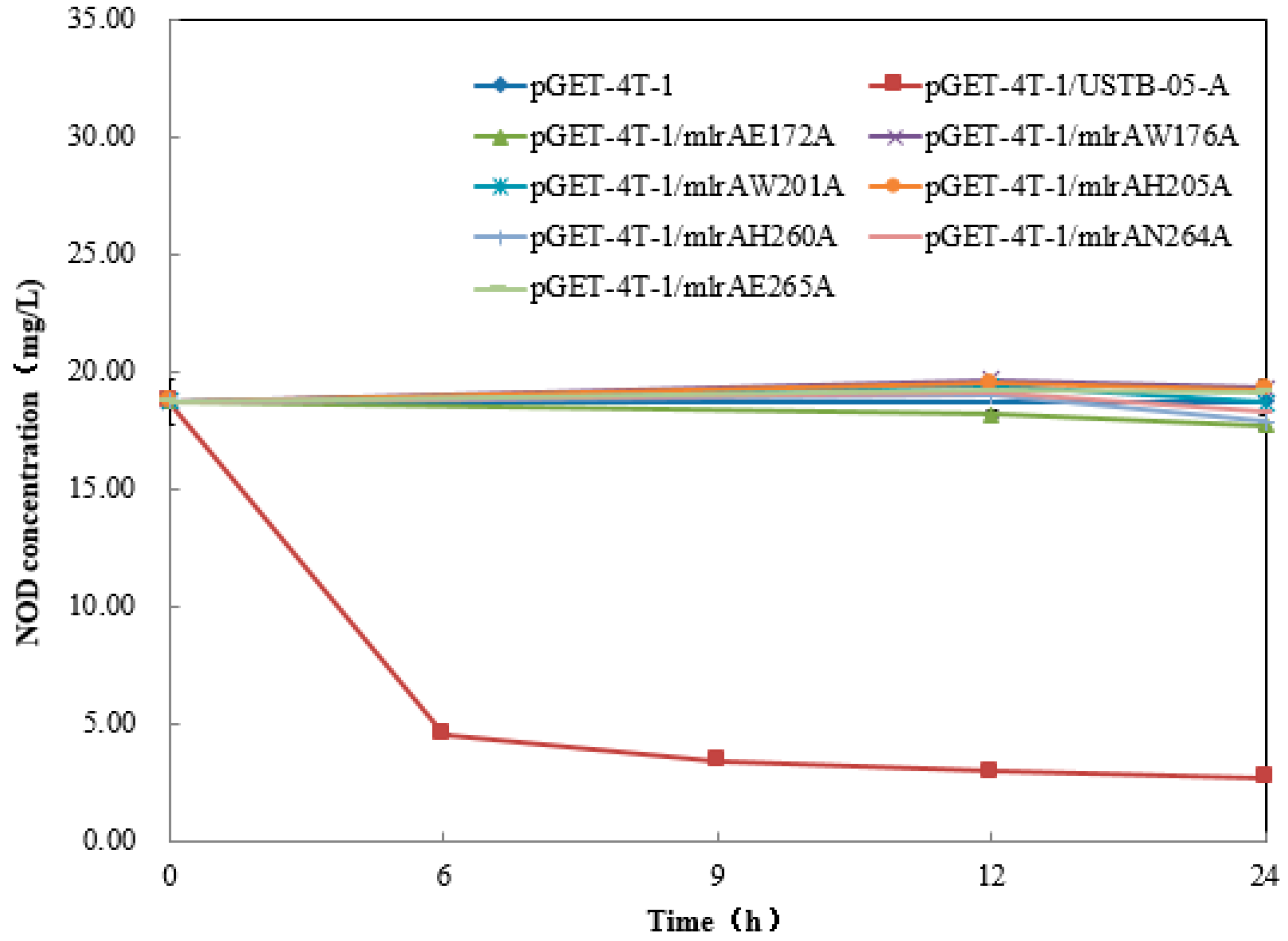
2.3. Recombinant USTB-05-A and Its Mutants
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains, Culture Conditions and Reagents
5.2. Expression of Gene USTB-05-A and USTB-05-C
5.3. Enzymatic Activity of Recombinant USTB-05-A and USTB-05-C
- Control groups for treatment A and treatment C: the CEs of recombinant bacteria pGEX-4T-1/BL21(DE3) without protein USTB-05-A and recombinant bacteria pET30a(+)/BL21(DE3) without protein USTB-05-C were added into two tubes with PBS containing NOD, respectively.
- Treatment A: the CE of recombinant bacteria pGEX-4T-1/USTB-05-A/BL21(DE3) containing crude protein USTB-05-A was added into PBS containing NOD.
- Treatment C: the CE of recombinant bacteria pET30a(+)/USTB-05-C/BL21(DE3) containing crude protein USTB-05-C was added into PBS containing NOD.
- Treatment AC: at 0 h, the CE containing crude protein USTB-05-A was added into PBS containing NOD. At 12 h, the CE containing crude protein USTB-05-C was added to the above solution.
- Control group for treatment A and treatment C: at 0 h, the CE containing crude protein USTB-05-A was added into the test tubes with PBS containing NOD. At 12 h, the CE of pET30a(+)/BL21(DE3) without protein USTB-05-C was added to the above solution.
5.4. Site-Directed Mutagenesis and Enzymatic Activity Comparison
5.5. Analysis Methods
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Heresztyn, T.; Nicholson, B. Nodularin concentrations in Lakes Alexandrina and Albert, South Australia, during a bloom of the cyanobacterium (blue-green alga) Nodularia spumigena and degradation of the toxin. Environ. Toxicol. Water Qual. 1997, 12, 273–282. [Google Scholar] [CrossRef]
- Nehring, S. Mortality of dogs associated with a mass development of Nodularia spumigena (Cyanophyceae) in a brackish lake at the German North Sea coast. J. Plankton. Res. 1993, 15, 867–872. [Google Scholar] [CrossRef]
- Graham, J.L.; Loftin, K.A.; Meyer, M.T.; Ziegler, A.C. Cyanotoxin mixtures and taste-and-odor compounds in cyanobacterial blooms from the midwestern United States. Environ. Sci. Technol. 2010, 44, 7361–7368. [Google Scholar] [CrossRef] [PubMed]
- Neilan, B.A.; Pearson, L.A.; Muenchhoff, J.; Moffitt, M.C.; Dittmann, E. Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ. Microbiol. 2013, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Zanchett, G.; Oliveira-Filho, E.C. Cyanobacteria and cyanotoxins: From impacts on aquatic ecosystems and human health to anticarcinogenic effects. Toxins 2013, 5, 1896–1917. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W.; Eschedor, J.T.; Patterson, G.M.; Moore, R.E. Toxicity and partial structure of a hepatotoxic peptide produced by the cyanobacterium Nodularia spumigena Mertens emend. L575 from New Zealand. Appl. Environ. Microb. 1988, 54, 2257–2263. [Google Scholar] [CrossRef]
- Harding, W.R.; Rowe, N.; Wessels, J.C.; Beattie, K.A.; Codd, G.A. Death of a dog attributed to the cyanobacterial (blue-green algal) hepatotoxin nodularin in South Africa. J. S. Afr. Vet. Assoc. 1995, 66, 256–259. [Google Scholar] [PubMed]
- Repka, S.; Meyerhofer, M.; von Brockel, K.; Sivonen, K. Associations of cyanobacterial toxin, nodularin, with environmental factors and zooplankton in the Baltic Sea. Microb. Ecol. 2004, 47, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Mariani, M.A.; Padedda, B.M.; Kastovsky, J.; Buscarinu, P.; Sechi, N.; Virdis, T.; Luglie, A. Effects of trophic status on microcystin production and the dominance of cyanobacteria in the phytoplankton assemblage of Mediterranean reservoirs. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Merel, S.; Walker, D.; Chicana, R.; Snyder, S.; Baurès, E.; Thomas, O. State of knowledge and concerns on cyanobacterial blooms and cyanotoxins. Environ. Int. 2013, 59, 303–327. [Google Scholar] [CrossRef] [PubMed]
- Miles, C.O.; Melanson, J.E.; Ballot, A. Sulfide oxidations for LC-MS analysis of methionine-containing microcystins in dolichospermum flos-aquae NIVA-CYA 656. Environ. Sci. Technol. 2014, 48, 13307–13315. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.C.; Lee, A.K.; Yates, R.S.; Liang, S.; Rochelle, P.A. Analysis of microcystins in drinking water by ELISA and LC/MS/MS. J. Am. Water Works Assoc. 2017, 109, 13–25. [Google Scholar] [CrossRef]
- Codd, G.; Bell, S.; Kaya, K.; Ward, C.; Beattie, K.; Metcalf, J. Cyanobacterial toxins, exposure routes and human health. Eur. J. Phycol. 1999, 34, 405–415. [Google Scholar] [CrossRef]
- Beattie, K.A.; Kaya, K.; Codd, G.A. The cyanobacterium Nodularia in PCC 7804, of freshwater origin, produces [L-Har2]nodularin. Phytochemistry 2000, 54, 57–61. [Google Scholar] [CrossRef]
- Van Halderen, A.; Harding, W.R.; Wessels, J.C.; Schneider, D.J.; Heine, E.W.; Van der Merwe, J.; Fourie, J.M. Cyanobacterial (blue-green algae) poisoning of livestock in the western Cape Province of South Africa. J. S. Afr. Vet. Assoc. 1995, 66, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Stewart, I.; Seawright, A.A.; Shaw, G.R. Cyanobacterial poisoning in livestock, wild mammals and birds—An overview. In Cyanobacterial Harmful Algal Blooms: State of the Science and Research Needs; Hudnell, H.K., Ed.; Springer: New York, NY, USA, 2008; Volume 619, pp. 613–637. [Google Scholar]
- Li, G.; Yan, W.; Dang, Y.; Li, J.; Liu, C.; Wang, J. The role of calcineurin signaling in microcystin-LR triggered neuronal toxicity. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Dong, X.; Zeng, S.; Bai, F.; Li, D.; He, M. Extracellular microcystin prediction based on toxigenic Microcystis detection in a eutrophic lake. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Liang, H.; Zhou, W.; Zhang, Y.; Qiao, Q.; Zhang, X. Are fish fed with cyanobacteria safe, nutritious and delicious? A laboratory study. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Backer, L.C.; Manassaram-Baptiste, D.; LePrell, R.; Bolton, B. Cyanobacteria and algae blooms: Review of health and environmental data from the harmful algal bloom-related Illness surveillance system (HABISS) 2007–2011. Toxins 2015, 7, 1048–1064. [Google Scholar] [CrossRef]
- Francis, G. Poisonous Australian lake. Nature 1878, 18, 11–12. [Google Scholar] [CrossRef]
- Pearson, L.; Mihali, T.; Moffitt, M.; Kellmann, R.; Neilan, B. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 2010, 8, 1650–1680. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Sueoka, E.; Iida, N.; Komori, A.; Suganuma, M.; Nishiwaki, R.; Tatematsu, M.; Kim, S.J.; Carmichael, W.W.; Fujiki, H. Nodularin, a potent inhibitor of protein phosphatases 1 and 2A, is a new environmental carcinogen in male F344 rat liver. Cancer Res. 1994, 54, 6402–6406. [Google Scholar] [PubMed]
- Hjornevik, L.V.; Fismen, L.; Young, F.M.; Solstad, T.; Fladmark, K.E. Nodularin exposure induces SOD1 phosphorylation and disrupts SOD1 co-localization with actin filaments. Toxins 2012, 4, 1482–1499. [Google Scholar] [CrossRef]
- Torunska, A.; Bolalek, J.; Plinski, M.; Mazur-Marzec, H. Biodegradation and sorption of nodularin (NOD) in fine-grained sediments. Chemosphere 2008, 70, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Merel, S.; Clement, M.; Thomas, O. State of the art on cyanotoxins in water and their behaviour towards chlorine. Toxicon 2010, 55, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Jones, G.J.; Blakeley, R.L.; Jones, A.; Negri, A.P.; Riddles, P. Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin-LR. Appl. Environ. Microb. 1996, 62, 4086–4094. [Google Scholar]
- Takenaka, S.; Watanabe, M.F. Microcystin LR degradation by Pseudomonas aeruginosa alkaline protease. Chemosphere 1997, 34, 749–757. [Google Scholar] [CrossRef]
- Chen, J.; Hu, L.B.; Zhou, W.; Yan, S.H.; Yang, J.D.; Xue, Y.F.; Shi, Z.Q. Degradation of microcystin-LR and RR by a Stenotrophomonas sp. strain EMS Isolated from Lake Taihu, China. Int. J. Mol. Sci. 2010, 11, 896–911. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wang, H.; Wang, J.; Yin, C.; Ma, S.; Liu, X.; Yin, X. Cloning and expression of the first gene for biodegrading microcystin LR by Sphingopyxis sp. USTB-05. J. Environ. Sci. 2012, 24, 1816–1822. [Google Scholar] [CrossRef]
- Yan, H.; Wang, J.; Chen, J.; Wei, W.; Wang, H.; Wang, H. Characterization of the first step involved in enzymatic pathway for microcystin-RR biodegraded by Sphingopyxis sp. USTB-05. Chemosphere 2012, 87, 12–18. [Google Scholar] [CrossRef]
- Dziga, D.; Wasylewski, M.; Szetela, A.; Bochenska, O.; Wladyka, B. Verification of the role of MlrC in microcystin biodegradation by studies using a heterologously expressed enzyme. Chem. Res. Toxicol. 2012, 25, 1192–1194. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.; Graham, D.; Fowler, N.; Lawton, L.A. Biodegradation of microcystins and nodularin in freshwaters. Chemosphere 2008, 73, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Hoefel, D.; Palazot, S.; Sawade, E.; Newcombe, G.; Saint, C.P.; Brookes, J.D. Investigations into the biodegradation of microcystin-LR in wastewaters. J. Hazard. Mater. 2010, 180, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Riddles, P.; Jones, G.J.; Smith, W.; Blakeley, R.L. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin-LR. Environ. Toxicol. 2001, 16, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Dziga, D.; Wladyka, B.; Zielińska, G.; Meriluoto, J.; Wasylewski, M. Heterologous expression and characterisation of microcystinase. Toxicon 2012, 59, 578–586. [Google Scholar] [CrossRef]
- Lawton, L.A.; Welgamage, A.; Manage, P.M.; Edwards, C. Novel bacterial strains for the removal of microcystins from drinking water. Water Sci. Technol. 2011, 63, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Rapala, J.; Berg, K.A.; Lyra, C.; Niemi, R.M.; Manz, W.; Suomalainen, S.; Paulin, L.; Lahti, K. Paucibacter toxinivorans gen. nov., sp nov., a bacterium that degrades cyclic cyanobacterial hepatotoxins microcystins and nodularin. Int. J. Syst. Evol. Microbiol. 2005, 55, 1563–1568. [Google Scholar] [CrossRef]
- Imanishi, S.; Kato, H.; Mizuno, M.; Tsuji, K.; Harada, K. Bacterial degradation of microcystins and nodularin. Chem. Res. Toxicol. 2005, 18, 591–598. [Google Scholar] [CrossRef]
- Torunska-Sitarz, A.; Kotlarskab, E.; Mazur-Marzec, H. Biodegradation of nodularin and other nonribosomal peptides by the Baltic bacteria. Int. Biodeterior. Biodegrad. 2018, 134, 48–57. [Google Scholar] [CrossRef]
- Wang, J.; Pengfei, W.; Jian, C.; Hai, Y. Biodegradation of Microcystin-RR by a New Isolated Sphingopyxis sp. USTB-05. Chin. J. Chem. Eng. 2010, 18, 108–112. [Google Scholar] [CrossRef]
- Xiao, C.; Yan, H.; Wang, J.; Wei, W.; Ning, J.; Pan, G. Microcystin-LR biodegradation by Sphingopyxis sp. USTB-05. Front. Environ. Sci. Eng. China 2011, 5, 526–532. [Google Scholar] [CrossRef]
- Xu, H.; Wang, H.; Xu, Q.; Lv, L.; Yin, C.; Liu, X.; Du, H.; Yan, H. Pathway for biodegrading microcystin-YR by Sphingopyxis sp. USTB-05. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Nan, F.; Fan, Y.; Hai, Y.; Chunhua, Y.; Xiaolu, L.; Haiyang, Z.; Qianqian, X.; Le, L.; Huasheng, W. Pathway for Biodegrading Nodularin (NOD) by Sphingopyxis sp. USTB-05. Toxins 2016, 8, 116. [Google Scholar] [CrossRef]
- Xu, Q.; Fan, J.; Yan, H.; Ahmad, S.; Zhao, Z.; Yin, C.; Liu, X.; Liu, Y.; Zhang, H. Structural basis of microcystinase activity for biodegrading microcystin-LR. Chemosphere 2019, 236, 124281. [Google Scholar] [CrossRef] [PubMed]
- Gehringer, M.M.; Adler, L.; Roberts, A.A.; Moffitt, M.C.; Mihali, T.K.; Mills, T.J.T.; Fieker, C.; Neilan, B.A. Nodularin, a cyanobacterial toxin, is synthesized in planta by symbiotic Nostoc sp. ISME J. 2012, 6, 1834–1847. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.J.; Orr, P.T. Release and degradation of microcystin following algicide treatment of a Microcystis aeruginosa bloom in a recreational lake, as determined by HPLC and protein phosphatase inhibition assay. Water Res. 1994, 28, 871–876. [Google Scholar] [CrossRef]
- Hyenstrand, P.; Rohrlack, T.; Beattie, K.A.; Metcalf, J.S.; Codd, G.A.; Christoffersen, K. Laboratory studies of dissolved radiolabelled microcystin-LR in lake water. Water Res. 2003, 37, 3299–3306. [Google Scholar] [CrossRef]
- Wang, H.; Yan, H.; Ma, S.; Liu, X.; Yin, C.; Wang, H.; Xu, Q.; Lv, L. Characterization of the second and third steps in the enzymatic pathway for microcystin-RR biodegradation by Sphingopyxis sp. USTB-05. Ann. Microbiol. 2015, 65, 495–502. [Google Scholar] [CrossRef]
- Kato, H.; Imanishi, S.Y.; Tsuji, K.; Harada, K. Microbial degradation of cyanobacterial cyclic peptides. Water Res. 2007, 41, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Mazur-Marzec, H.; Torunska, A.; Blonska, M.J.; Moskot, M.; Plinski, M.; Jakobkiewicz-Banecka, J.; Wegrzyn, G. Biodegradation of nodularin and effects of the toxin on bacterial isolates from the Gulf of Gdansk. Water Res. 2009, 43, 2801–2810. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification ofmicrogram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Treatment | Experiment Condition |
|---|---|
| Control group for treatment A | NOD + the CEs of pGEX-4T-1/BL21(DE3)* |
| Control group for treatment C | NOD + the CEs of pET30a(+)/BL21(DE3)** |
| Treatment A | NOD + USTB-05-A |
| Treatment C | NOD + USTB-05-C |
| Treatment AC | NOD + USTB-05-A, 12 h later, + USTB-05-C |
| Control group for treatment AC | NOD + USTB-05-A, 12 h later, + the CEs of pET30a(+)/BL21(DE3) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Ma, H.; Fan, J.; Yan, H.; Zhang, H.; Yin, C.; Liu, X.; Liu, Y.; Wang, H. Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05. Toxins 2019, 11, 549. https://doi.org/10.3390/toxins11100549
Xu Q, Ma H, Fan J, Yan H, Zhang H, Yin C, Liu X, Liu Y, Wang H. Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05. Toxins. 2019; 11(10):549. https://doi.org/10.3390/toxins11100549
Chicago/Turabian StyleXu, Qianqian, Hongfei Ma, Jinhui Fan, Hai Yan, Haiyang Zhang, Chunhua Yin, Xiaolu Liu, Yang Liu, and Huasheng Wang. 2019. "Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05" Toxins 11, no. 10: 549. https://doi.org/10.3390/toxins11100549
APA StyleXu, Q., Ma, H., Fan, J., Yan, H., Zhang, H., Yin, C., Liu, X., Liu, Y., & Wang, H. (2019). Cloning and Expression of Genes for Biodegrading Nodularin by Sphingopyxis sp. USTB-05. Toxins, 11(10), 549. https://doi.org/10.3390/toxins11100549






