Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors
Abstract
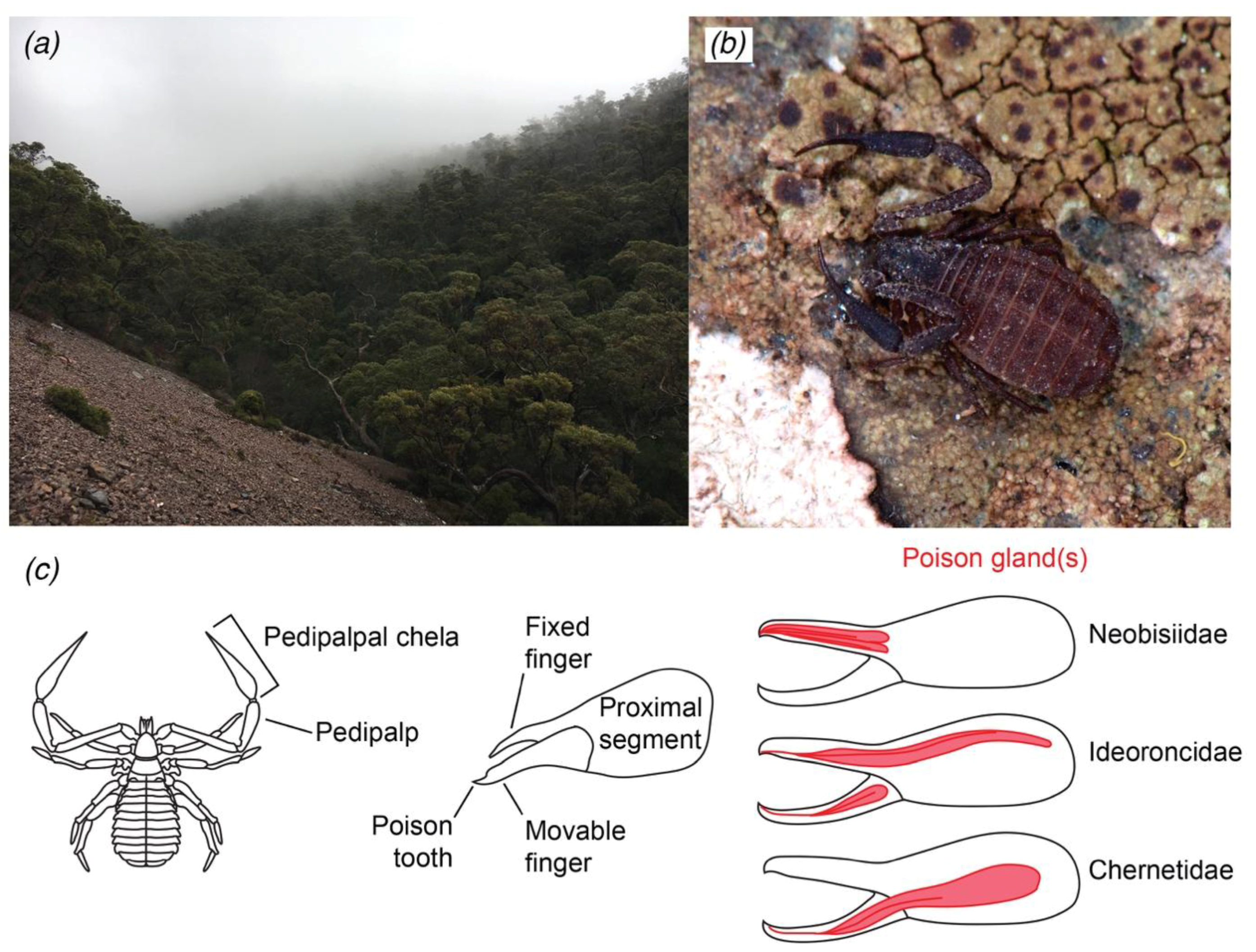
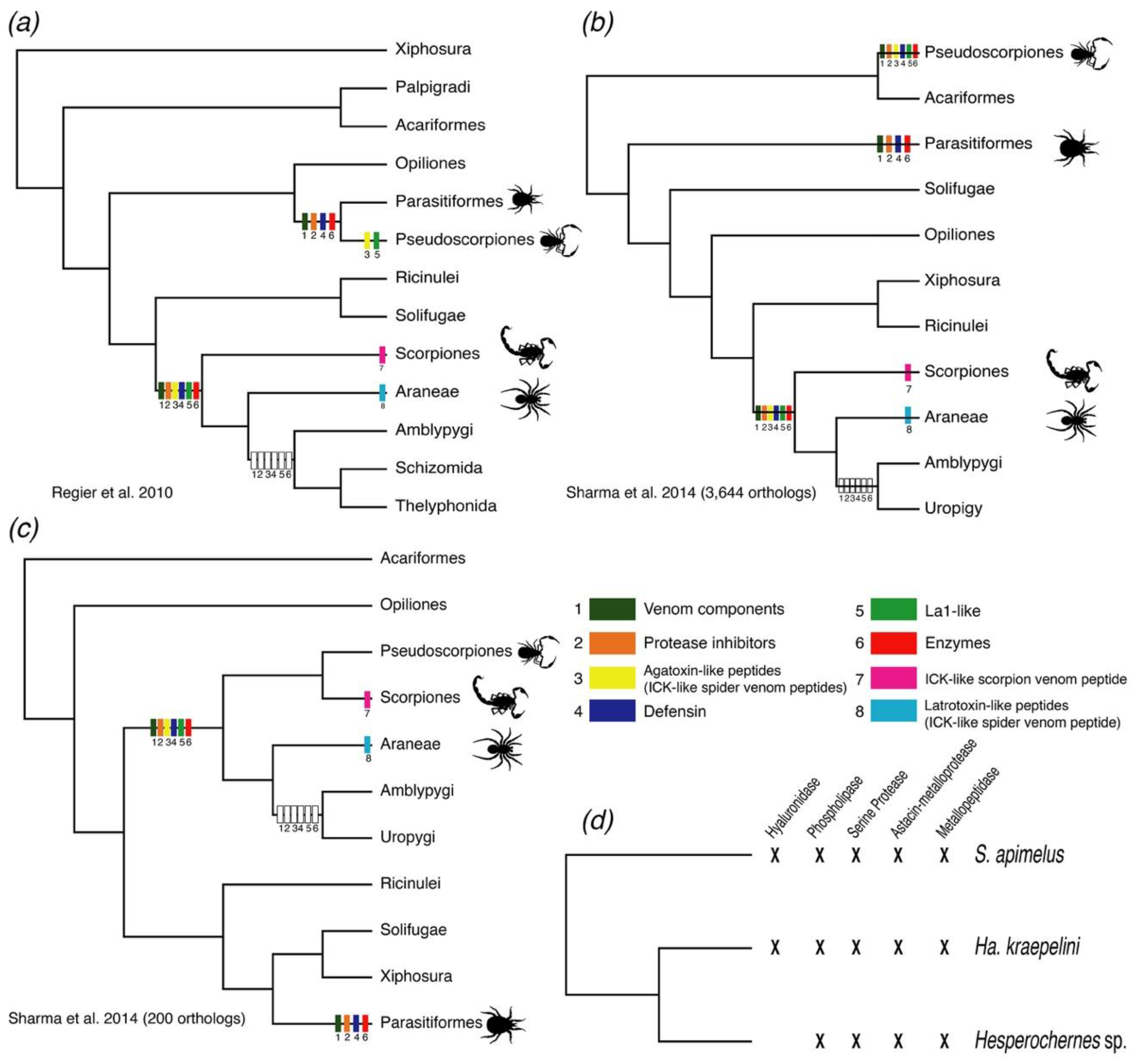
1. Introduction
2. Results
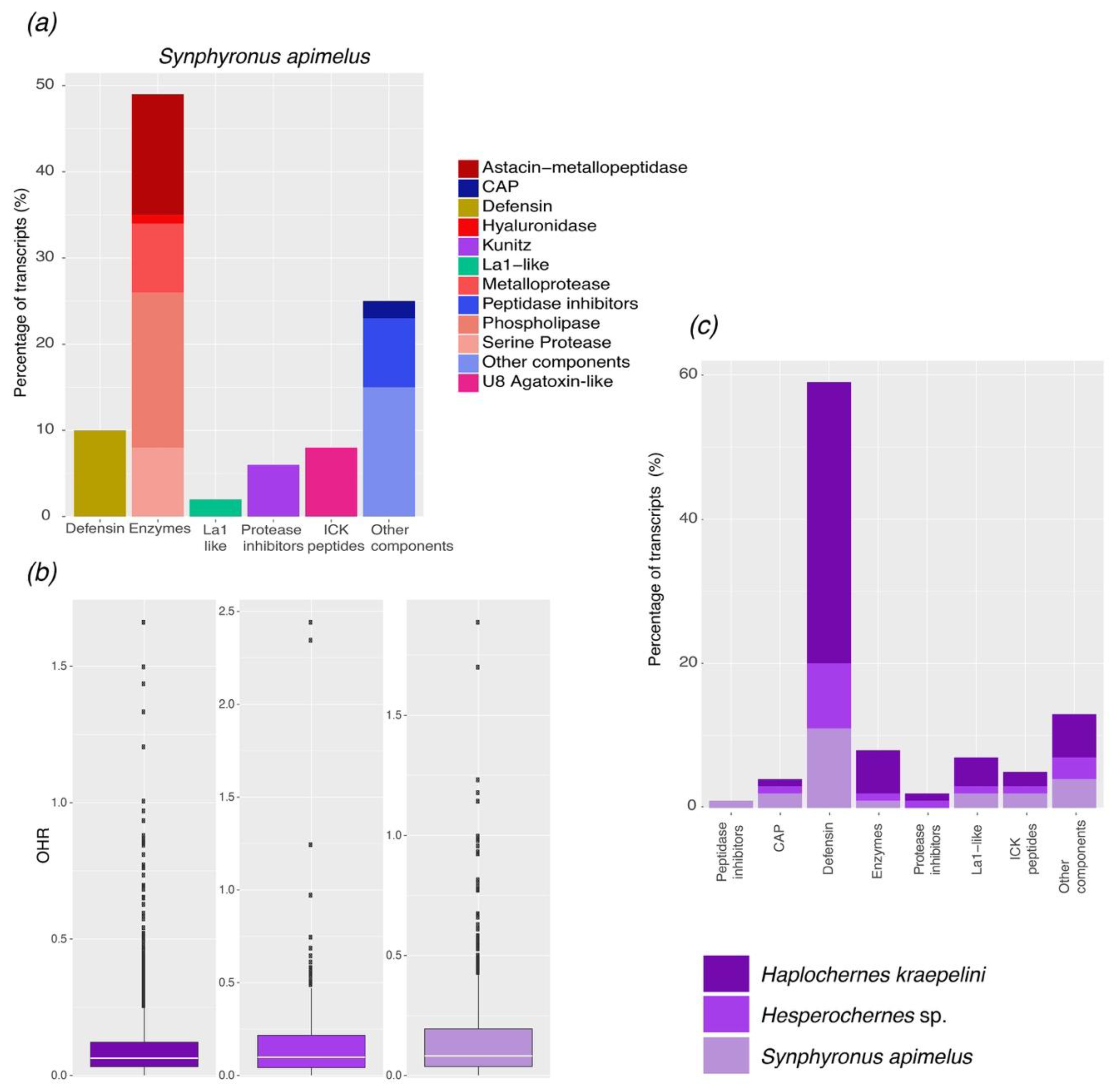
2.1. Transcriptomic Analysis
2.1.1. ICK-Like Spider Venom Peptides
2.1.2. Protease Inhibitors
2.1.3. Enzymes
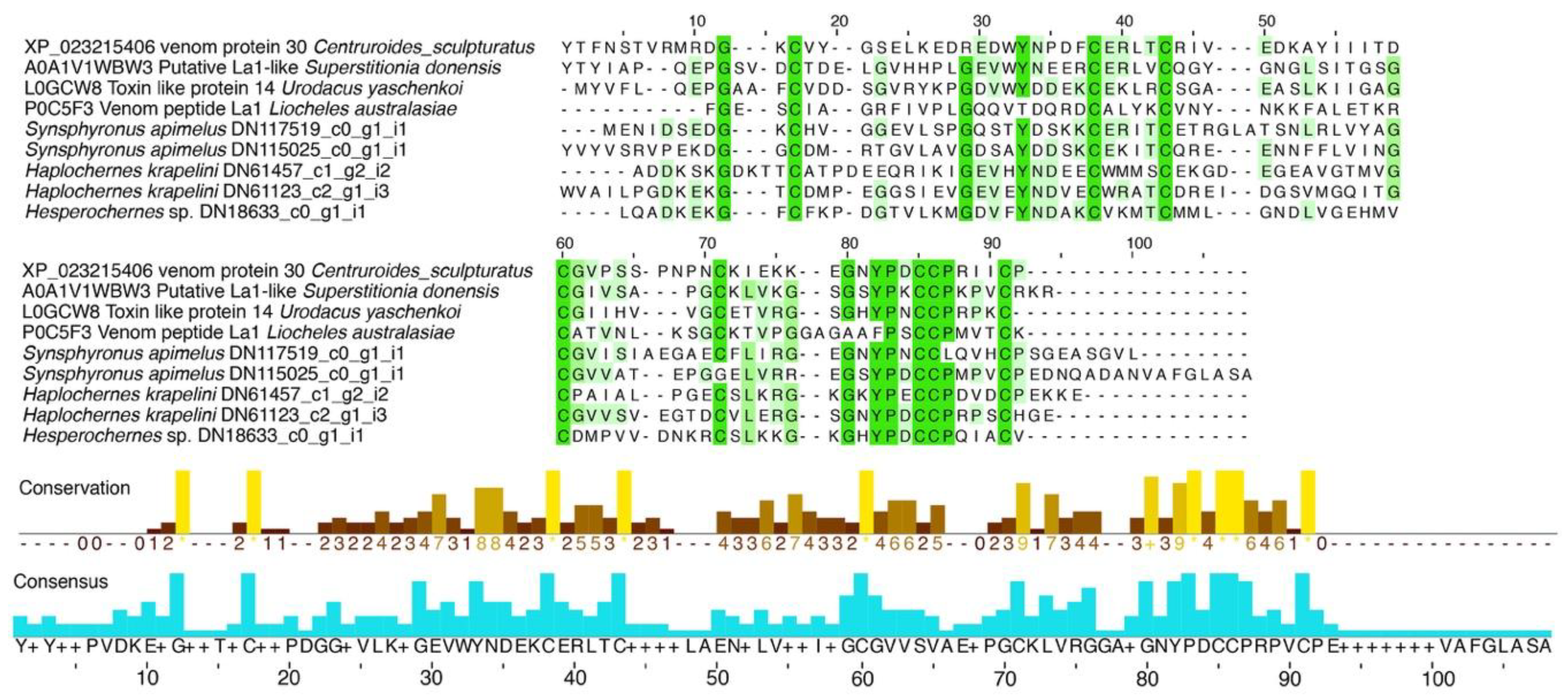
2.1.4. Single Domain von Willebrand Factor Type C Peptides (La1-Like Peptides)
2.1.5. Defensins
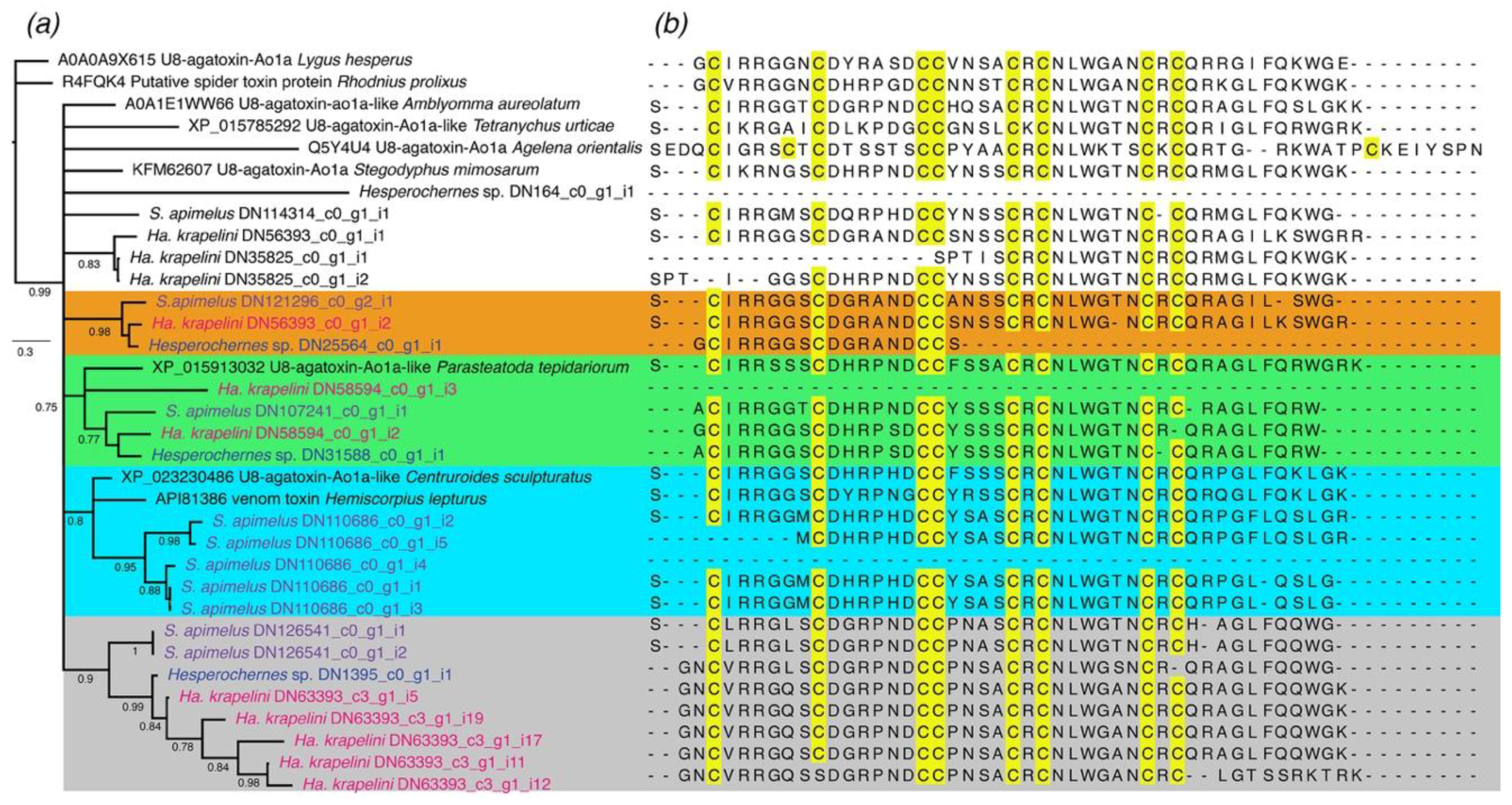
2.1.6. Other Components
2.2. Comparative Analysis of the Repertoire of Venom-Specific Transcripts in S. apimelus
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harvey, M.S. Pseudoscorpions of the World, version 3.0; Western Australian Museum: Perth, Australia, 2013. Available online: http://www.museum.wa.gov.au/catalogues-beta/pseudoscorpions (accessed on 18 April 2018).
- Murienne, J.; Harvey, M.S.; Giribet, G. First molecular phylogeny of the major clades of Pseudoscorpiones (Arthropoda: Chelicerata). Mol. Phylogenet. Evol. 2008, 49, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Harms, D.; Dunlop, J.A. The fossil history of Pseudoscorpions (Arachnida: Pseudoscorpiones). Foss. Rec. 2017, 20, 215–238. [Google Scholar] [CrossRef]
- Wheeler, W.C.; Hayashi, C.Y. The phylogeny of the extant chelicerate orders. Cladistics 1998, 14, 173–192. [Google Scholar] [CrossRef]
- Giribet, G.; Edgecombe, G.D.; Wheeler, W.C.; Babbitt, C. Phylogeny and Systematic Position of Opiliones: A Combined Analysis of Chelicerate Relationships Using Morphological and Molecular Data. Cladistics 2002, 18, 5–70. [Google Scholar] [CrossRef] [PubMed]
- Shultz, J.W. A phylogenetic analysis of the arachnid orders based on morphological characters. Zool. J. Linn. Soc. 2007, 150, 221–265. [Google Scholar] [CrossRef]
- Regier, J.C.; Shultz, J.W.; Zwick, A.; Hussey, A.; Ball, B.; Wetzer, R.; Martin, J.W.; Cunningham, C.W. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 2010, 463, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Borner, J.; Rehm, P.; Schill, R.O.; Ebersberger, I.; Burmester, T. A transcriptome approach to ecdysozoan phylogeny. Mol. Phylogenet. Evol. 2014, 80, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.P.; Kaluziak, S.T.; Pérez-Porro, A.R.; González, V.L.; Hormiga, G.; Wheeler, W.C.; Giribet, G. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 2014, 31, 2963–2984. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.S. The phylogeny and classification of the Pseudoscorpionida (Chelicerata: Arachnida). Invertebr. Syst. 1992, 6, 1373–1435. [Google Scholar] [CrossRef]
- Santos dos, W.F.; Coutinho-Netto, J. Effects of the Paratemnus elongatus pseudoscorpion venom in the uptake and binding of the L-glutamate and GABA from rat cerebral cortex. J. Biochem. Mol. Toxicol. 2006, 20, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnakone, P.; Possani, L.D.; Schwartz, E.F.; Rodriguez de la Vega, R.C. Scorpion Venoms; Springer: Berlin, Germany, 2015. [Google Scholar]
- Gopalakrishnakone, P.; Corzo, G.; de Lima, M.E.; Diego-Garcia, E. Spider Venoms; Springer: Berlin, Germany, 2016. [Google Scholar]
- Pessini, A.C.; Takao, T.T.; Cavalheiro, E.C.; Vichnewski, W.; Sampaio, S.V.; Giglio, J.R.; Arantes, E.C. A hyaluronidase from Tityus serrulatus scorpion venom: Isolation, characterization and inhibition by flavonoids. Toxicon 2001, 39, 1495–1504. [Google Scholar] [CrossRef]
- Ferrer, V.P.; de Mari, T.L.; Gremski, L.H.; Trevisan Silva, D.; da Silveira, R.B.; Gremski, W.; Chaim, O.M.; Senff-Ribeiro, A.; Nader, H.B.; Veiga, S.S. A Novel Hyaluronidase from Brown Spider (Loxosceles intermedia) Venom (Dietrich’s Hyaluronidase): From Cloning to Functional Characterization. PLoS Negl. Trop. Dis. 2013, 7, e2206. [Google Scholar] [CrossRef] [PubMed]
- Bordon, K.C.F.; Wiezel, G.A.; Amorim, F.G.; Arantes, E.C. Arthropod venom Hyaluronidases: Biochemical properties and potential applications in medicine and biotechnology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, D.M.; Zobel-Thropp, P.A.; Kumirov, V.K.; Bandarian, V.; Binford, G.J.; Cordes, M.H.J. Phospholipase D Toxins of Brown Spider Venom Convert Lysophosphatidylcholine and Sphingomyelin to Cyclic Phosphates. PLoS ONE 2013, 8, e72372. [Google Scholar] [CrossRef] [PubMed]
- Incamnoi, P.; Patramanon, R.; Thammasirirak, S.; Chaveerach, A.; Uawonggul, N.; Sukprasert, S.; Rungsa, P.; Daduang, J.; Daduang, S. Heteromtoxin (HmTx), a novel heterodimeric phospholipase A2 from Heterometrus laoticus scorpion venom. Toxicon 2013, 61, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Cruz, A.; Valdés, J.J. Are ticks venomous animals? Front. Zool. 2014, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Perner, J.; Provazník, J.; Schrenková, J.; Urbanová, V.; Ribeiro, J.M.C.; Kopáček, P. RNA-seq analyses of the midgut from blood- and serum-fed Ixodes ricinus ticks. Sci. Rep. 2016, 6, 36695. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Valle, M.; Moolhuijzen, P.; Barrero, R.A.; Ong, C.T.; Busch, G.; Karbanowicz, T.; Booth, M.; Clark, R.; Koehbach, J.; Ijaz, H.; et al. Transcriptome and toxin family analysis of the paralysis tick, Ixodes holocyclus. Int. J. Parasitol. 2018, 48, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Vachon, M. Ordre des pseudoscorpions. In Grassé, Traité de Zoologie; Masson: Paris, France, 1949; Volume 6, pp. 437–481. [Google Scholar]
- Weygoldt, P. The Biology of Pseudoscorpions; Harvard University Press: Cambridge, MA, USA, 1969. [Google Scholar]
- Garb, J.E.; Sharma, P.P.; Ayoub, N.A. Recent progress and prospects for advancing arachnid genomics. Curr. Opin. Insect Sci. 2018, 25, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, 258D–261D. [Google Scholar]
- Hodgson, E. Toxins and Venoms. Prog. Mol. Biol. Transl. Sci. 2012, 112, 373–415. [Google Scholar] [PubMed]
- Chen, Z.; Wang, B.; Hu, J.; Yang, W.; Cao, Z.; Zhuo, R.; Li, W.; Wu, Y. SjAPI, the first functionally characterized Ascaris-type protease inhibitor from animal venoms. PLoS ONE 2013, 8, e57529. [Google Scholar] [CrossRef] [PubMed]
- Mourão, C.; Schwartz, E. Protease inhibitors from marine venomous animals and their counterparts in terrestrial venomous animals. Mar. Drugs 2013, 11, 2069–2112. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Lee, K.S.; Kim, B.Y.; Zou, F.M.; Yoon, H.J.; Je, Y.H.; Li, J.; Jin, B.R. A Spider-derived Kunitz-Type Serine Protease Inhibitor that acts as a Plasmin Inhibitor and an Elastase Inhibitor. PLoS ONE 2013, 8, e53343. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-Y.; Hu, Y.-T.; Yang, W.-S.; He, Y.-W.; Feng, J.; Wang, B.; Zhao, R.-M.; Ding, J.-P.; Cao, Z.-J.; Li, W.-X.; et al. Hg1, Novel Peptide Inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type Potassium channel toxin family. J. Biol. Chem. 2012, 287, 13813–13821. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Otsuki, J.; Hanai, Y.; Nakagawa, Y.; Miyagawa, H. Characterization of peptide components in the venom of the scorpion Liocheles australasiae (Hemiscorpiidae). Toxicon 2007, 50, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.-C.; Nie, Y.; Luo, X.; Wu, S.; Shi, W.; Zhang, L.; Liu, Y.; Cao, H.; Yang, Y.; Zhou, J. Molecular and Bioinformatical characterization of a novel superfamily of cysteine-rich peptides from arthropods. Peptides 2013, 41, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.; Cid-Uribe, J.; Batista, C.; Ortiz, E.; Possani, L. Venom gland transcriptomic and proteomic analyses of the enigmatic scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with insights on the evolution of its venom components. Toxins 2016, 8, 367. [Google Scholar] [CrossRef] [PubMed]
- Shafee, T.M.A.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The Defensins consist of two independent, convergent protein superfamilies. Mol. Biol. Evol. 2016, 33, 2345–2356. [Google Scholar] [CrossRef] [PubMed]
- White, S.H.; Wimley, W.C.; Selsted, M.E. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 1995, 5, 521–527. [Google Scholar] [CrossRef]
- Froy, O.; Gurevitz, M. Arthropod defensins illuminate the divergence of scorpion neurotoxins. J. Pept. Sci. 2004, 10, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Peigneur, S.; Gao, B.; Umetsu, Y.; Ohki, S.; Tytgat, J. Experimental conversion of a defensin into a neurotoxin: Implications for origin of toxic function. Mol. Biol. Evol. 2014, 31, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.; Malyavka, A.; McCutchen, B.; Lu, A.; Schepers, E.; Herrmann, R.; Grishin, E. A novel strategy for the identification of toxinlike structures in spider venom. Proteins 2005, 59, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.E. Agatoxins: Ion channel specific toxins from the American funnel web spider, Agelenopsis aperta. Toxicon 2004, 43, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Strømgaard, K.; Jensen, L.S.; Vogensen, S.B. Polyamine toxins: Development of selective ligands for ionotropic receptors. Toxicon 2005, 45, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Quistad, G.B.; Suwanrumpha, S.; Jarema, M.A.; Shapiro, M.J.; Skinner, W.S.; Jamieson, G.C.; Lui, A.; Fu, E.W. Structure of paralytic acylpolyamines from the spider Agelenopsis aperta. Biochem. Biophys. Res. Commun. 1990, 169, 51–56. [Google Scholar] [CrossRef]
- Foelix, R. Biology of Spiders; OUP: New York, NY, USA, 2011. [Google Scholar]
- Dowell, N.L.; Giorgianni, M.W.; Kassner, V.A.; Selegue, J.E.; Sanchez, E.E.; Carroll, S.B. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 2016, 26, 2434–2445. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 December 2017).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, S.T.; Dzurisin, J.D.K.; Carmichael, R.D.; Lobo, N.F.; Emrich, S.J.; Hellmann, J.J. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC Genom. 2010, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Ewen-Campen, B.; Shaner, N.; Panfilio, K.A.; Suzuki, Y.; Roth, S.; Extavour, C.G. The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC Genom. 2011, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Riesgo, A.; Andrade, S.C.S.; Sharma, P.P.; Novo, M.; Pérez-Porro, A.R.; Vahtera, V.; González, V.L.; Kawauchi, G.Y.; Giribet, G. Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Front. Zool. 2012, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Hormiga, G. A new orthology assessment method for phylogenomic data: Unrooted Phylogenetic Orthology. Mol. Biol. Evol. 2016, 33, 2117–2134. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; Wood, D.L.A.; Newell, F.; Chaumeil, P.A.; Kaas, Q.; Binford, G.J.; Nicholson, G.M.; Gorse, D.; King, G.F. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2010, 39, D653–D657. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple sequence alignment software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santibáñez-López, C.E.; Ontano, A.Z.; Harvey, M.S.; Sharma, P.P. Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors. Toxins 2018, 10, 207. https://doi.org/10.3390/toxins10050207
Santibáñez-López CE, Ontano AZ, Harvey MS, Sharma PP. Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors. Toxins. 2018; 10(5):207. https://doi.org/10.3390/toxins10050207
Chicago/Turabian StyleSantibáñez-López, Carlos E., Andrew Z. Ontano, Mark S. Harvey, and Prashant P. Sharma. 2018. "Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors" Toxins 10, no. 5: 207. https://doi.org/10.3390/toxins10050207
APA StyleSantibáñez-López, C. E., Ontano, A. Z., Harvey, M. S., & Sharma, P. P. (2018). Transcriptomic Analysis of Pseudoscorpion Venom Reveals a Unique Cocktail Dominated by Enzymes and Protease Inhibitors. Toxins, 10(5), 207. https://doi.org/10.3390/toxins10050207





