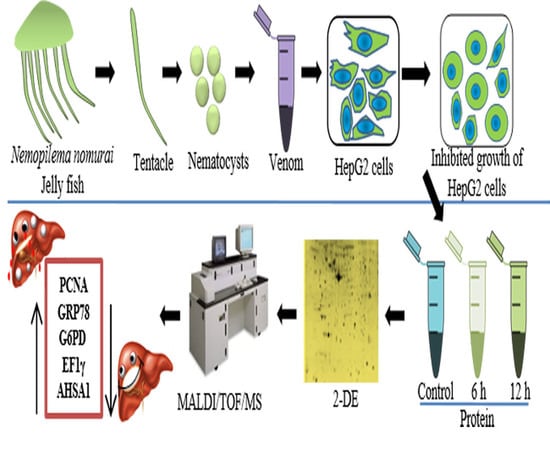
Proteomic Investigation to Identify Anticancer Targets of Nemopilema nomurai Jellyfish Venom in Human Hepatocarcinoma HepG2 Cells
Abstract
1. Introduction
2. Results and Discussion
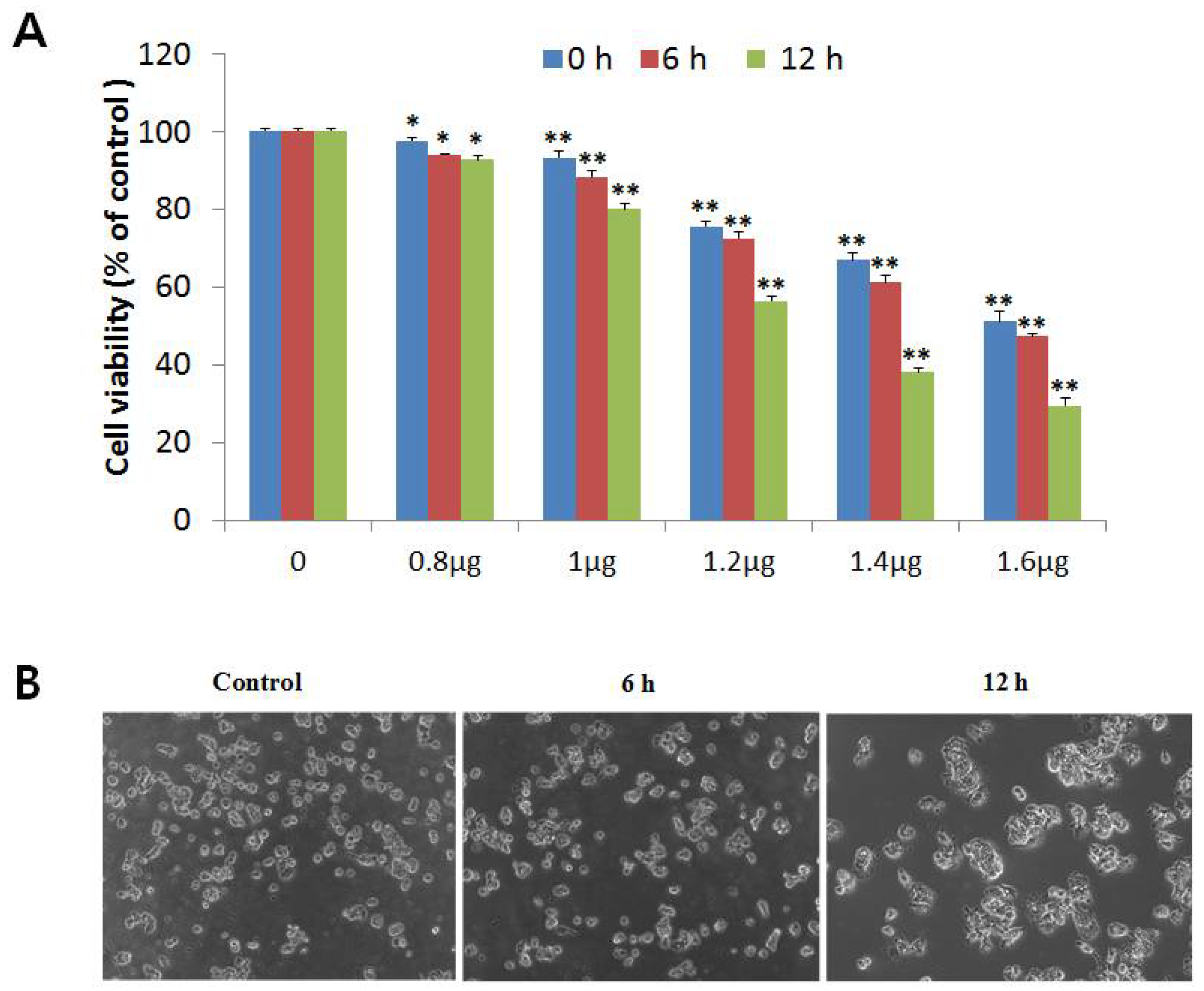
2.1. NnV Induces Cytotoxicity in HepG2 Cell Lines
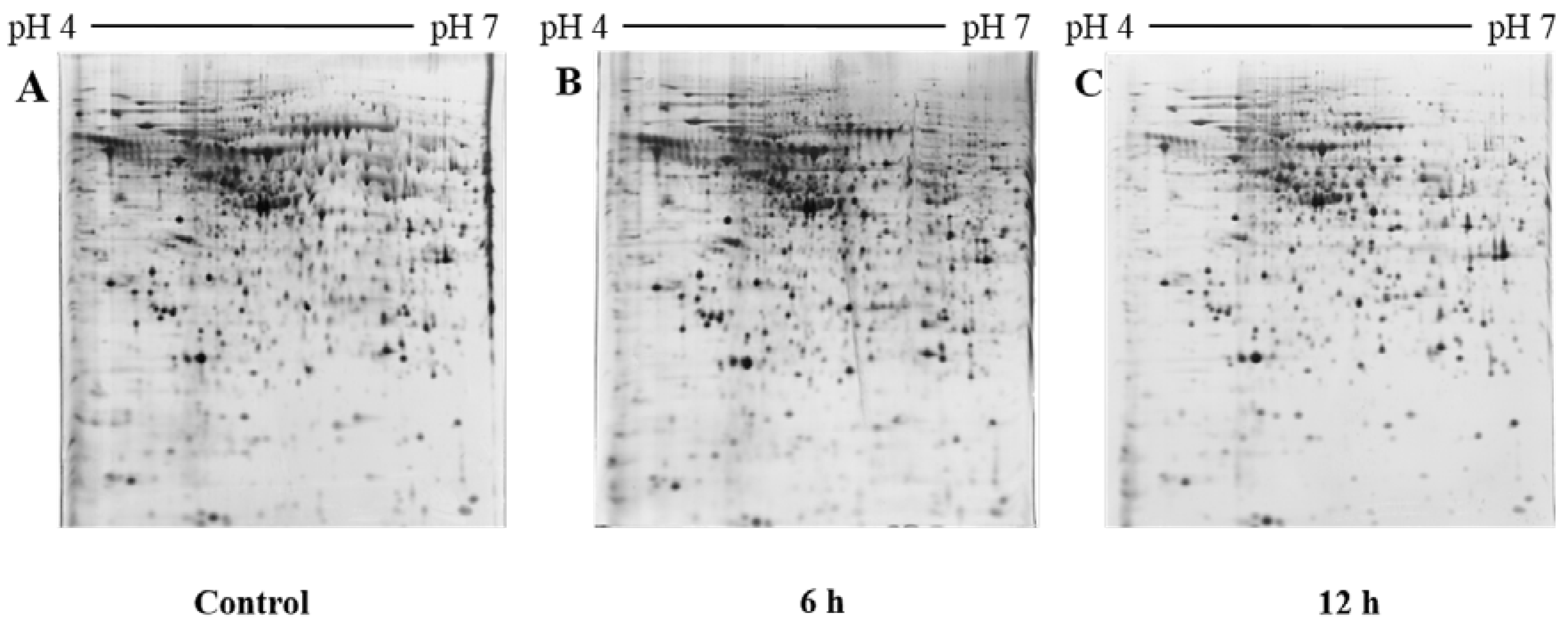
2.2. 2D PAGE Analysis of Untreated and NnV Treated H9c2 Cells
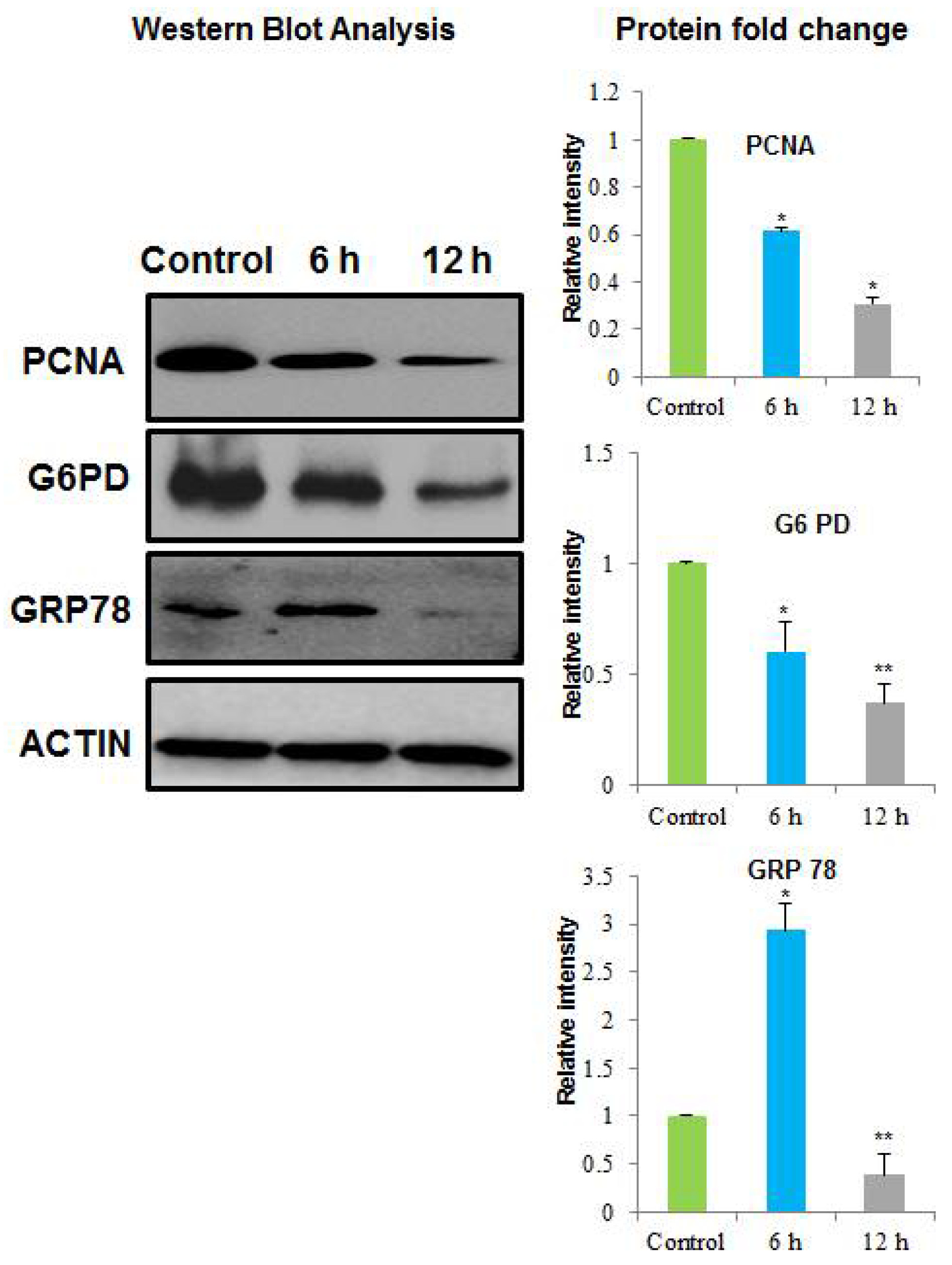
2.3. Identification of the Differential Abundant Proteins by MALDI/TOF/MS and Western Blotting
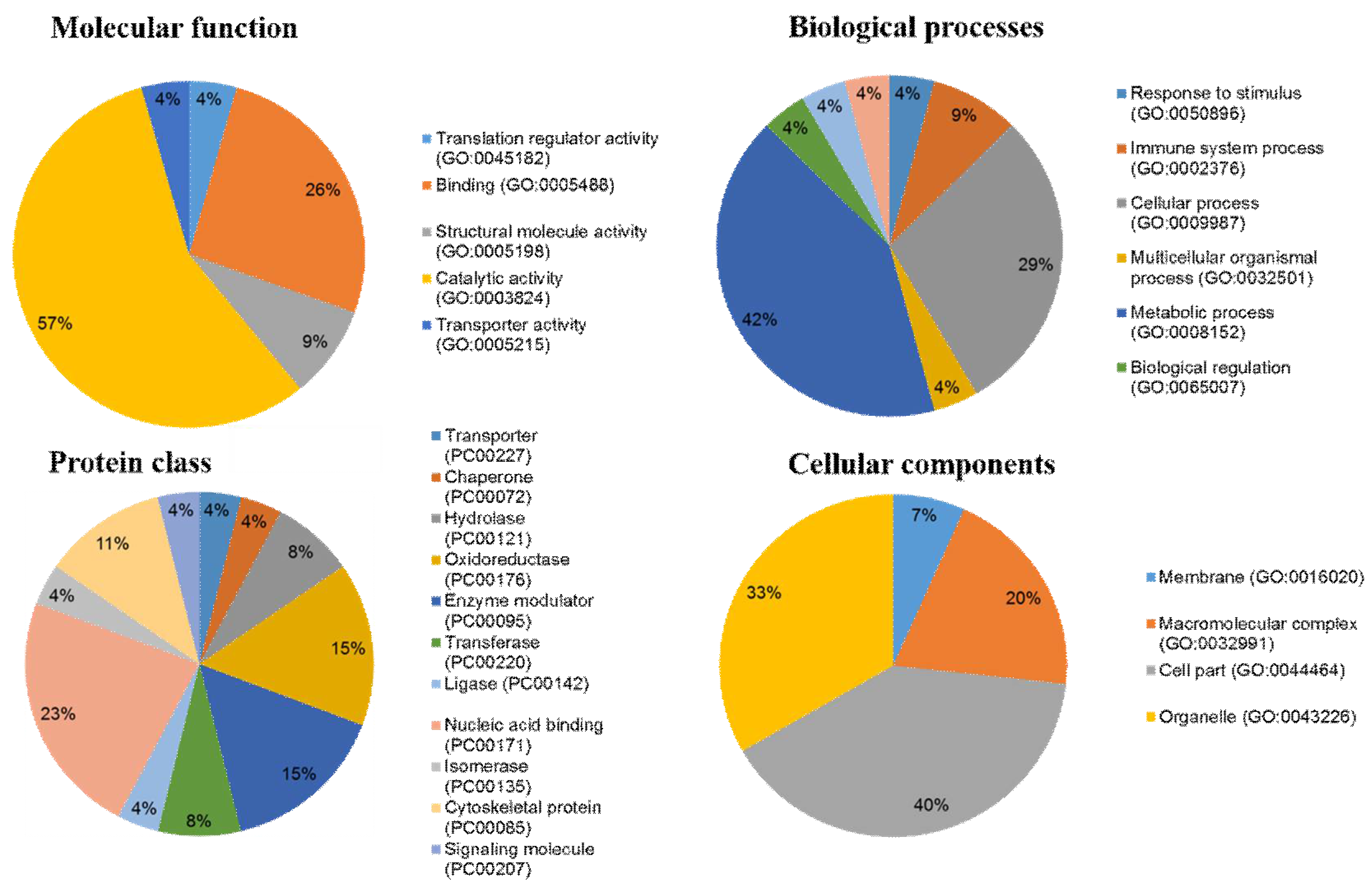
2.4. Ontological Classification of Differentially Abundant Proteins
3. Materials and Methods
3.1. Sample Collection and Preparation
3.2. Venom Extraction and Preparation
3.3. Cell Culture
3.4. MTT Assay for Cell Viability
3.5. Protein Extraction and Sample Preparation
3.6. Two-Dimensional Gel Electrophoresis and Image Analysis
3.7. In-Gel Digestion
3.8. MALDI/TOF/MS Analysis and Database Searching
3.9. Western Blotting
3.10. Statistical and Bioinformatics Analysis of Protein Identified by MALDI/TOF/MS
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Rocha, L. Analysis of molecular structures and mechanisms for toxins derived from venomous animals. Comput. Biol. Chem. 2016, 61, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Rash, L.D.; Hodgson, W.C. Pharmacology and biochemistry of spider venoms. Toxicon 2002, 40, 225–254. [Google Scholar] [CrossRef]
- Ponce, D.; Brinkman, D.L.; Luna-Ramírez, K.; Wright, C.E.; Dorantes-Aranda, J.J. Comparative study of the toxic effects of Chrysaora quinquecirrha (cnidaria: Scyphozoa) and Chironex fleckeri (cnidaria: Cubozoa) venoms using cell-based assays. Toxicon 2015, 106, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Beilei, W.; Ying, L.; Qianqian, W.; Sihua, L.; Yang, W.; Guoyan, L.; Jia, L.; Xuting, Y.; Liming, Z. Cardiovascular effect is independent of hemolytic toxicity of tentacle-only extract from the jellyfish Cyanea capillata. PLoS ONE 2012, 7, e43096. [Google Scholar] [CrossRef] [PubMed]
- Ayed, Y.; Chayma, B.; Hayla, A.; Abid, S.; Bacha, H. Is cell death induced by nematocysts extract of medusa Pelagia noctiluca related to oxidative stress? Environ. Toxicol. 2013, 28, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, I.; Lee, H.; Pyo, M.-J.; Heo, Y.; Bae, S.K.; Kwon, Y.C.; Yoon, W.D.; Kang, C.; Kim, E. Proteomics approach to examine the cardiotoxic effects of Nemopilema nomurai jellyfish venom. J. Proteom. 2015, 128, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Uye, S.-i.; Ohtsu, K.; Iizumi, H. Unusual population explosion of the giant jellyfish Nemopilema nomurai (scyphozoa: Rhizostomeae) in East Asian waters. Mar. Ecol. Prog. Ser. 2006, 307, 161–173. [Google Scholar] [CrossRef]
- Dong, Z.; Liu, D.; Keesing, J.K. Jellyfish blooms in china: Dominant species, causes and consequences. Mar. Pollut. Bull. 2010, 60, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Uye, S.; Burnett, J.; Mianzan, H. Stings of edible jellyfish (Rhopilema hispidum, Rhopilema esculentum and Nemopilema nomurai) in Japanese waters. Toxicon 2006, 48, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Munawir, A.; Cha, M.; Sohn, E.-T.; Lee, H.; Kim, J.-S.; Yoon, W.D.; Lim, D.; Kim, E. Cytotoxicity and hemolytic activity of jellyfish Nemopilema nomurai (scyphozoa: Rhizostomeae) venom. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 150, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Heinen, T.E.; da Veiga, A.B.G. Arthropod venoms and cancer. Toxicon 2011, 57, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Ménez, A.; Stöcklin, R.; Mebs, D. ‘Venomics’ or: The Venomous Systems Genome Project; Pergamon: Oxford, UK, 2006. [Google Scholar]
- Zargan, J.; Sajad, M.; Umar, S.; Naime, M.; Ali, S.; Khan, H.A. Scorpion (Odontobuthus doriae) venom induces apoptosis and inhibits DNA synthesis in human neuroblastoma cells. Mol. Cell. Biochem. 2011, 348, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Ramezanpour, M.; Da Silva, K.B.; Barbara, J.; Sanderson, S. Venom present in sea anemone (Heteractis magnifica) induces apoptosis in non-small-cell lung cancer a549 cells through activation of mitochondria-mediated pathway. Biotechnol. Lett. 2014, 36, 489. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Son, D.J.; Kwak, D.H.; Song, H.S.; Oh, K.-W.; Yoo, H.-S.; Lee, Y.M.; Song, M.J.; Hong, J.T. Snake venom toxin inhibits cell growth through induction of apoptosis in neuroblastoma cells. Arch. Pharm. Res. 2009, 32, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Abu-Sinna, G.; Esmat, A.Y.; Al-Zahaby, A.-A.S.; Soliman, N.A.; Ibrahim, T.M. Fractionation and characterization of Cerastes cerastes cerastes snake venom and the antitumor action of its lethal and non-lethal fractions. Toxicon 2003, 42, 207–215. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, Y.; Li, J.; Xu, S.; Liu, C.; Zhu, Y.; Liang, S. The venom of the spider Macrothele raveni induces apoptosis in the myelogenous leukemia k562 cell line. Leuk. Res. 2012, 36, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-W.; Chaturvedi, P.K.; Chun, S.N.; Lee, Y.G.; Ahn, W.S. Honeybee venom possesses anticancer and antiviral effects by differential inhibition of hpv e6 and e7 expression on cervical cancer cell line. Oncol. Rep. 2015, 33, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, Y.; Hada, N.; Kaneda, T.; Suzuki, T.; Ohshio, T.; Takeda, T.; Kasahara, T. A synthetic glycosphingolipid-induced antiproliferative effect in melanoma cells is associated with suppression of FAK, Akt, and Erk activation. Biol. Pharm. Bull. 2008, 31, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Wang, W.; Smith, D.; Chan, S.C. Effects of the anti-bacterial peptide cecropin b and its analogs, cecropins b-1 and b-2, on liposomes, bacteria, and cancer cells. Biochim. Biophys. Acta BBA-Gener. Subj. 1997, 1336, 171–179. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, X.-Y.; Shou, Q.-Y.; Yan, J.-F.; Chen, L.; Fu, H.-Y.; Wang, J.-C. Bufalin inhibits pancreatic cancer by inducing cell cycle arrest via the c-myc/nf-κb pathway. J. Ethnopharmacol. 2016, 193, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, E.; Reddy, B.V.; Menon, V.P. Antitumor and antioxidant role of Chrysaora quinquecirrha (sea nettle) nematocyst venom peptide against ehrlich ascites carcinoma in swiss albino mice. Mol. Cell. Biochem. 2010, 338, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Maeda, T.; Hasegawa, Y.; Tokunaga, T.; Ogawa, S.; Fukuda, K.; Nagatsuka, N.; Nagao, K.; Ueno, S. Antioxidant activity of the giant jellyfish Nemopilema nomurai measured by the oxygen radical absorbance capacity and hydroxyl radical averting capacity methods. Mol. Med. Rep. 2011, 4, 919–922. [Google Scholar] [PubMed]
- Morishige, H.; Sugahara, T.; Nishimoto, S.; Muranaka, A.; Ohno, F.; Shiraishi, R.; Doi, M. Immunostimulatory effects of collagen from jellyfish in vivo. Cytotechnology 2011, 63, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Armani, A.; Tinacci, L.; Giusti, A.; Castigliego, L.; Gianfaldoni, D.; Guidi, A. What is inside the jar? Forensically informative nucleotide sequencing (fins) of a short mitochondrial coi gene fragment reveals a high percentage of mislabeling in jellyfish food products. Food Res. Int. 2013, 54, 1383–1393. [Google Scholar] [CrossRef]
- Ohta, N.; Sato, M.; Ushida, K.; Kokubo, M.; Baba, T.; Taniguchi, K.; Urai, M.; Kihira, K.; Mochida, J. Jellyfish mucin may have potential disease-modifying effects on osteoarthritis. BMC Biotechnol. 2009, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Bae, S.K.; Kim, M.; Pyo, M.J.; Kim, M.; Yang, S.; Won, C.-K.; Yoon, W.D.; Han, C.H.; Kang, C.; et al. Anticancer effect of Nemopilema nomurai jellyfish venom on HepG2 cells and a tumor xenograft animal model. Evid.-Based Complement. Altern. Med. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. Pcna controls establishment of sister chromatid cohesion during s phase. Mol. Cell 2006, 23, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Lee, H. Lys-110 is essential for targeting pcna to replication and repair foci, and the k110a mutant activates apoptosis. Biol. Cell 2008, 100, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Stoimenov, I.; Helleday, T. Pcna on the Crossroad of Cancer; Portland Press Limited: London, UK, 2009. [Google Scholar]
- Kato, T.; Kameoka, S.; Kimura, T.; Nishikawa, T.; Kobayashi, M. C-erbb-2 and pcna as prognostic indicators of long-term survival in breast cancer. Anticancer Res. 2002, 22, 1097–1103. [Google Scholar] [PubMed]
- Kuang, R.-G.; Wu, H.-X.; Hao, G.-X.; Wang, J.-W.; Zhou, C.-J. Expression and significance of igf-2, pcna, mmp-7, and alpha-actin in gastric carcinoma with lauren classification. Turk. J. Gastroenterol. 2013, 24, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Kimos, M.C.; Wang, S.; Borkowski, A.; Yang, G.Y.; Yang, C.S.; Perry, K.; Olaru, A.; Deacu, E.; Sterian, A.; Cottrell, J. Esophagin and proliferating cell nuclear antigen (pcna) are biomarkers of human esophageal neoplastic progression. Int. J. Cancer 2004, 111, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Dong, Z.R.; Guo, Z.Y.; Wang, C.H.; Zhi, X.T.; Zhou, J.W.; Li, D.K.; Chen, Z.T.; Chen, Z.Q.; Hu, S.Y. Mannose-mediated inhibitory effects of pa-msha on invasion and metastasis of hepatocellular carcinoma via egfr/akt/iκbβ/nf-κb pathway. Liver Int. 2015, 35, 1416–1429. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Wortman, M.; Dillehay, K.L.; Seibel, W.L.; Evelyn, C.R.; Smith, S.J.; Malkas, L.H.; Zheng, Y.; Lu, S.; Dong, Z. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol. Pharmacol. 2012, 81, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, K.; Li, Z. Unfolded protein response in cancer: The physician’s perspective. J. Hematol. Oncol. 2011, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S. The er chaperone and signaling regulator grp78/bip as a monitor of endoplasmic reticulum stress. Methods 2005, 35, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lee, A.S. Stress induction of grp78/bip and its role in cancer. Curr. Mol. Med. 2006, 6, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z. Glucose regulated protein 78: A critical link between tumor microenvironment and cancer hallmarks. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2012, 1826, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S. Grp78 induction in cancer: Therapeutic and prognostic implications. Cancer Res. 2007, 67, 3496–3499. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, S.; Liu, J.; Wang, X.; Ji, J.; Cao, Y.; Lu, K.; Wang, J.; Gao, Y. Expression of grp78 predicts taxane-based therapeutic resistance and recurrence of human gastric cancer. Exp. Mol. Pathol. 2014, 96, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, H.; Hyman, G.; Evans, S.; Manglik, V.; Gwebu, E.; Banerjee, A.; Vaughan, D.; Medley, J.; Krauss, C.; Wilkins, J. Identification of the transmembrane glucose regulated protein 78 as a biomarker for the brain cancer glioblastoma multiforme by gene expression and proteomic studies. J. Membr. Sci. Technol. 2014, 4. [Google Scholar] [CrossRef]
- Shimizu, A.; Kaira, K.; Yasuda, M.; Asao, T.; Ishikawa, O. Clinical and pathological significance of er stress marker (bip/grp78 and perk) expression in malignant melanoma. Pathol. Oncol. Res. 2017, 23, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Kaira, K.; Toyoda, M.; Shimizu, A.; Mori, K.; Shino, M.; Sakakura, K.; Takayasu, Y.; Takahashi, K.; Oyama, T.; Asao, T. Expression of er stress markers (grp78/bip and perk) in patients with tongue cancer. Neoplasma 2015, 63, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Lee, A.S. Glucose regulated proteins in cancer progression, drug resistance and immunotherapy. Cancer Biol. Ther. 2006, 5, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, Y.; Jia, Z.; Li, Q.; Gong, W.; Wang, L.; Wei, D.; Yao, J.; Fang, S.; Xie, K. Association of elevated grp78 expression with increased lymph node metastasis and poor prognosis in patients with gastric cancer. Clin. Exp. Metast. 2006, 23, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Boros, L.G.; Puigjaner, J.; Cascante, M.; Lee, W.-N.P.; Brandes, J.L.; Bassilian, S.; Yusuf, F.I.; Williams, R.D.; Muscarella, P.; Melvin, W.S. Oxythiamine and dehydroepiandrosterone inhibit the nonoxidative synthesis of ribose and tumor cell proliferation. Cancer Res. 1997, 57, 4242–4248. [Google Scholar] [PubMed]
- Furuta, E.; Okuda, H.; Kobayashi, A.; Watabe, K. Metabolic genes in cancer: Their roles in tumor progression and clinical implications. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2010, 1805, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Duan, X.; Mao, W.; Li, X.; Li, Z.; Li, Q.; Zheng, Z.; Xu, H.; Chen, M.; Wang, P.G. O-glcnacylation of g6pd promotes the pentose phosphate pathway and tumor growth. Nat. Commun. 2015, 6, 8468. [Google Scholar] [CrossRef] [PubMed]
- Koudstaal, J.; Makkink, B.; Overdiep, S.H. Enzyme histochemical pattern in human tumours—ii. Oxidoreductases in carcinoma of the colon and the breast. Eur. J. Cancer 1975, 11, 111–114. [Google Scholar] [CrossRef]
- Batetta, B.; Pulisci, D.; Bonatesta, R.; Sanna, F.; Piras, S.; Mulas, M.; Spano, O.; Putzolu, M.; Broccia, G.; Dessı̀, S. G6pd activity and gene expression in leukemic cells from g6pd-deficient subjects. Cancer Lett. 1999, 140, 53–58. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, C.; Tang, Q.; Su, Y.; Li, B.; Chen, L.; Zhang, Z.; Cai, T.; Zhu, Y. Variant g6pd levels promote tumor cell proliferation or apoptosis via the stat3/5 pathway in the human melanoma xenograft mouse model. BMC Cancer 2013, 13, 251. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Li, Y.-S.; Chen, B.; Chang, Y.-F.; Liu, G.-C.; Hong, Y.; Chen, H.-L.; Xiyang, Y.-B. Elevated glucose-6-phosphate dehydrogenase expression in the cervical cancer cases is associated with the cancerigenic event of high-risk human papillomaviruses. Exp. Biol. Med. 2015, 240, 1287–1297. [Google Scholar] [CrossRef] [PubMed]
- Nna, E.; Tothill, I.E.; Ludeman, L.; Bailey, T. Endogenous control genes in prostate cells: Evaluation of gene expression using ‘real-time’quantitative polymerase chain reaction. Med. Princ. Pract. 2010, 19, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Hughes, E.C. The effect of enzymes upon metabolism, storage, and release of carbohydrates in normal and abnormal endometria. Cancer 1976, 38, 487–502. [Google Scholar] [CrossRef]
- Kuo, W.-Y.; Tang, T.K. Effects of g6pd overexpression in nih3t3 cells treated with tert-butyl hydroperoxide or paraquat. Free Radic. Biol. Med. 1998, 24, 1130–1138. [Google Scholar] [CrossRef]
- Kuo, W.Y.; Lin, J.Y.; Tang, T.K. Human glucose-6-phosphate dehydrogenase (g6pd) gene transforms NIH 3t3 cells and induces tumors in nude mice. Int. J. Cancer 2000, 85, 857–864. [Google Scholar] [CrossRef]
- Hu, H.; Ding, X.; Yang, Y.; Zhang, H.; Li, H.; Tong, S.; An, X.; Zhong, Q.; Liu, X.; Ma, L. Changes in glucose-6-phosphate dehydrogenase expression results in altered behavior of HBV-associated liver cancer cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2014, 307, G611–G622. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Jiang, C.; Feng, Y.; Chen, R.; Lin, X.; Zhang, Z.; Han, L.; Chen, X.; Li, H.; Guo, Y. Effects of g6pd activity inhibition on the viability, ROS generation and mechanical properties of cervical cancer cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Riis, B.; Rattan, S.I.; Clark, B.F.; Merrick, W.C. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990, 15, 420–424. [Google Scholar] [CrossRef]
- Mathur, S.; Cleary, K.R.; Inamdar, N.; Kim, Y.H.; Steck, P.; Frazier, M.L. Overexpression of elongation factor-1γ protein in colorectal carcinoma. Cancer 1998, 82, 816–821. [Google Scholar] [CrossRef]
- Al-Maghrebi, M.; Anim, J.T.; Olalu, A.A. Up-regulation of eukaryotic elongation factor-1 subunits in breast carcinoma. Anticancer Res. 2005, 25, 2573–2577. [Google Scholar] [PubMed]
- Mimori, K.; Mori, M.; Akiyoshi, T.; Tanaka, S.; Sugimachi, K. The overexpression of elongation factor 1 gamma mrna in gastric carcinoma. Cancer 1995, 75, 1446–1449. [Google Scholar] [CrossRef]
- Shuda, M.; Kondoh, N.; Tanaka, K.; Ryo, A.; Wakatsuki, T.; Hada, A.; Goseki, N.; Igari, T.; Hatsuse, K.; Aihara, T. Enhanced expression of translation factor mRNAs in hepatocellular carcinoma. Anticancer Res. 2000, 20, 2489–2494. [Google Scholar] [PubMed]
- Lew, Y.; Jones, D.V.; Mars, W.M.; Evans, D.; Byrd, D.; Frazier, M.L. Expression of elongation factor-1 gamma-related sequence in human pancreatic cancer. Pancreas 1992, 7, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Mimori, K.; Mori, M.; Inoue, H.; Ueo, H.; Mafune, K.; Akiyoshi, T.; Sugimachi, K. Elongation factor 1 gamma mRNA expression in oesophageal carcinoma. Gut 1996, 38, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Wang, J.; Van Houten, B. The Role of Dynamin-Related Protein 1 in Cancer Growth: A Promising Therapeutic Target? Taylor & Francis: Abingdon, UK, 2013. [Google Scholar]
- Inoue-Yamauchi, A.; Oda, H. Depletion of mitochondrial fission factor drp1 causes increased apoptosis in human colon cancer cells. Biochem. Biophys. Res. Commun. 2012, 421, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, J.; Yu, M.; Xie, Y.; Huang, Y.; Wolff, D.W.; Abel, P.W.; Tu, Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013, 32, 4814–4824. [Google Scholar] [CrossRef] [PubMed]
- Rehman, J.; Zhang, H.J.; Toth, P.T.; Zhang, Y.; Marsboom, G.; Hong, Z.; Salgia, R.; Husain, A.N.; Wietholt, C.; Archer, S.L. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012, 26, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Parone, P.A.; Da Cruz, S.; Tondera, D.; Mattenberger, Y.; James, D.I.; Maechler, P.; Barja, F.; Martinou, J.-C. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE 2008, 3, e3257. [Google Scholar] [CrossRef] [PubMed]
- Raemaekers, T.; Ribbeck, K.; Beaudouin, J.; Annaert, W.; Van Camp, M.; Stockmans, I.; Smets, N.; Bouillon, R.; Ellenberg, J.; Carmeliet, G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 2003, 162, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Iyer, J.; Moghe, S.; Furukawa, M.; Tsai, M.Y. What’s Nu(SAP) in mitosis and cancer? Cell. Signal. 2011, 23, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Satow, R.; Shitashige, M.; Kanai, Y.; Takeshita, F.; Ojima, H.; Jigami, T.; Honda, K.; Kosuge, T.; Ochiya, T.; Hirohashi, S.; et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin. Cancer Res. 2010, 16, 2518–2528. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.T.; Nasir, A.; Culhane, A.; Venkataramu, C.; Fulp, W.; Rubio, R.; Wang, T.; Agrawal, D.; McCarthy, S.M.; Gruidl, M.; et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res. Treat. 2010, 119, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ryu, B.; Kim, D.S.; Deluca, A.M.; Alani, R.M. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE 2007, 2, e594. [Google Scholar] [CrossRef] [PubMed]
- Kokkinakis, D.M.; Liu, X.; Neuner, R.D. Modulation of cell cycle and gene expression in pancreatic tumor cell lines by methionine deprivation (methionine stress): Implications to the therapy of pancreatic adenocarcinoma. Mol. Cancer Ther. 2005, 4, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Wang, H.; He, F.; Huang, H. NuSAP1 is essential for neural crest cell migration in zebrafish. Protein Cell 2010, 1, 259–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okamoto, A.; Higo, M.; Shiiba, M.; Nakashima, D.; Koyama, T.; Miyamoto, I.; Kasama, H.; Kasamatsu, A.; Ogawara, K.; Yokoe, H.; et al. Down-regulation of nucleolar and spindle-associated protein 1 (NuSAP1) expression suppresses tumor and cell proliferation and enhances anti-tumor effect of paclitaxel in oral squamous cell carcinoma. PLoS ONE 2015, 10, e0142252. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Wang, L.; Zhong, C.; Qi, R.; Li, Y. AHSA1 regulates proliferation, apoptosis, migration, and invasion of osteosarcoma. Biomed. Pharmacother. 2016, 77, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Panaretou, B.; Siligardi, G.; Meyer, P.; Maloney, A.; Sullivan, J.K.; Singh, S.; Millson, S.H.; Clarke, P.A.; Naaby-Hansen, S.; Stein, R.; et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 2002, 10, 1307–1318. [Google Scholar] [CrossRef]
- Shen, H.; Cai, M.; Zhao, S.; Wang, H.; Li, M.; Yao, S.; Jiang, N. Overexpression of rfc3 is correlated with ovarian tumor development and poor prognosis. Tumour Biol. 2014, 35, 10259–10266. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Kondoh, N.; Imazeki, N.; Hada, A.; Hatsuse, K.; Matsubara, O.; Yamamoto, M. The knockdown of endogenous replication factor c4 decreases the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Liver Int. 2009, 29, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.M.; Choi, S.J.; Kim, J.K. Expression profiles of sv40-immortalization-associated genes upregulated in various human cancers. J. Cell. Biochem. 2009, 106, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Nadin, S.B.; Sottile, M.L.; Montt-Guevara, M.M.; Gauna, G.V.; Daguerre, P.; Leuzzi, M.; Gago, F.E.; Ibarra, J.; Cuello-Carrion, F.D.; Ciocca, D.R.; et al. Prognostic implication of hspa (hsp70) in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Cell Stress Chaperones 2014, 19, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Flores, I.L.; Kawahara, R.; Miguel, M.C.; Granato, D.C.; Domingues, R.R.; Macedo, C.C.; Carnielli, C.M.; Yokoo, S.; Rodrigues, P.C.; Monteiro, B.V.; et al. Eef1d modulates proliferation and epithelial-mesenchymal transition in oral squamous cell carcinoma. Clin. Sci. 2016, 130, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Utsunomiya, T.; Mimori, K.; Tanaka, Y.; Tanaka, F.; Inoue, H.; Murayama, S.; Mori, M. Clinical significance of elongation factor-1 delta mRNA expression in oesophageal carcinoma. Br. J. Cancer 2004, 91, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.A.; Burnett, J.W.; Alderslade, P. Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon 1998, 36, 1075–1085. [Google Scholar] [CrossRef]
- Carrette, T.; Seymour, J. A rapid and repeatable method for venom extraction from cubozoan nematocysts. Toxicon 2004, 44, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Finney, D.J.; Tattersfield, F. Probit Analysis, 2nd ed.; Finney, D.J., Ed.; Cambridge University Press: New York, NY, USA, 1952. [Google Scholar]
- Mortz, E.; Krogh, T.N.; Vorum, H.; Gorg, A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 2001, 1, 1359–1363. [Google Scholar] [CrossRef]
- Shevchenko, A.; Jensen, O.N.; Podtelejnikov, A.V.; Sagliocco, F.; Wilm, M.; Vorm, O.; Mortensen, P.; Shevchenko, A.; Boucherie, H.; Mann, M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 1996, 93, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the panther classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
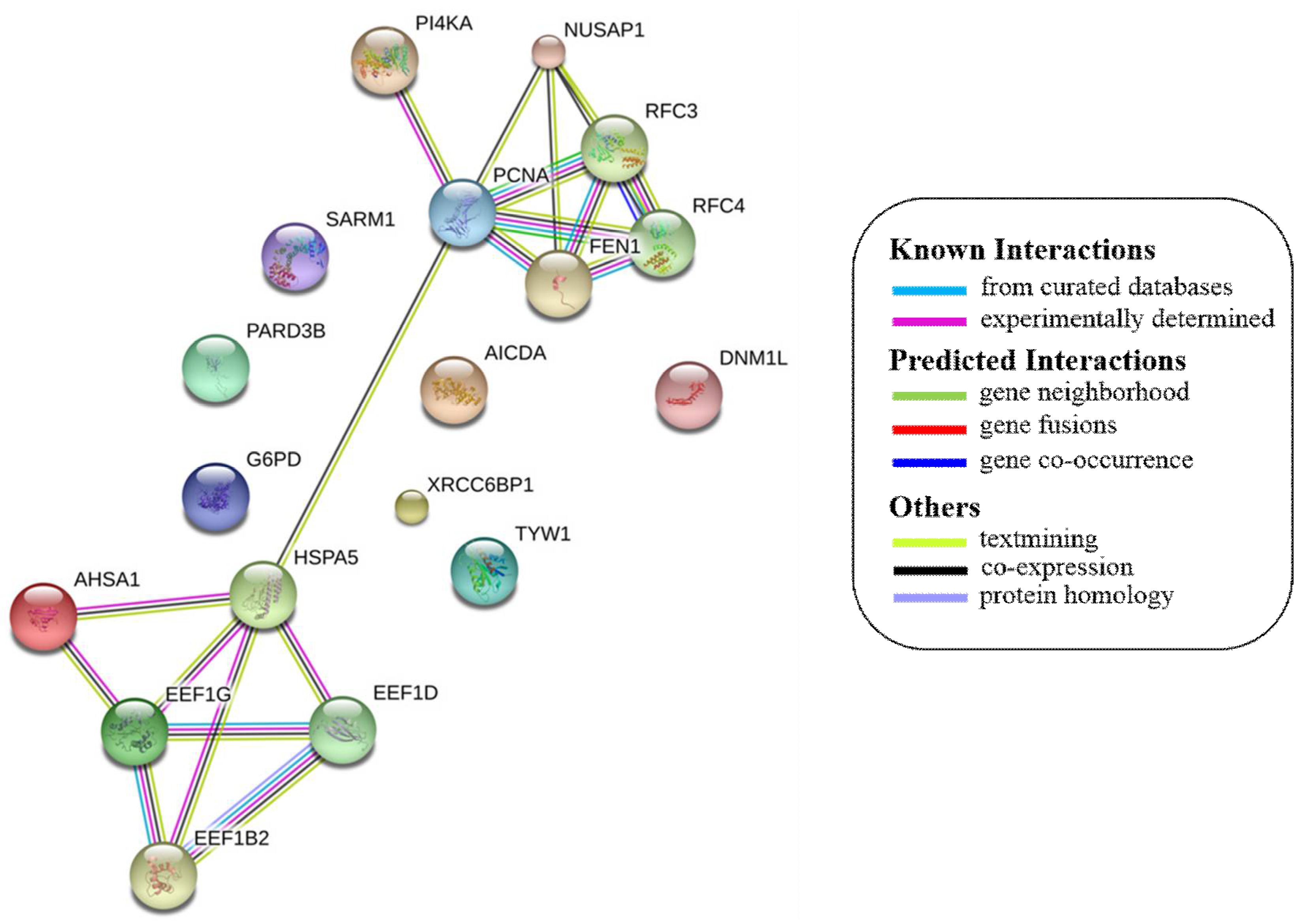
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The string database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
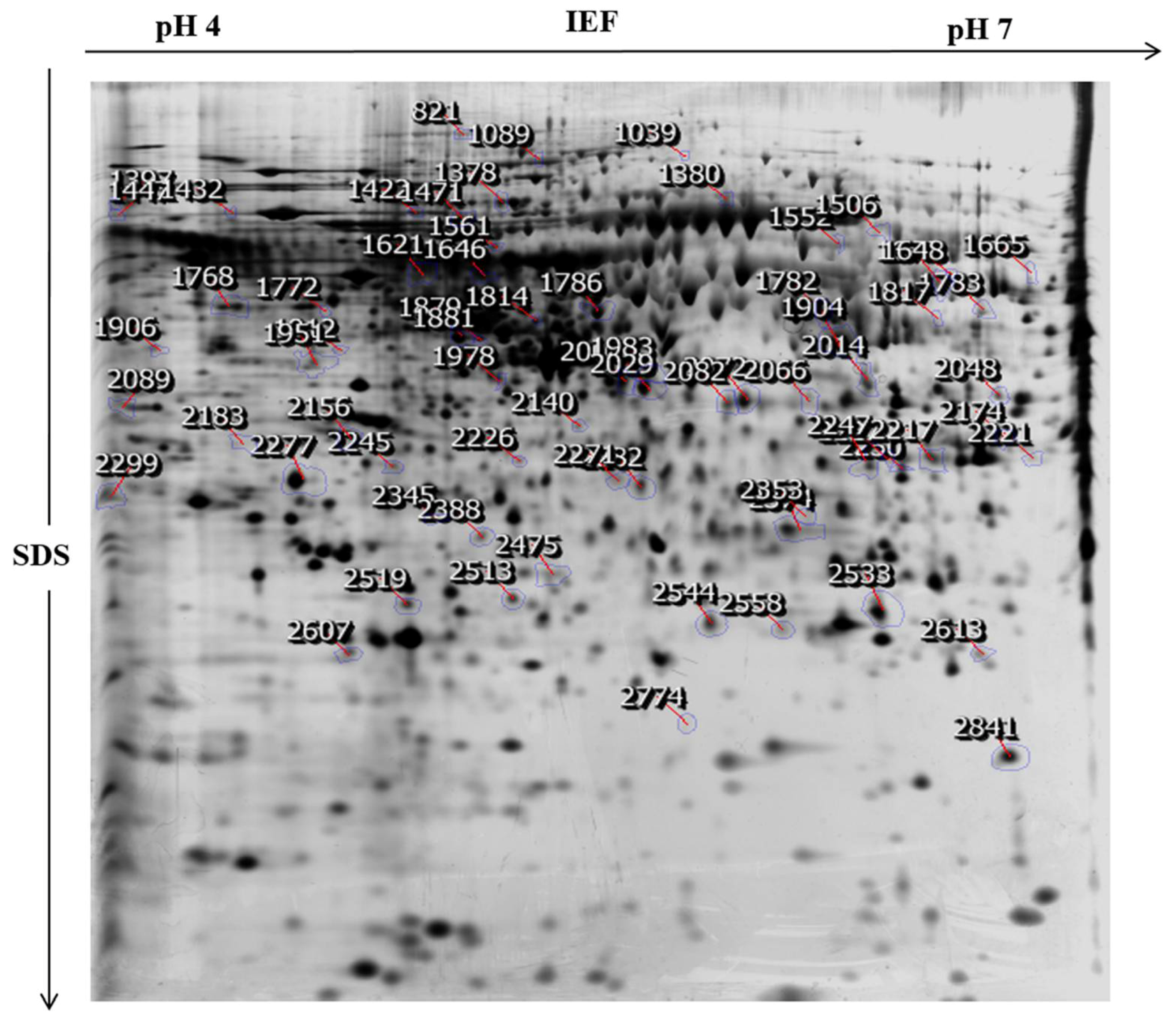
| Spot No 1 | Accession Number 2 | Protein Name | Theoretical MW/Pi 3 | Mascot Score 4 | Coverage % 5 | Matched Peptides 6 | Biological Features |
|---|---|---|---|---|---|---|---|
| Down-regulated Proteins | |||||||
| 2277 | PCNA_HUMAN | Proliferating cell nuclear antigen | 29,092/4.57 | 144 | 72% | 16 | DNA repair, DNA Regulation, and mismatch repair. |
| 1648 | KIF28_HUMAN | Kinesin like protein KIF 28P | 109,098/8.68 | 58 | 9% | 8 | Mitochondrian organization and organelle transport along microtubule. |
| 2299 | GRP78_HUMAN | 78 kDa glucose -regulated protein | 72,402/5.07 | 69 | 17% | 9 | Negative regulation of the apoptotic process and negative regulation of transforming growth factor beta receptor signaling pathway. |
| 2008 | AHSA1_HUMAN | Activator of 90 kDa heat shock protein ATPase homolog 1 | 38,421/5.41 | 72 | 28% | 9 | Positive regulation of ATPase activity. |
| 1768 | NUSAP_HUMAN | Nucleolar and spindle-associated protein 1 | 49,593/9.92 | 61 | 32% | 12 | Establishment of mitotic spindle localization, mitotic chromosome condensation, and positive regulation of mitosis. |
| 2245 | NDUF7_HUMAN | NADH dehydrogenase [ubiquinone] complex 1, assembly factor 7 | 49,435/8.47 | 68 | 11% | 5 | Methylation, mitochondrial respiratory complex 1 assembly, and methyltransferase activity. |
| 1978 | DNM1L_HUMAN | Dynamin-1-like protein | 82,339/6.37 | 57 | 14% | 6 | Positive regulation of the apoptotic process and positive regulation of intrinsic apoptotic signaling pathway. |
| 2183 | ATLA3_HUMAN | Atlastin-3 | 60,960/5.43 | 58 | 10% | 6 | GTP catabolic process, golgi organization, endoplasmic reticulum organization and, homooligomerization. |
| 1772 | HNRH1_HUMAN | Heterogeneous nuclear ribonucleoprotein H | 49,484/5.89 | 74 | 33% | 10 | RNA Processing, regulation of RNA splicing, POLY(A) RNA binding, and poly(U) RNA binding. |
| 1817 | EF1G_HUMAN | Elongation factor 1-gamma | 50,429/6.25 | 58 | 15% | 6 | Translational elongation, cellular protein metabolic process, translation, and gene expression. |
| Up-regulated Proteins | |||||||
| 1665 | G6PD_HUMAN | Glucose-6-phosphate 1-dehydrogenase | 59,675/6.39 | 82 | 16% | 8 | Pentose-phosphate shunt, oxidative branch, Glucose 6-phosphate metabolic process, NADP metabolic process, and NADPH (nicotinamide adenine dinucleotide phosphate-oxidase) regeneration. |
| 2245 | ATP23_HUMAN | Mitochondrial inner membrane protease ATP23 homolog | 28,690/8.30 | 57 | 29% | 4 | Double strand break via non homologous end joining and protein phosphorylation. |
| 2089 | DNM3A_HUMAN | DNA (Cytosine-5)-methyltransferase 3A | 103,390/6.19 | 58 | 18% | 16 | DNA methylation, S-adenosylhomocysteine metabolic process, regulation of transcription from RNA poly II promoter, and regulation of gene expression by genetic imprinting. |
| 1648 | TYW1_HUMAN | S-adenosyl-l-methionine-dependent tRNA 4-demethylwyosine synthase | 84,732/6.42 | 57 | 7% | 7 | tRNA processing. |
| 1646 | CP21A_HUMAN | Steroid 21-hydroxylase | 56,251/7.71 | 57 | 11% | 5 | Glucocorticoid biosynthetic process and steroid metabolic process. |
| 2353 | AICDA_HUMAN | Single-stranded DNA cytosine deaminase | 24,337/9.50 | 68 | 47% | 9 | mRNA processing, DNA demethylation, cell differentiation, and protein binding. |
| 1906 | UBP15_HUMAN | Ubiquitin carboxyl-terminal hydrolase 15 | 113,602/5.06 | 56 | 18% | 10 | Transforming growth factor beta receptor signaling pathway, BMP signaling pathway, and protein deubiquitination. |
| 2029 | PI4KA_HUMAN | Phosphatidylinositol 4-kinase alpha | 233,622/6.43 | 70 | 28% | 62 | Signal transduction, phosphatidylinositol-mediated signaling, and phospholipid metabolic process. |
| 2014 | SYHC_HUMAN | Histidine-tRNA ligase | 57,944/5.72 | 64 | 34% | 17 | Histidyl-tRNA aminoacylation, cellular metabolism, tRNA amino acylation for protein translation, and protein biosynthesis. |
| 821 | DHB13_HUMAN | 17-beta-hydroxysteroid dehydrogenase 13 | 33,976/9.14 | 65 | 46% | 10 | Oxidoreductase activity. |
| 2841 | SARM1_HUMAN | Sterile alpha and TIR motif-containing protein 1 | 80,365/6.14 | 57 | 20% | 9 | Toll like receptor signaling pathway, regulation of dendrite morphogenesis, and regulation of neuron death. |
| 2282 | ZSC31_HUMAN | Zinc finger and SCAN domain-containing protein 31 | 48,233/6.42 | 63 | 14% | 5 | Transcription and transcription regulation. |
| 2217 | PAR3L_HUMAN | Partitioning defective 3 homolog B | 133,097/8.54 | 72 | 11% | 11 | Cell cycle and cell division. |
| 1378 | GARL3_HUMAN | GTPase-activating Rap/Ran-GAP domain-like protein 3 | 113,808/7.57 | 60 | 9% | 8 | Regulation of small GTPase mediated signal transduction. |
| S.NO | Protein Name | String Interactions | KEGG Pathways | |
|---|---|---|---|---|
| 1 | PCNA | 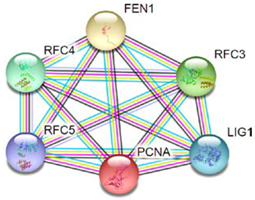 | DNA replication, mismatch repair, nucleotide excision repair, and base excision repair. | |
| 2 | HSPA5/GRP78 | 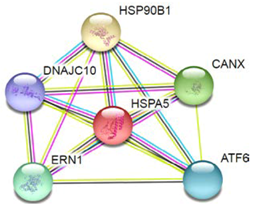 | Protein processing in endoplasmic reticulum, thyroid hormone synthesis, antigen processing, and presentation. | |
| 3 | G6P8 | 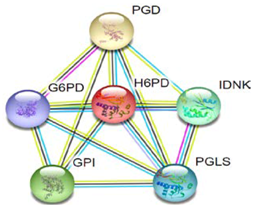 | Microbial metabolism in diverse environments, carbon metabolism, glycolysis/gluconeogenesis, amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, pentose phosphate pathway, biosynthesis of amino acids, and pyruvate metabolism. | |
| 4 | EEF1G | 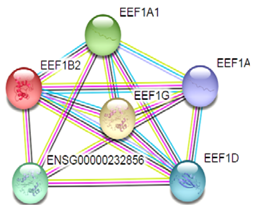 | Ribosome Legionellosis and Legionellosis. | |
| 5 | DNM1L |  | TNF signaling pathway. | |
| 6 | NUSAP1 | 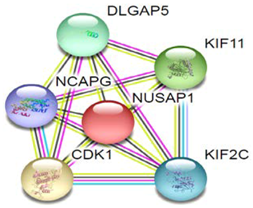 | Mitotic sister chromatid, segregation, mitotic nuclear division, and cell division. | |
| 7 | HSP90 | 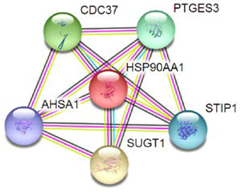 | Protein processing in endoplasmic reticulum, antigen processing and presentation, NOD-like receptor signaling pathway, progesterone-mediated oocyte, prostate cancer, and estrogen signaling pathway. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choudhary, I.; Lee, H.; Pyo, M.J.; Heo, Y.; Chae, J.; Yum, S.S.; Kang, C.; Kim, E. Proteomic Investigation to Identify Anticancer Targets of Nemopilema nomurai Jellyfish Venom in Human Hepatocarcinoma HepG2 Cells. Toxins 2018, 10, 194. https://doi.org/10.3390/toxins10050194
Choudhary I, Lee H, Pyo MJ, Heo Y, Chae J, Yum SS, Kang C, Kim E. Proteomic Investigation to Identify Anticancer Targets of Nemopilema nomurai Jellyfish Venom in Human Hepatocarcinoma HepG2 Cells. Toxins. 2018; 10(5):194. https://doi.org/10.3390/toxins10050194
Chicago/Turabian StyleChoudhary, Indu, Hyunkyoung Lee, Min Jung Pyo, Yunwi Heo, Jinho Chae, Seung Shic Yum, Changkeun Kang, and Euikyung Kim. 2018. "Proteomic Investigation to Identify Anticancer Targets of Nemopilema nomurai Jellyfish Venom in Human Hepatocarcinoma HepG2 Cells" Toxins 10, no. 5: 194. https://doi.org/10.3390/toxins10050194
APA StyleChoudhary, I., Lee, H., Pyo, M. J., Heo, Y., Chae, J., Yum, S. S., Kang, C., & Kim, E. (2018). Proteomic Investigation to Identify Anticancer Targets of Nemopilema nomurai Jellyfish Venom in Human Hepatocarcinoma HepG2 Cells. Toxins, 10(5), 194. https://doi.org/10.3390/toxins10050194