Human Monoclonal scFvs that Neutralize Fribrinogenolytic Activity of Kaouthiagin, a Zinc-Metalloproteinase in Cobra (Naja kaouthia) Venom
Abstract
1. Introduction
2. Results
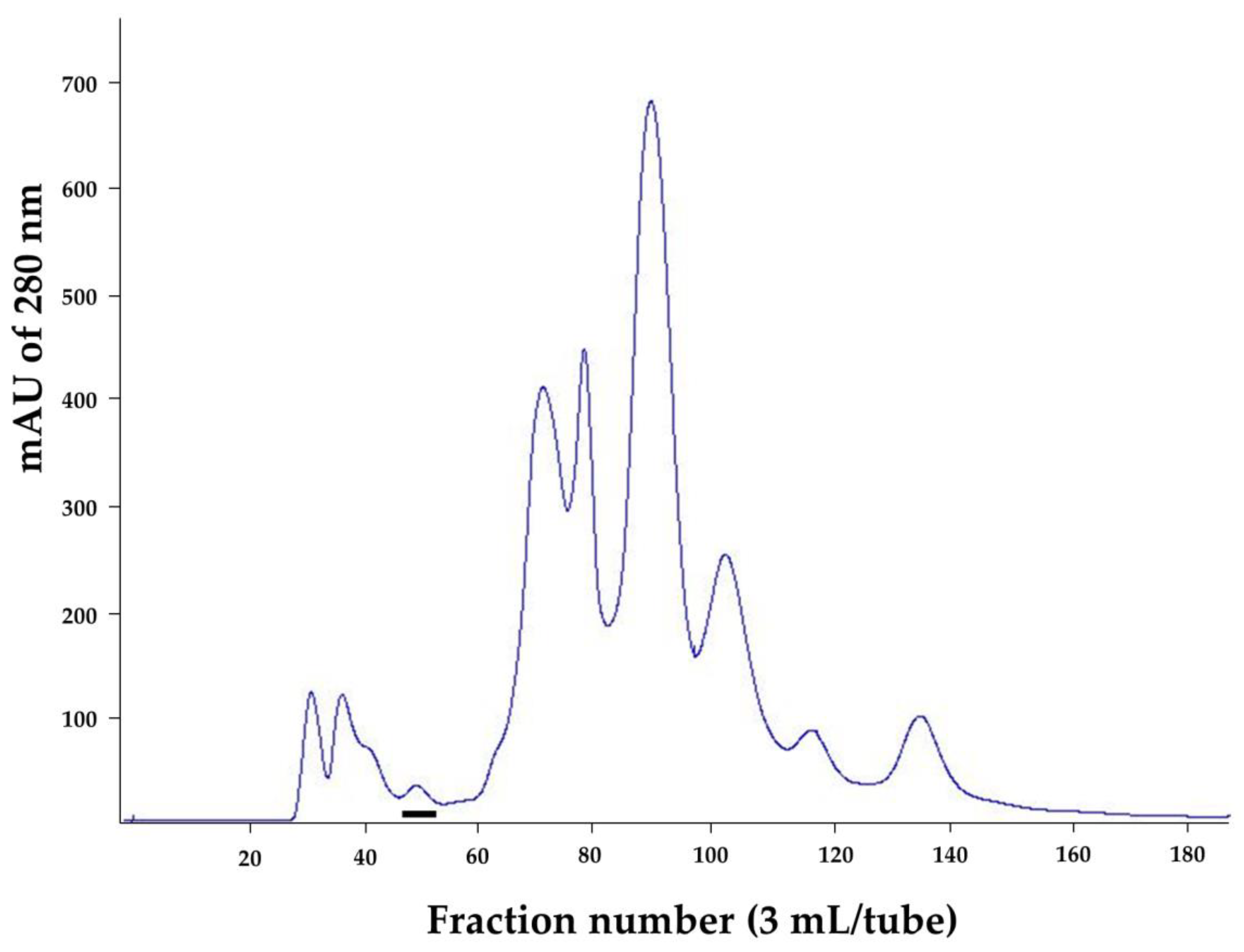
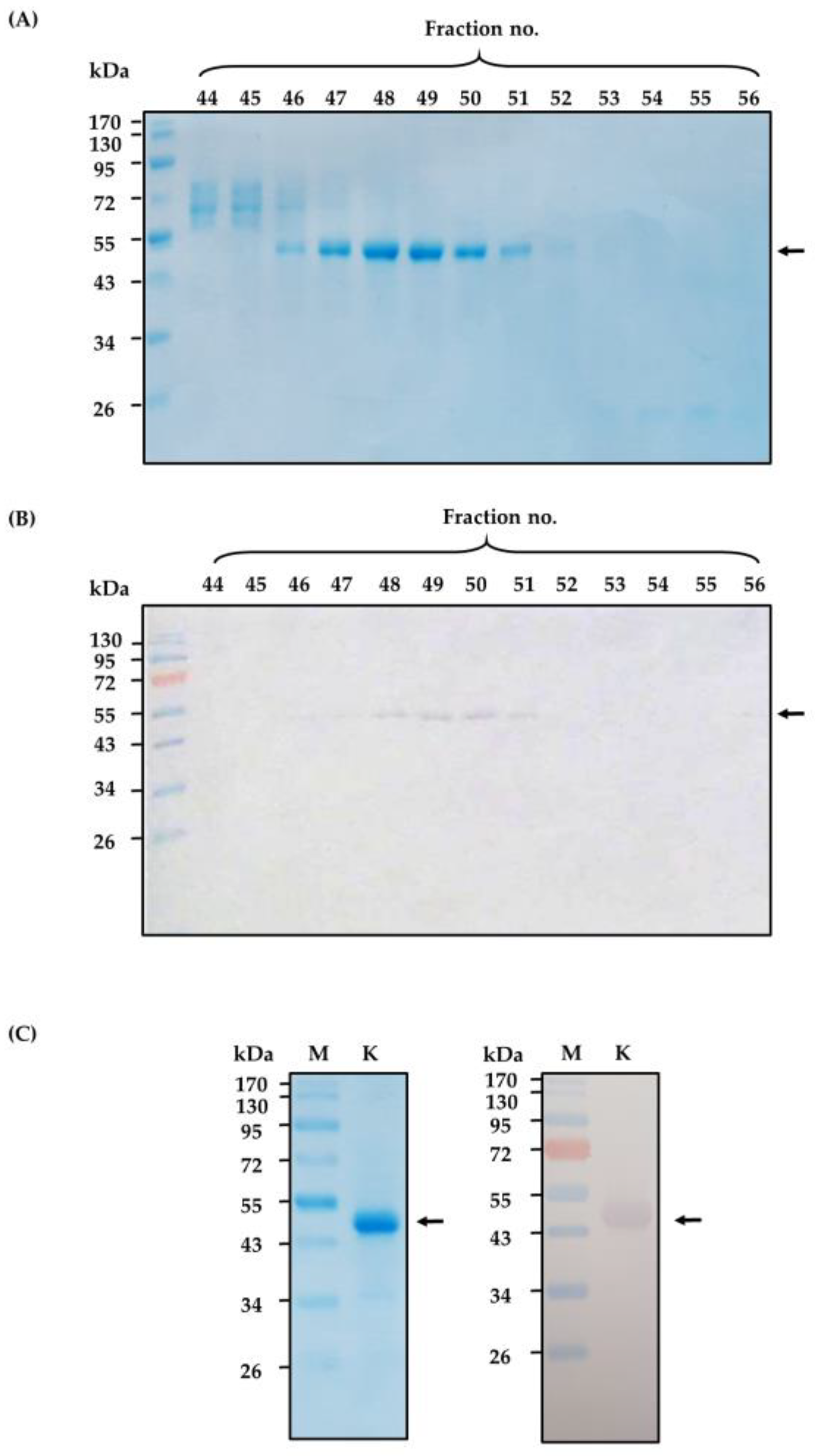
2.1. Purified Cobra Kaouthiagin
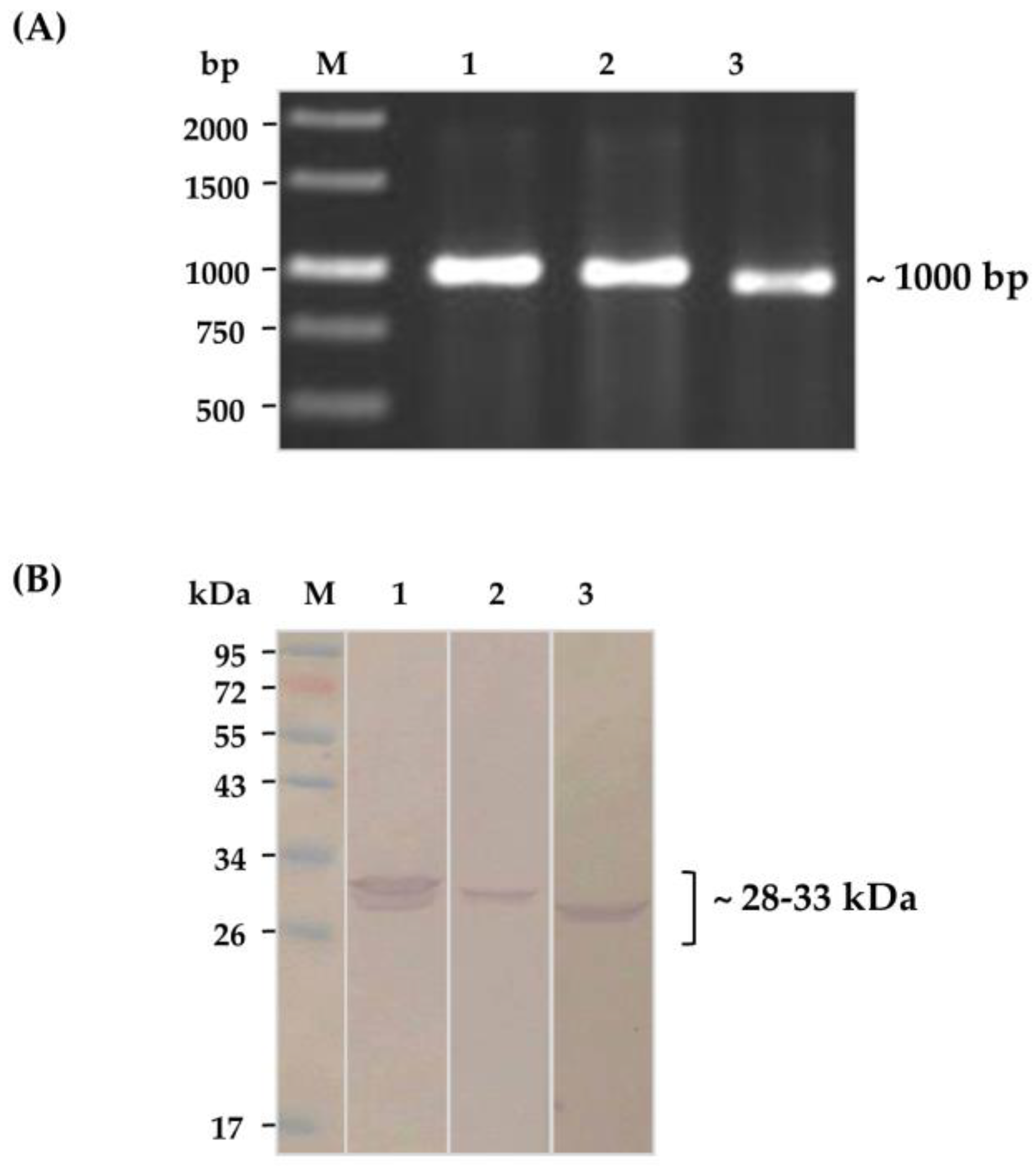
2.2. Soluble HuscFvs That Bound to Kaouthiagin
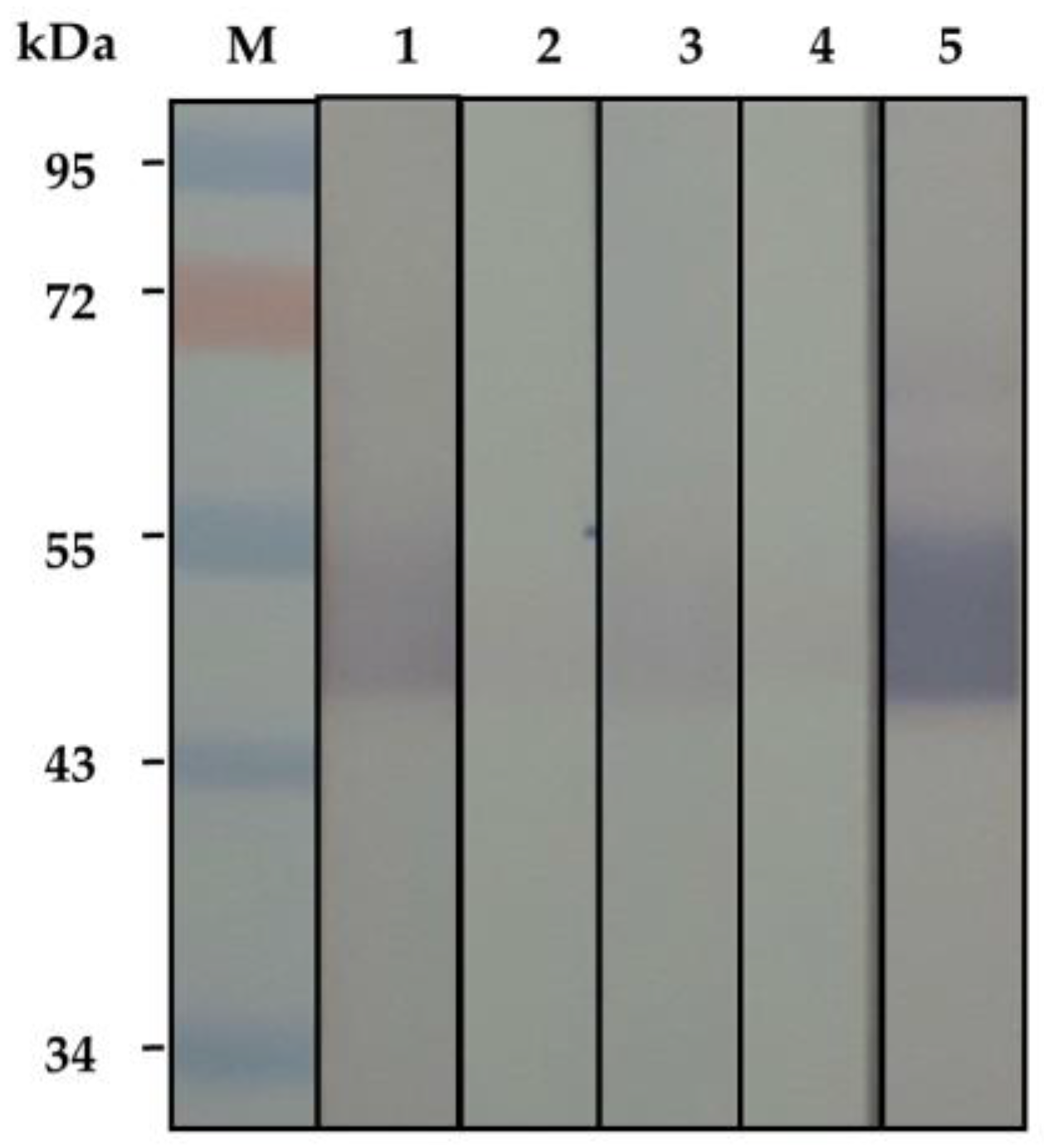
2.3. Binding of the Soluble HuscFvs to Kaouthiagin
2.4. HuscFvs-Mediated Inhibition of Binding of Kaouthiagin to vWF
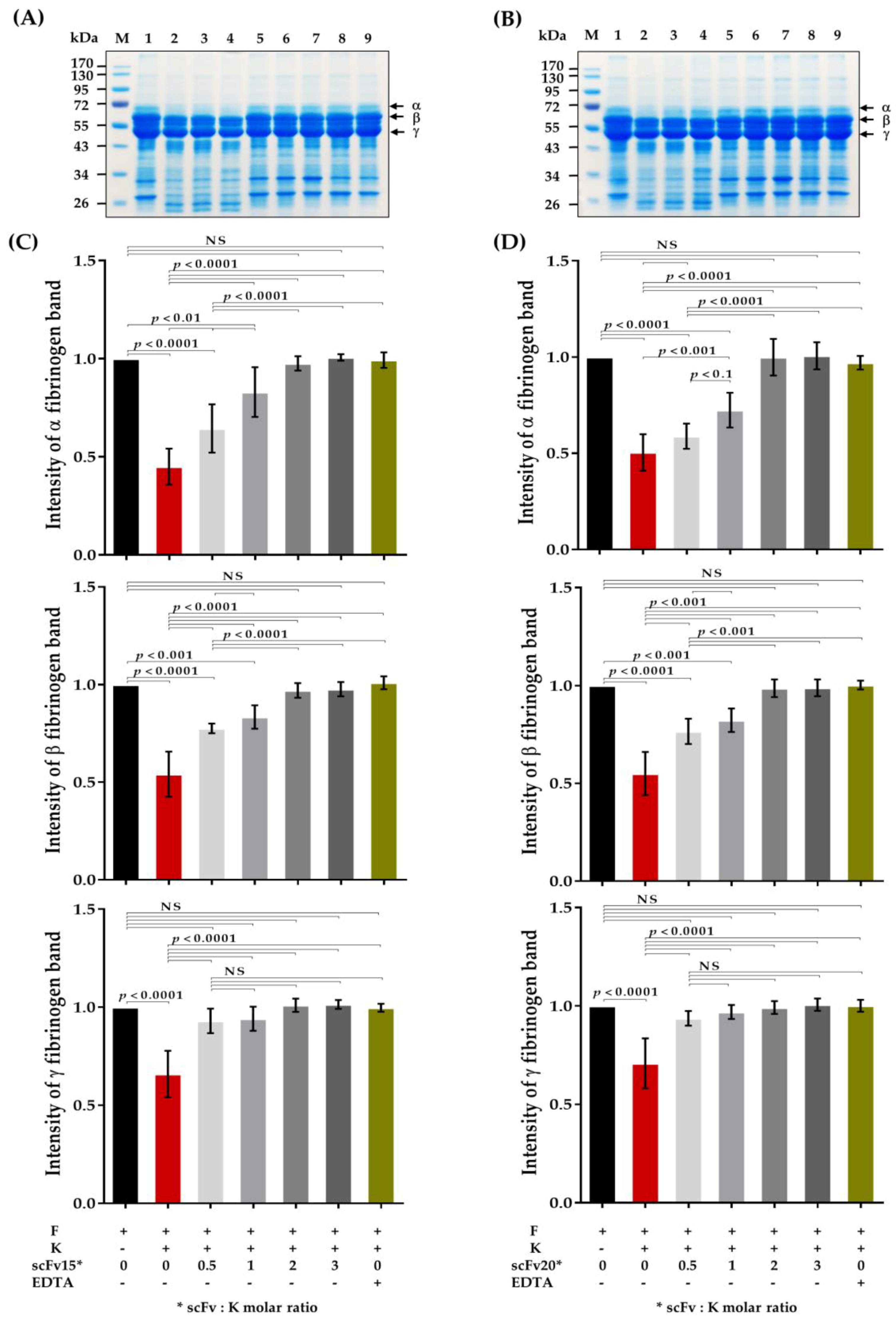
2.5. HuscFv-Mediated Inhibition of Fibrinogenolytic Activity of Kaouthiagin
2.6. Residues and Regions of the Kaouthiagin That Were Bound by the HuscFvs
3. Discussion
4. Materials and Methods
4.1. N. kaouthia Holovenom and Kaouthiagin Purification
4.2. Production of Kaouthiagin-Bound HuscFvs
4.3. Binding of the HuscFvs to Kaouthiagin
4.4. HuscFv-Mediated Inhibition of Kaouthiagin Binding to Human vWF (Binding Inhibition Assay)
4.5. Inhibition of Fribrinogenolytic Activity of Kaouthiagin by HuscFvs
4.6. Computerized Simulation to Predict the Kaouthiagin Regions and Residues Bound by the HuscFvs
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Snakebite Envenoming: Prevalence of Snake Envenoming; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Bureau of Epidemiology, Ministry of Public Health, Thailand. Available online: www.boe.moph.go.th/ (accessed on 30 October 2018).
- Chahome, L.; Jintakune, P.; Wilde, H.; Cox, M.J. Venomous snake husbandry in Thailand. Wilderness Environ. Med. 2001, 12, 17–23. [Google Scholar] [CrossRef]
- Kulkeaw, K.; Chaicumpa, W.; Sakolvaree, Y.; Tongtawe, P.; Tapchaisri, P. Proteome and immunome of the venom of the Thai cobra, Naja kaouthia. Toxicon 2007, 49, 1026–1041. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.; Amberg, H.; Eaker, D. Isolation of the principal neurotoxins of two Naja naja subspecies. Eur. J. Biochem. 1971, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K. Corelation between the phospholipid domains of the target cell membrane and the extentce of distinct catalytic and cytotoxic sites in PLA2 molecules. Biocim. Biophys. Acta 2007, 1770, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Chavanayarn, C.; Thanongsaksrikul, J.; Thueng-in, K.; Bangphoomi, K.; Sookrung, N.; Chaicumpa, W. Humanized-single domain antibodies (VH/VHH) that bound specifically to Naja kaouthia phospholipase A2 and neutralized the enzymatic activity. Toxins 2012, 4, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Eggertsen, P.; Lind, J.; Sjoquist, J. Molecular characterization of the complement activating protein in the venom of the Indian cobra (Naja naja siamensis). Mol. Immunol. 1981, 18, 125–133. [Google Scholar] [CrossRef]
- Kumar, T.K.; Jayaraman, G.; Lee, C.S.; Arunkumar, A.I.; Sivaraman, T.; Samuel, D.; Yu, C. Snake venom cardiotoxins-structure, dynamics, function and folding. J. Biomol. Struct. Dyn. 1997, 15, 431–463. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, J.B.; Fox, J.W. Snake venom metalloendopeptidases: Reprolysins. Methods Enzymol. 1995, 248, 345–368. [Google Scholar]
- Kouassi, K.J.; M’bra, K.I.; Sery, B.J.; Yao, L.B.; Krah, K.L.; Lohourou, G.F.; Kouassi, A.A.; Traore, I.; Asséré, Y.A.; Kodo, M. Amputation of a limb secondary to snakebite in a child. Arch. Pediatr. 2017, 24, 350–352. [Google Scholar] [CrossRef]
- Kamiguti, A.S.; Hay, C.R.M.; Theakston, R.D.G.; Zuzel, M. Insight into the mechanism of haemorrhage caused by snake venom metalloproteinases. Toxicon 1996, 34, 627–642. [Google Scholar] [CrossRef]
- Gutie´rrez, J.M.; Rucavado, A.; Escalante, T.; Dı´az, C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 2005, 45, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Bode, W.; Gomis-Ruth, F.X.; Stockler, W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993, 331, 134–140. [Google Scholar] [CrossRef]
- Markland, F.S., Jr.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 2005, 45, 969–985. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Takeya, H.; Iwanaga, S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta 2012, 1824, 164–176. [Google Scholar] [CrossRef]
- Clemetson, K.J.; Morita, T.; Kini, R.M. Scientific and standardization committee communication: Classification and nomenclature of snake venom C-type lectins and related proteins. J. Thromb. Haemost. 2009, 7, 360. [Google Scholar] [CrossRef]
- Baramova, E.N.; Shannon, J.D.; Bjarnason, J.B.; Fox, J.W. Degradation of extracellular matrix proteins by hemorrhagic metalloproteinases. Arch. Biochem. Biophys. 1989, 275, 63–71. [Google Scholar] [CrossRef]
- Morita, T.; Iwanaga, S. [24] Prothrombin activator from Echis carinatus venom. Methods Enzymol. 1981, 80, 303–311. [Google Scholar]
- Hamako, J.; Matsui, T.; Nishida, S.; Nomura, S.; Fujimura, Y.; Ito, M.; Ozeki, Y.; Titani, K. Purification and characterization of kaouthiagin, a von Willebrand factor-binding and -cleaving metalloproteinase from Naja kaouthia cobra venom. Thromb. Haemost. 1998, 80, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Kamiguti, A.S.; Slupsky, J.R.; Zuzel, M.; Hay, C.R. Properties of fibrinogen cleaved by Jararhagin, a metalloproteinase from the venom of Bothrops jararaca. Thromb. Haemost. 1994, 72, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Kulkeaw, K.; Sakolvaree, Y.; Srimanote, P.; Tongtawe, P.; Maneewatch, S.; Sookrung, N.; Tungtrongchitr, A.; Tapchaisri, P.; Kurazono, H.; Chaicumpa, W. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, neurotoxin. J. Proteom. 2009, 72, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Muniz, J.R.; Ambrosio, A.L.; Selistre-de-Araujo, H.S.; Cominetti, M.R.; Mourada-Silva, A.M.; Oliva, G.; Garratt, S.C.; Souza, D.H. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: Insights for a new classification of snakevenom metalloprotease subgroups. Toxicon 2008, 52, 807–816. [Google Scholar] [CrossRef] [PubMed]
- De Roodt, A.R.; Litwin, S.; Vidal, J.C. Hemorrhagic activity of Bothrops venoms determined by two different methods and relationship with proteolytic activity on gelatin and lethality. Toxicon 2003, 41, 949–958. [Google Scholar] [CrossRef]
- Panfoli, I.; Ravera, S.; Calzia, D.; Dazzi, E.; Gandolfo, S.; Pepe, I.M.; Morelli, A. Inactivation of phospholipase A2 and metalloproteinase from Crotalus atrox venom by direct current. J. Biochem. Mol. Toxicol. 2007, 21, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Pitthayanukul, P.; Leanpolchareanchai, J.; Saparpakorn, P. Molecular docking studies and anti-snake venom metalloproteinase activity of Thai mango seed kernel extract. Molecules 2009, 14, 3198–3213. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, L.; Shannon, J.D.; Valente, R.H.; Rucavado, A.; Alape-Giron, A.; Kamigut, A.S.; Theakston, R.D.; Fox, J.W.; Guteirrez, J.M.; Arni, R.K. Amino acid sequence and crystal structure of BAP1, a metalloproteinase from Bothrops asper snake venom that exerts multiple tissue-damaging activities. Protein Sci. 2003, 12, 2273–2281. [Google Scholar] [CrossRef]
- Wallnoefer, H.G.; Lingott, T.; Gutierrez, J.M.; Merfort, L.; Liedi, K.R. Backbone flexibility controls the activity and specificity of a protein-protein interface: Specificity of snake venom metalloproteinases. J. Am. Chem. Soc. 2010, 132, 10330–10337. [Google Scholar] [CrossRef]
- Kumar, K.R.; Vennila, R.; Kanchana, S.; Arumugam, M.; Balasubramaniam, T. Fibrinogenolytic and anticoagulant activities in the tissue covering the stingers of marine stingrays Dasyatis sephen and Aetobatis narinari. J. Thromb. Thrombolysis 2011, 31, 464–471. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]

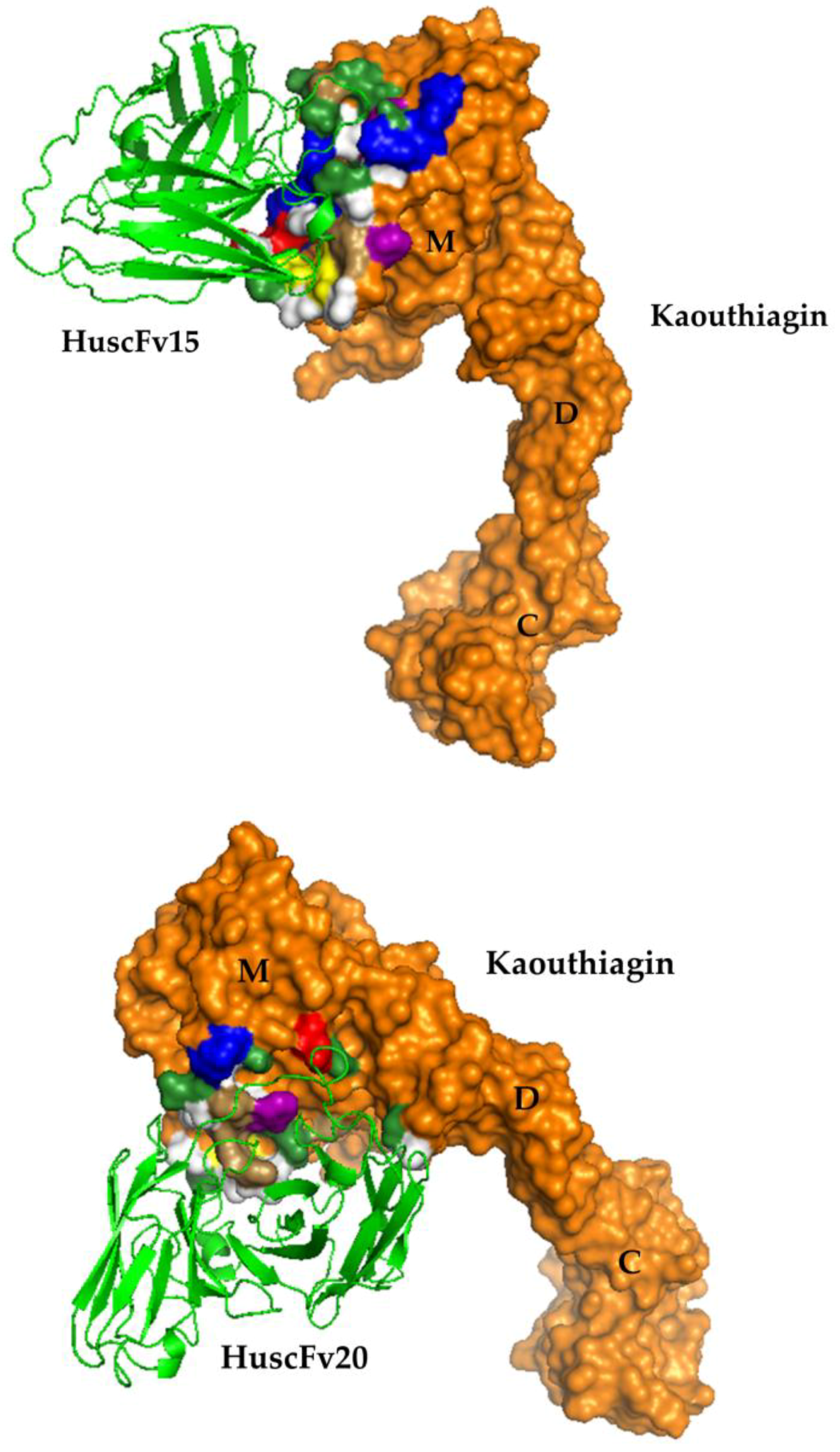
| Kaouthiagin Residue | Amino Acid(s) | Domain(s) | Interactive Bond(s) |
|---|---|---|---|
| HuscFv15 | |||
| N112 | S193 | VL-CDR3 | Hydrogen |
| V116 | Y101/ I103 | VH-CDR3/CDR3 | Hydrophobic/hydrophobic |
| Y136 | Y101 | VH-CDR3 | Hydrogen, hydrophobic and aromatic-aromatic |
| N137 | I103 | VH-CDR3 | Hydrogen |
| R139 | D102 | VH-CDR3 | Hydrogen and ionic |
| L142 | Y101 | VH-CDR3 | Hydrophobic |
| T146 | Y101 | VH-CDR3 | Hydrogen |
| H159 | S197 | VL-FR3 | Hydrogen |
| A162 | I115 | VH-CDR3 | Hydrophobic |
| C166 | G25 | VH-CDR1 | Hydrogen |
| P170 | F26 | VH-CDR1 | Hydrophobic |
| L174 | F26/Y31/I115 | VH-CDR1/CDR1/CDR3 | Hydrophobic/hydrophobic/hydrophobic |
| K176 | D110/D114/Y101 | VH-CDR3/CDR3 | Hydrogen and ionic/hydrogen/hydrogen and cationic-π |
| T178 | Y31/S99 | VH-CDR1/CDR3 | Hydrogen/hydrogen |
| HuscFv20 | |||
| E43 | K64 | VH-FR3 | Hydrogen and ionic |
| A162 | S107/I174 | VH-CDR3/VL-CDR1 | Hydrogen |
| S163 | S107 | VH-CDR3 | Hydrophobic |
| C166 | R31 | VH-CDR1 | Hydrogen |
| I167 | F52/R100/W239 | VH-CDR2/CDR3/VL-CDR3 | Hydrophobic/hydrogen/hydrophobic |
| P168 | F52/Y236/W239 | VH-CDR2/VL-CDR3/CDR3 | Hydrophobic/hydrophobic/hydrogen and hydrophobic |
| C169 | Y236 | VL-CDR3 | Hydrogen |
| P170 | W239/P240 | VL-CDR3/CDR3 | Hydrogen/hydrophobic |
| A181 | W239 | VL-CDR3 | Hydrophobic |
| F182 | W239 | VL-CDR3 | Hydrophobic and aromatic-aromatic |
| Q183 | E56 | VH-CDR2 | Hydrogen |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanongnoi, J.; Phanthong, S.; Reamtong, O.; Tungtronchitr, A.; Chaicumpa, W.; Sookrung, N. Human Monoclonal scFvs that Neutralize Fribrinogenolytic Activity of Kaouthiagin, a Zinc-Metalloproteinase in Cobra (Naja kaouthia) Venom. Toxins 2018, 10, 509. https://doi.org/10.3390/toxins10120509
Khanongnoi J, Phanthong S, Reamtong O, Tungtronchitr A, Chaicumpa W, Sookrung N. Human Monoclonal scFvs that Neutralize Fribrinogenolytic Activity of Kaouthiagin, a Zinc-Metalloproteinase in Cobra (Naja kaouthia) Venom. Toxins. 2018; 10(12):509. https://doi.org/10.3390/toxins10120509
Chicago/Turabian StyleKhanongnoi, Jirawat, Siratcha Phanthong, Onrapak Reamtong, Anchalee Tungtronchitr, Wanpen Chaicumpa, and Nitat Sookrung. 2018. "Human Monoclonal scFvs that Neutralize Fribrinogenolytic Activity of Kaouthiagin, a Zinc-Metalloproteinase in Cobra (Naja kaouthia) Venom" Toxins 10, no. 12: 509. https://doi.org/10.3390/toxins10120509
APA StyleKhanongnoi, J., Phanthong, S., Reamtong, O., Tungtronchitr, A., Chaicumpa, W., & Sookrung, N. (2018). Human Monoclonal scFvs that Neutralize Fribrinogenolytic Activity of Kaouthiagin, a Zinc-Metalloproteinase in Cobra (Naja kaouthia) Venom. Toxins, 10(12), 509. https://doi.org/10.3390/toxins10120509





