Abstract
Mucus provides protective functions in the gastrointestinal tract and plays an important role in the adhesion of microorganisms to host surfaces. Mucin glycoproteins polymerize, forming a framework to which certain microbial populations can adhere, including probiotic Lactobacillus species. Numerous mechanisms for adhesion to mucus have been discovered in lactobacilli, including partially characterized mucus binding proteins. These mechanisms vary in importance with the in vitro models studied, which could significantly affect the perceived probiotic potential of the organisms. Understanding the nature of mucus-microbe interactions could be the key to elucidating the mechanisms of probiotic adhesion within the host.
1. Introduction
Lactobacilli are of significant importance to food industries due to their involvement in the production of various fermented dairy, meat, and vegetable foods. Also important to the health industry, lactobacilli are used as probiotics due to their health-promoting effects when consumed. One of the frequently exploited activities used to screen probiotic candidates is adhesion to the host gut, which is presumed to be requisite for sufficient host-interaction to confer health benefits [1]. Various in vitro models are used for the study of bacterial adhesion because of the complexity of studying the in vivo system. However, these models are simplifications of in vivo situations, resulting in limited conclusions. One significant aspect of the gastrointestinal (GI) tract that is easy to overlook when studying bacterial adhesion, and choosing models with which to measure adhesion, is the presence of a mucus layer between the lumen and GI epithelial cells. The presence of mucus is particularly relevant in the colon, where mucus is thickest and microorganisms are most abundant.
The layer of mucus bound to GI epithelia is formed from a continuous gel matrix composed primarily of complex glycoproteins that acts as a barrier to protect the host from harmful antigens and promote luminal motility. This layer of mucus is the first physical barrier to host-cell stimulation by bacteria in the gut. Adhesion to this mucus is therefore the first step required for probiotic organisms to interact with host cells and elicit any particular response. In the human intestinal tract, the layer of mucus may vary in thickness from about 30 to 300 µm, generally increasing in thickness from the small intestine to the rectum, but the layer of mucus most closely bound to the epithelial layer rarely contains any bacteria at all [2,3].
Numerous studies have characterized interactions between bacteria and host epithelia that induce alterations in host mucosal response [4,5,6] but how changes in mucus composition affect adhesion by gut microorganisms is not well understood. Likewise, exposure to mucus during growth has been shown to affect bacterial gene expression [7], but resulting changes to adhesion are not well recognized. Additionally, existing studies for bacterial adhesion show great variability due to a lack of standardization, complicating the interpretation of data from the current literature [8].
In this review, we will examine the composition of the mucus layers protecting GI epithelial tissue, which is considered to be the primary location of host-probiotic interaction [9]. Our focus will be on its relevance to Lactobacillus species, commensal bacteria of the human gut that are used extensively in commercial probiotic supplements and contain the most widely studied probiotic species in scientific literature. Our goal is to provide a framework for a better understanding of the role that mucus plays in probiotic-host interactions.
2. Intestinal Mucus
The epithelial tissue that forms the lining of the intestine is composed of various columnar cell types. Scattered across the length of the intestine, and all mucosal tissues, are goblet cells. These cells are unicellular glands that produce glycoproteins called mucins, which give mucus its characteristic viscoelastic physical properties. Secreted mucins polymerize to form the matrix that provides the structural foundation of the mucus layer resulting in protection from pathogens, enzymes, toxins, dehydration and abrasion [10]. Goblet cells produce secretory mucin at a basal constitutive level under normal physiological conditions to maintain this protective layer of mucus, which is exposed to the harsh luminal environment and constantly eroded by luminal particulates and intestinal peristalsis [11].
Table 1 shows a reported 21 MUC genes code for the protein cores of mucins in humans. Gastrointestinal mucins are either translocated to the membrane surface or secreted into the mucous gel. Mucins are also either neutral or acidic, depending on their glycosyl modification. These categories can be further subdivided to account for greater variation in mucin structure [12].

Table 1.
Known human MUC genes, their functions and locations.
| Gene | Organisms with known homologues 1 | Function 2 | GeneAtlas location of highest expression 2 | Type | Selected references |
|---|---|---|---|---|---|
| MUC1 | Dog, cow, mouse, rat, rabbit | Cellular signal transduction, barrier activity | Lungs | Membrane | [13,14] |
| MUC2 | Chimpanzee, dog, chicken | Primary extracellular matrix constituent in colon, lubricant activity | Colon | Secretory | [14,15] |
| MUC3A | Rat, mouse | Involved in epithelial cell protection, adhesion modulation, and signaling | Various | Membrane | [16] |
| MUC3B | Rat, mouse | Unknown, possibly cellular signal transduction | Various | Membrane | [16] |
| MUC4 | Many mammals, chicken, frog, platypus | Involved in intestinal epithelial cell differentiation, renewal, lubrication | Colon | Membrane | [17,18] |
| MUC5B (MUC9) | Chimpanzee, zebrafish, mouse, chicken, more | Unknown, primarily lubricant | Various | Secretory | [19,20] |
| MUC5AC | Chimpanzee, rat, zebrafish | Major component of airway mucus involved in intestinal epithelial cell differentiation | Trachea, Lungs | Secretory | [21,22] |
| MUC6 | Chimpanzee, dog, mouse, chicken | Unknown, involved in renal morphogenesis processes | Pancreas, digestive and reproductive systems | Secretory | [22,23,24] |
| MUC7 | Chimpanzee, cow, rat | Facilitating the clearance of oral bacteria | Salivary Gland | Secretory | [25,26] |
| MUC8 | Unknown | Unknown | Trachea | Secretory | [27] |
| MUC12 (MUC11) | Cow, M.grisea, N. crassa, rice | May be involved in epithelial cell regulation | Colon | Membrane | [28] |
| MUC13 | Chimpanzee, dog, mouse, rat | Barrier function in epithelial tissues | Pancreas, small intestine, colon | Membrane | [29] |
| EMCN (MUC14) | Dog, cow, mouse, rat, chicken | Interferes with the assembly of focal adhesion complexes | Fetal lung, uterus, thyroid | Membrane | [30] |
| MUC15 | Chimpanzee, cow, mouse, rat | Barrier function in epithelial tissues | Testis leydig cell | Membrane | [31] |
| MUC16 (CA125) | Chimpanzee, dog, mouse, chicken | Unknown, plays a role in ovarian cancer | Lymph nodes, respiratory tract | Membrane | [32,33] |
| MUC17 (MUC3) | Chimpanzee, S. pombe, S. cerevisiae, and K. lactis | Extracellular matrix constituent, lubricant activity | Small intestine, stomach | Membrane | [34,35] |
| MCAM (MUC18, CD146) | Chimpanzee, dog, mouse, rat, zebrafish | AKA “melanoma cell adhesion molecule”, cell-cell adhesion | Various | Membrane | [36,37] |
| MUC19 | Chimpanzee, dog, mouse, rat, frog | Major gel-forming mucin in the human middle ear | Secretory cells of the ears and eyes | Secretory | [38] |
| MUC20 | Chimpanzee, dog, cow, mouse, rat | Cellular signal transduction | Intestine, respiratory and urinary tract | Membrane | [39] |
| MUC21 | Chimpanzee, cow, mosquito, and A. thaliana | Unknown, mediates cell adhesion | Unknown | Membrane | [40,41] |
| CD164 (MUC24) | Chimpanzee, dog, cow, mouse, rat, chicken, zebrafish | Regulates stem cell localization to the bone marrow | Thyroid, placenta, intestine, immune cells | Membrane | [42] |
1 Via HomoloGene [43] database; 2 via GenAtlas [44] and BioGPS [45] databases.
Mucin Genes and Modifications
Gastrointestinal MUC proteins contain characteristic tandem repeats of threonine, proline, and serine residues, where O-glycosidic linkages occur between the protein core and N-acetylgalactosamine (GalNAc) termini of oligosaccharides [46,47]. Neither the amino nor the carboxy termini of secretory mucins are generally glycosylated [48], but contain cysteine rich regions that promote intermolecular disulfide bonding (as shown in Figure 1). The dense glycosylation of the protein core and intermolecular bonding of the terminal regions effectively protects the mucin polymers from protease activity, preserving the protective structural matrix [49].
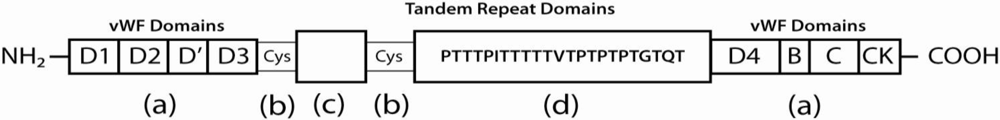
Figure 1.
Diagram of the MUC2 protein core. The protein termini contain cysteine-rich regions homologous to von Willebrand Factor (vWF) domains (a); The N-terminal regions of MUC2 proteins contain vWF domain homologs D1, D2, D′, and D3 and the C-terminal regions contain vWF domain homologs D4, B, C, and CK. These terminal domains are responsible for the extensive polymerization between mucin monomers, along with the cysteine rich interruptions between glycosylated tandem repeats (b); The first of two repetitive domains (c) contains 21 repeats of an irregular motif, whereas the second domain (d) is formed of 50-115 tandem 23aa motifs (PTTTPITTTTTVTPTPTPTGTQT). Threonines in the repeats are O-glycosylated, forming a densely packed envelope of short, branched carbohydrate chains surrounding these regions.
The predominant genes expressing membrane-bound mucins in human colonic goblet cells are MUC1, MUC3A/B, MUC4, and MUC12. Membrane-bound mucins could play a role in immunomodulatory effects of bacterial interactions with the epithelial membrane when the secretory mucin matrix is bypassed [50], however bacteria more frequently come in contact with secretory mucins considering the majority of bacteria only inhabit the outer portions of the mucus layers [51]. MUC2 is the principal secretory mucin gene expressed in the colon, comprising the majority of the mucous gel protecting the underlying tissue [52]. The role and mechanisms of mucin in innate immunity is reviewed more thoroughly by Dharmani et al. [53] and for a more detailed structural analysis of MUC2, see Allenet al. [15].
Oligosaccharide chains are affixed to MUC proteins by membrane-bound transferases in the Golgi apparatus and endoplasmic reticulum of goblet cells. GalNAc is affixed to the mucin protein from a sugar-nucleotide donor and a collection of specific glycosyltransferases continues to add residues, resulting in an oligosaccharide with a particular structure and terminus [54,55]. Glycosylation biosynthesis pathways are highly complex; glycosyltransferase gene expression levels, variability in spatiotemporal concentrations of enzymes, cofactors, and substrates, as well as the number of branching configurations possible all contribute to the wide range of potential protein-modifications [55]. This leads to glycoproteins forming from the same mucin gene product that will vary in glycan modification with location or tissue.
The oligosaccharide modifications can comprise up to 80% of the weight of a mucin and vary in length and structure. Secreted colonic mucins commonly contain side-chains of 4-15 monosaccharides with galactose and GalNAc backbones and branched chains terminating with GalNAc, fucose, or sialic (neuraminic) acid to varying degrees [56,57].
The predominance of acidic mucin subtypes, those with side-chains containing terminal ester sulfates and sialic acid groups, varies by location in the GI tract from species to species, as does the type of acidic modification most heavily expressed [58]. The presence of acidic side-chains can result in greater inhibition of bacterial growth in vitro [59] and reduced enzymatic degradation [60,61], but what causes the prevalence of these modifications in different parts of GI tissues is likely due to the tissue-dependence of specific collections of glycosyltransferase enzymes [62].
intestinal mucin polymers are considered nutritive glycans for commensal bacteria in the promotion of their residence and associated benefits [63,64]. Host glycosylation patterns in the gut may have coevolved with intestinal microbiota to accommodate the filling of niches beneficial to the host [65,66]. Host provision of mucin oligosaccharides specific to particular bacterial enzymes could provide a nutritional advantage to bacteria with those enzymes and differential expression of mucin oligosaccharides by tissue could hypothetically regulate host-microbe interactions to direct certain microbial populations to fill particular host-niches. So-called host “legislation” of glycosylation to promote particular microbial populations is evaluated in greater depth in a review by Patsos and Corfield [67]. Whether this plays a crucial role in Lactobacillus adhesion or is primarily a mechanism of promoting maintenance of other commensal microbiota is currently unclear.
One broad example of host legislation comes from the analysis of mucin oligosaccharide composition along the human intestinal tract, which showed that certain glycosylation patterns were conserved regionally despite inter-individual variation [68]. A gradient of sialylated mucin concentration was observed, decreasing from the ileum to the colon, running against an increasing gradient of more heavily fucosylated mucin.
A more specific example of microbial legislation by hosts lies in the presence of O-glycans on mucin that exhibit Lewis type or blood group ABO antigens. The secretor genes that determine host blood type also control the specificity of the ABO blood group type terminal glycosides of certain mucin oligosaccharide chains [69]. The glycosyltransferase responsible for blood group antigen precursors has been identified in secretory tissues producing mucins and glycoproteins [70]. There is evidence that populations of bacteria that produce specific blood type antigen-degrading glycosidases are present at levels 50,000 times greater in individuals with that particular blood type [71].
While evidence of mucin oligosaccharide degradation by bacteria is fairly well established [64,72,73,74,75], the dramatic impact of blood type on the composition of enteric microbial populations could imply that there is some degree of binding preference at play with host glycan legislation as well. This is supported by evidence of bacteria binding with human milk oligosaccharides, which can exhibit structural similarities with mucin oligosaccharides and blood type antigens [76].
Glycosidases of lactic acid bacteria have been fairly well characterized in terms of oligosaccharide breakdown and metabolism [77,78], but knowledge of glycoconjugate adhesion remains poorly described. Figure 2 displays a model of molecular binding mechanisms that may play a role in host-bacteria interactions. Elucidation of these binding mechanisms may be the key to understanding adhesion of lactobacilli in the gut.
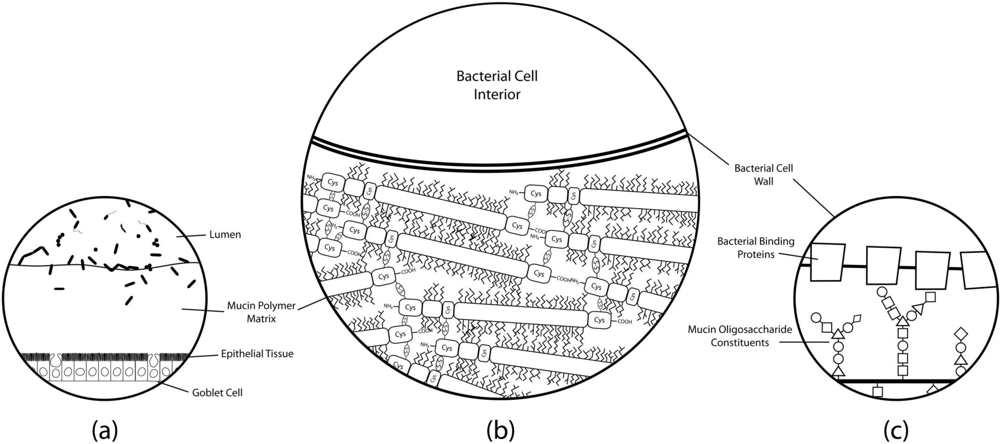
Figure 2.
Simplified histological cross-section of microbial adhesion to the colonic mucosal surface at various magnifications. (a) The layer of mucus atop colonic epithelial villi. Goblet cells can be seen interspersed throughout the columnar enterocytes, producing secretory mucin that makes up the gel matrix. The microbial communities residing in and on top of the mucus layer can only be found at substantial concentrations in the outermost regions of the mucus; (b) The mucus-bacteria interface. The mucin molecules polymerize to form the mucus layer matrix to which cells adhere. Extensive disulfide bonding between cysteine-rich regions of the mucin protein cores creates the characteristic viscoelastic properties of mucus. Oligosaccharide modifications of mucin protein cores form “bottle-brush” regions providing substrate for adhesion to binding proteins on bacterial cell surfaces; (c) A proposed molecular mechanism of adhesion. Evidence suggests that putative mucin-binding proteins anchored to the bacterial cell wall may interact with the glycosyl modifications of the mucin proteins to promote adhesion of the cell to the mucus layer. Mucin oligosaccharide structures vary due to tissue and cell-specific glycosyltransferase expression levels, so the specificity of particular oligosaccharide moieties may lead to preferential binding of particular bacteria to different host niches.
For a more detailed characterization of mucin production, structure, and host function see the review by Lindén et al. [79]. Further information on the study of mucin glycomics, including identified O-linked glycan modifications characteristic of GI mucin and their biosynthetic pathways, can be obtained from glycobiology resources such as the Kyoto Encyclopedia of Genes and Genomes [80] and the Consortium for Functional Glycomics [81].
3. Adhesion
Most clinical studies of probiotic persistence and colonization show that probiotic organisms do not permanently colonize the GI tract and continue providing their hosts benefits only for brief periods after they have stopped being administered [82,83]. Little is known of what makes probiotic organisms so transient relative to commensals, so it is important to consider the factors influencing their capability to adhere and persist in the gut when studying and manipulating probiotic organisms. Bacteria adhere initially to GI surfaces by nonspecific physical interactions, such as steric and hydrophobic interactions, which result in reversible attachment. Several researchers have reported that there is a degree of correlation between hydrophobicity and adhesion to the hydrophobic mucosal surface [84,85,86,87]. However, other studies indicated that there was no correlation between cell surface hydrophobicity and adhesion to intestinal mucus [88,89]. In these studies, highly adhesive bacteria demonstrated fairly low surface hydrophobicities. This has suggested that cell surface hydrophobicity is not an accurate measure of adhesive potential.
While adhesive characteristics of lactobacilli vary considerably among strains and species [90,91,92], many have large surface proteins with highly repetitive structures that are involved in mucus adhesion. Though specific mechanisms are not yet well understood, evidence suggests that carbohydrate-protein interactions play a key role in the adhesion of these proteins to mucin-bound oligosaccharides, especially considering numerous mucus-binding proteins contain regions homologous with binding domains of other known proteins such as lectins. The evolution of lectin-like adhesins in endosymbiotic bacteria may have been favored by the presence of multivalent substrates such as the mucins found in the GI tract. Affinities of lectins for multivalent glycoproteins can be 50-fold to 106-fold greater than for individual glycan moieties [93]. Recently, a number of mucus-binding proteins have been isolated, some of which have been shown to display lectin-like interactions, and some of which may be conserved in numerous Lactobacillus species.
3.1. Mucus Binding Proteins
Several lactobacilli proteins have been shown to promote mucus adhesion (Table 2). The most studied example of mucus-targeting bacterial adhesins is the mucus-binding protein, MUB, produced by L. reuteri [82,94]. The MUB protein contains repeated functional domains, referred to by the authors as Mub domains, which are responsible for the protein’s adhesive properties. The Mub domain has since been designated a member of the MucBP domain family (Pfam PF06458). Numerous MUB homologues and MucBP domain containing proteins have been found, but almost exclusively in lactic acid bacteria and predominantly in lactobacilli found naturally in intestinal niches (Table 3). This suggests that MucBP domain containing proteins play an important role in establishing host-microbial interactions in the gut and promoted the evolution of the species as primarily GI organisms [93,95,96,97].

Table 2.
Adhesion promoting proteins in Lactobacillus spp.
| Protein | Info. | Species | References |
|---|---|---|---|
| MUB | Demonstrates binding to mucus in vitro | L. reuteri | [95] |
| MucBP Domain Containing Proteins | Contain MucBP domains, implicated in mucus adhesion | 13 known Lactobacillus spp. | [98] |
| Pili | Pilin subunit SpaC binds to mucus in vitro | L. johnsonii, L. rhamnosus | [99,100,101] |
| 32-Mmubp | Demonstrates binding to mucus in vitro | L. fermentum | [102] |
| SlpA | Knockouts show diminished adhesion to mucus in vitro | L. acidophilus | [103] |
| Msa | Demonstrates binding of mannose in vitro | L. plantarum | [104] |
| MapA | Demonstrates binding to mucus in vitro | L. reuteri | [105,106] |
| EF-Tu | Expression upregulated in the presence of mucus | L. johnsonii | [107,108,109,110,111] |

Table 3.
MucBP domain containing sequences in available Lactobacillus genomes.
| Currently available whole genomes | Accession# | Gene | # of domains | Size |
|---|---|---|---|---|
| Lactobacillus acidophilus NCFM | Q5FKK8 | LBA0909 | 1 | 508aa |
| Q5FKA8 | LBA1017 | 1 | 294aa | |
| Q5FKA7 | LBA1018 | 1 | 346aa | |
| Q5FKA6 | LBA1019 | 7 | 2650aa | |
| Q5FKA5 | LBA1020 | 5 | 2310aa | |
| Q5FJS1 | LBA1218 | 1 | 697aa | |
| Q5FJC2 | LBA1377 | 2 | 1017aa | |
| Q5FJA7 | LBA1392 | 17 | 4326aa | |
| Q5FJ43 | LBA1460 | 2 | 339aa | |
| Q5FIQ0 | LBA1609 | 2 | 643aa | |
| Q5FIL0 | LBA1652 | 3 | 1174aa | |
| Q5FIF3 | LBA1709 | 3 | 1208aa | |
| Lactobacillus brevis ATCC 367 | Q03U29 | LVIS_0122 | 2 | 912aa |
| Q03T21 | LVIS_0493 | 3 | 1519aa | |
| Q03P66 | LVIS_1947 | 1 | 1111aa | |
| Q03NB2 | LVIS_2262 | 1 | 422aa | |
| Lactobacillus crispatus ST1 | D5H0E1 | LCRIS_00029 | 3 | 1232aa |
| D5H2Y1 | LCRIS_00919 | 7 | 2935aa | |
| D5GXR1 | LCRIS_01123 | 1 | 304aa | |
| D5GZ92 | LCRIS_01654 | 2 | 3552aa | |
| Lactobacillus fermentum IFO 3956 | B2GFA4 | LAF_0157 | 1 | 208aa |
| B2GBH7 | LAF_0673 | 2 | 1059aa | |
| Lactobacillus gasseri ATCC 33323 | Q047B3 | LGAS_0044 | 4 | 873aa |
| Q047B2 | LGAS_0045 | 11 | 3692aa | |
| Q047B1 | LGAS_0046 | 4 | 985aa | |
| Q046R7 | LGAS_0143 | 6 | 2823aa | |
| Q045Q7 | LGAS_0410 | 5 | 2457aa | |
| Q043P5 | LGAS_0939 | 2 | 615aa | |
| Q043P2 | LGAS_0942 | 10 | 2833aa | |
| Q043P0 | LGAS_0944 | 1 | 524aa | |
| Q041C4 | LGAS_1655 | 2 | 1425aa | |
| Q041B7 | LGAS_1663 | 6 | 2449aa | |
| Q041A9 | LGAS_1671 | 4 | 2552aa | |
| Q040V9 | LGAS_1725 | 6 | 1993aa | |
| Lactobacillus helveticus DPC 4571 | A8YTV1 | lhv_0494 | 1 | 155aa |
| A8YTV2 | lhv_0495 | 1 | 178aa | |
| A8YUX0 | lhv_0973 | 1 | 278aa | |
| A8YUX3 | lhv_0979 | 1 | 858aa | |
| Lactobacillus johnsonii FI9785 | D0R4C3 | FI9785_1070 | 6 | 3401aa |
| D0R5H6 | FI9785_1482 | 5 | 1356aa | |
| Lactobacillus johnsonii NCC 533 | Q74LY7 | LJ_0046 | 4 | 870aa |
| Q74LY6 | LJ_0047 | 6 | 2139aa | |
| Q74LY5 | LJ_0048 | 4 | 983aa | |
| Q74L43 | LJ_0382 | 4 | 3619aa | |
| Q74KU3 | LJ_0484 | 4 | 4037aa | |
| Q74HP3 | LJ_0574 | 5 | 1571aa | |
| Q74HU0 | LJ_0621 | 5 | 2789aa | |
| Q74HW0 | LJ_0641 | 3 | 1563aa | |
| Q74HA8 | LJ_1839 | 7 | 1814aa | |
| Lactobacillus plantarum JDM1 | C6VP10 | JDM1_1038 | 4 | 1082aa |
| C6VQ03 | JDM1_1381 | 6 | 2219aa | |
| C6VKM3 | JDM1_2438 | 4 | 1345aa | |
| C6VL52 | JDM1_2491 | 4 | 2037aa | |
| C6VL55 | JDM1_2494 | 1 | 750aa | |
| Lactobacillus plantarum WCFS1 | Q88Y49 | lp_0946 | 1 | 1189aa |
| Q88XH5 | lp_1229 | 3 | 1010aa | |
| Q88WI9 | lp_1643 | 6 | 2219aa | |
| Q88UJ0 | lp_2486 | 2 | 917aa | |
| Q88TB8 | lp_3059 | 4 | 1356aa | |
| Q88T70 | lp_3114 | 4 | 2032aa | |
| Q88T67 | lp_3117 | 1 | 750aa | |
| Lactobacillus reuteri DSM 20016 | A5VKZ1 | Lreu_1258 | 1 | 745aa |
| Lactobacillus reuteri JCM 1112 | B2G8C6 | LAR_1192 | 1 | 745aa |
| Lactobacillus salivarius CECT 5713 | D8IM74 | HN6_01114 | 4 | 785aa |
| Lactobacillus salivarius UCC118 | Q1WSI9 | LSL_1335 | 4 | 785aa |
Data gathered from the Pfam [112] and Uniprot [113] databases; Databases contained no MucBP domain containing sequences in Lactobacillus delbrueckii subsp. bulgaricus strains ATCC 11842 and ATCC BAA-365, Lactobacillus fermentum CECT 5716, Lactobacillus casei strains Zhang, BL23, and ATCC334, Lactobacillusplantarum subsp. plantarum ST-III, Lactobacillusrhamnosus strains GG and Lc 705, and Lactobacillussakei subsp. sakei 23K.
MUB and most MucBP domain containing proteins exhibit characteristics typical of Gram-positive cell surface proteins; a C-terminal sortase recognition motif (LPXTG) for anchoring the protein to peptidoglycan, repeated functional domains and an N-terminal region signaling the protein for secretion [94,95].
Roos and Jonsson’s competitive adhesion study showed that the binding of MUB to mucus was inhibited by the glycoproteins fetuin and asialofetuin as well as fucose, suggesting that MUB interacts with specific muco-oligosaccharides [94]. The study also demonstrated equivalent adhesion to mucus from different hosts indicating that MUB binding has little to no host specificity regarding mucus components. The recent resolution of the crystal structure of a MucBP domain in MUB, dubbed Mub2 [92], and subsequent discovery of immunoglobulin binding, provides further evidence of a broad binding specificity. This suggests that binding specificities of MucBP domain containing proteins are dictated by multiple factors, not solely resulting from the presence of MucBP domains.
Fimbrial genes have been reported in L. johnsonii NCC533 [99], but the direct visualization of pili on Lactobacillus cells has only been shown for L. rhamnosus GG [100]. Fimbriae, also referred to as pili, are thin proteinaceous extensions from bacterial cells, predominantly in Gram-negative bacteria, that promote adhesion. In many pathogens, pili are virulence factors that promote attachment to the host [101]. Kankainen et al. [100] isolated a pilin subunit, SpaC, located within the pili structure and found at the pilus tip, which was concluded to be essential to the interaction of L. rhamnosus GG with host mucus. A mutant strain lacking spaC expression showed significantly reduced binding. While these genes are uncommon among lactobacilli, this study has shown for the first time that fimbrial interaction with mucus can mediate host adhesion in lactobacilli.
SlpA, an S-layer protein in L. acidophilus,has been implicated in promoting adhesion directly to the GI surface, because slpA knockouts showed decreased adhesion capability [103]. However, this could possibly be due to disruption of other surface proteins. S-layer proteins and glycoproteins can form a latticed monolayer coating the surface of bacterial cells [114]. S-layer components can vary widely by species, but function to protect the cell from enzymatic damage, low pH, bacteriophages and phagocytosis. While S-layers are present in only some Lactobacillus species, they are beginning to be studied for their adhesive functions. A number of studies have begun associating S-layer proteins in probiotic bacteria with competitive exclusion of pathogens and pathogen adhesion to mucus [115,116,117].
Certain other surface proteins are implicated in contributing to adhesive properties of lactobacilli but are otherwise not well characterized or their importance to adhesive mechanisms is poorly defined. For instance, a 32 kDa protein associated with adhesion to porcine mucus in L. fermentum, named 32-Mmubp, was identified as a homologue of the substrate binding domains of the OpuAC ABC-transport protein family [102]. A mannose-specific adhesin protein (Msa, a MucBP domain containing protein) is responsible for the binding of mannose by L. plantarum [104]. While this was initially discovered as a protein responsible for agglutinating Saccharomyces cerevisiae,the presence of L. plantarum in many intestinal niches suggests that the MucBP domains of Msa may also play a role in adhesion to non-mannosylated muco-oligosaccharides as well. Elongation Factor Tu is a guanosine binding protein that is important in protein synthesis in the cytoplasm, but has been identified as a membrane associated protein as well [107,108]. More recently it has been found on the cell surfaces of many lactobacilli [109,110] and the demonstration of its upregulation in the presence of mucus suggests it may play a role in adhesion to the GI tract [111]. Mucus adhesion-promoting protein (MapA) is reported to mediate the binding of L. reuteri and L. fermentum to mucus [105,106]. Interestingly, it is also degraded into an antimicrobial peptide, which lends the host anti-pathogenic properties and provides an example of how large surface proteins may exhibit evolutionarily beneficial pleiotropic effects [118].
3.2. Factors that Influence Binding in Vivo and in Vitro
Numerous factors have been shown to influence binding of lactobacilli to mucus in vitro. Certain aspects of experimental design in particular should be reviewed when choosing or comparing methods to study adhesion in vitro because of the direct effects they have on adhesion. Time allotted for incubation of bacteria on immobilized mucus can have a significant influence on observed adhesion if microbial sedimentation occurs and the substrate is saturated at an artificial level [119].
Ramiah et al. [111] showed that growth conditions mimicking the GI environment have significant effects on the expression of several mucus adhesins in vitro in L. plantarum. MapA was upregulated 6-8-fold when incubated in the presence of mucin and up to 25-fold when exposed to physiological concentrations of pancreatin and bile compared to MRS grown controls. It was also found that mapA was significantly downregulated in the presence of cysteine, and suggested that cysteine is an effector molecule that represses transcription of mapA. Mub was expressed 80-140-fold more in the presence of mucin, but was suppressed 7-30-fold under normal gut physiological conditions containing bile and pancreatin. EF-Tu was expressed 33-100 times greater in media containing mucus, but was not affected by bile or pancreatin concentrations. This may connote interplay between different mechanisms regulating adhesin expression to adapt to particular environments.
The possible mechanisms whereby food components affect the adhesion of probiotic organisms in vivo have not been investigated thoroughly. Exposure to milk and milk fatty acids has been observed to reduce the adhesive properties of some probiotic lactobacilli [120,121] to human intestinal mucus in vitro, which may also be relevant in vivo. It is also hypothesized that entrapment in food matrices in vivo, resulting in binding to or steric hindrance of adhesins, can decrease adhesion of bacteria to intestinal surfaces [119].
All bacterial adhesion in the gut is also likely inhibited to some degree by competitive exclusion of access to binding sites by commensal organisms, but quantification of these effects have yet to be studied thoroughly.
4. In Vitro Models
Adhesion to the GI tract has been widely used as a criterion for the selection of probiotic lactobacilli [122]. It has generally been assumed that probiotic efficacy is enhanced by adhesion to the GI tract, which increases residence time in vivo. This extends the period during which probiotic organisms can exert beneficial effects, such as immune stimulation from contact with the intestinal tract [123,124]. However, it is difficult to link adhesion, specifically, with probiotic efficacy. Studies with isogenic strains containing adhesion factor knockouts [125] demonstrate decreased adhesion to the gut, however it is not known how such a knockout would alter probiotic efficacy. Adhesion has been demonstrated as an important factor in the displacement of pathogens by probiotic bacteriain vitro [126,127,128], but isolating the influence of adhesionin vivo is complicated by various confounded factors. The effects of probiotic bacteria stem not only from adhesion to the GI tract and competition for binding sites with pathogens, but from competition for nutrients as well as the production of exogenous antimicrobial and immune-stimulating compounds. Some studies do correlate adhesive capacity with immune response [129,130], but it is uncertain to what extent confounded factors may influence observed probiotic activity. Understanding the molecular mechanisms behind microbial adhesion in the gut could help determine the degree of probiotic functionality imparted by adhesion alone.
The validity of the experimental models used in the measurement of probiotic adhesion may, however, be difficult to interpret. No standard model for in vitro adhesion exists so findings vary widely not only between strains and species, but between models as well [131]. The in vitro model determines the nature of adhesion sites in the system; some cell culture models will emphasize the measurement of direct host-microbe cellular contact, whereas mucus-secreting cultures or immobilized mucus models will emphasize mucus and muco-oligosaccharide interactions more than other models. As summarized in Table 4, there are advantages and disadvantages to various types of in vitro adhesion models. It may therefore be important to study adhesion in vitro via different methods for a more thorough understanding of the interaction mechanisms most important to probiotic adhesion.

Table 4.
Summary of in vitro adhesion models.
| Model | Description | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Immobilized mucus | Mucus preparations immobilized, usually in microtitre wells | Fast, isolates mucus-microbe interactions from other in vivo conditions | Difficult to separate mucus-specific from hydrophobic interactions | [91,131,132,133] |
| Cell culture | Polar monolayer of enterocytes resembling intestinal tissue | Provides conditions more similar to in vivo environment | Derived from cancer cells, could differ from healthy tissue. Not representative of cell-type ratios in mucosal epithelial tissues | |
| Caco-2/HT29 | Caco-2 and HT29 carcinoma cell lines | Simple, well established in literature | Does not account for mucus presence | [134,135,136,137] |
| HT29-MTX/FU | HT29 culture treated with methotrexate or fluoruracil to secret mucus of different types | Accounts for presence of mucus | May not represent appropriate MUC gene expression | [138,139,140,141,142,143,144] |
| Co-cultures | Mixed culture of secreting and mucus-secreting cells | Better represents cell-type ratio of mucosal epithelial tissues | Little literature for use in adhesion studies | [145,146,147] |
| Whole tissue | Whole, intact or excised tissue | Provides in vitro conditions most similar to in vivo environment | Costly, difficult to obtain | |
| Resected tissue | Fragments of tissue excised from host | Mucus, epithelial tissue, and commensal organisms accounted for in model | Only small fragments at a time available from living hosts | [148,149] |
| Organ culture | Whole organs maintained in vitro | Better maintains the architecture of the tissue | Prohibitively expensive, may not function in same manner as in vivo | [150,151] |
4.1. Mucus Adhesion Models
The simplest method to measure the adhesion of bacterial strains to mucus is by immobilizing commercially available mucin. Mucin is bound to microtitre well plates, bacterial culture is bound to the mucin and strains are compared thereafter in any number of methods, qualitatively or quantitatively [91,132,133]. The use of mucin alone in adhesion assays allows for the targeting of interactions between bacteria and particular mucins known to be expressed in a given host locations. It also isolates microbe-mucus interactions from other interactions, such as host cell-microbe interactions, which may or may not be desirable. One drawback of this model is the complication of microbial hydrophobic properties with mucus-binding properties. The comparison of hydrophobic binding interactions of bacteria to untreated polycarbonate wells with mucus binding interactions in treated wells in one study [131] showed that hydrophobic binding interactions are not easily separable from mucus binding interactions.
4.2. Cell-Culture Models
Cultures of human intestinal cell lines are often presumed to better represent the environment in vivo because of the presence of actual tissue. The availability of a simple in vitro intestinal tissue model, as in the Caco-2 cell line, has provided valuable insight into cellular interaction mechanisms that would have been much more difficult to obtain with more complex in vitro techniques or in vivo. Caco-2 and HT29 cells, the two most commonly used intestinal cell lines, can be grown in culture to form a homogeneous polar monolayer of mature enterocytes resembling the tissue of the small intestine [134]. These models were developed primarily for the study of absorption and permeability in the small intestine and are derived from intestinal carcinomas, so they may or may not be accurate models for adhesion to healthy colonic tissue [135]. Several studies have compared the extent of Caco-2 cell binding by potential probiotic bacteria to adhesionin vivo with mixed results [136,137]. Regardless, these cell lines do not take into account the omnipresent mucus layer found in the healthy intestinal tract. The HT29 cell line, however, can be treated with methotrexate (MTX) to differentiate the cells into mucin-secreting goblet cells [138,139]. The production of mucus by HT29-MTX cells increase adhesion of bacterial cells relative to Caco-2 or HT29 cells alone [140,141], further supporting the importance of the presence of mucus to bacterial adhesion.
The HT29-MTX line is composed primarily of goblet cells, which incorporates a mucus layer into the model, but it still does not accurately represent the enterocyte/goblet cell ratio of the gut epithelial layer. In response to this drawback, Caco-2/HT29-MTX co-cultures have been developed [145,146,147]. Unfortunately, HT29-MTX differentiated goblet cells express MUC5AC and MUC5B at a much greater rate than MUC2 [139], which could be a significant drawback when studying microbial adhesion to the colon, where MUC2 is prevalent. HT29 cells also differentiate in the presence of 5-fluorouracil (FU) to secrete MUC2 [142], and while this would emulate the colonic environment more closely, the HT29-FU model only seems to have been used in the study of pathogens thus far [139,140].
4.3. Whole Tissue Models
The disadvantage of many models is that they don’t take into account the presence of normal GI microbiota and the competitive exclusion that would take place in vivo between established commensal populations and exogenous microbes. For a more complete model of intestinal tissue in vitro, encompassing the mucus layer and epithelial tissue accurately, but also accounting for the presence of commensal microbiota, whole intestinal tissue fragments can be used [148,149]. Resected fragments of healthy colonic tissue may be difficult to obtain, but likely display characteristics closer to those probiotic bacteria would be expected to encounter in vivo than other models. Similarly, organ culture can be employed to maintain the viability and architecture of the tissue and has been used to assess adhesive properties of pathogens [150,151]. As of yet, it does not seem that organ culture has been used in the study of probiotic organisms; the expense of using organ culture is more easily justifiable with pathogenic organisms that could not otherwise be used in human models safely, unlike probiotic organisms.
5. Conclusion
As the field advances, discovery and selection of better probiotic organisms will become more sophisticated. Refinement of cell-culture techniques to better represent colonic environment could provide more accurate measures of adhesion, further aiding the selection of the best probiotic candidates for clinical trials. Printed glycan microarrays are beginning to be used to elaborate binding patterns of whole bacterial cells to different glycan structures [152]. Discoveries using similar techniques could promote the understanding of specific affinities for different binding proteins. Determining the structural characteristics and binding specificities of mucus-binding proteins improve our understanding of the mechanisms behind probiotic-host interactions. This could in turn lead to the development of better tools to select the most beneficial probiotic organisms, potentially opening the door for designer probiotics engineered or selected for desired host-responses. Likewise, a better understanding of host glycosyl legislation in the context of bacterial binding specificity could result in the development of probiotics targeted for specific hosts or host tissue.
Regardless of what future advances may come, knowledge of the limitations within the study of bacterial adhesion, as in any field, should help in the interpretation of current discovery as well as with the planning of further research.
Acknowledgements
The authors would like to acknowledge Rex Gaskins and members of the Miller lab for insightful discussion and technical help. Max Van Tassell was supported by the Bill and Agnes Brown Fellowship in Food Microbiology.
References
- Ouwehand, A.C.; Salminen, S.; Isolauri, E. Probiotics: An overview of beneficial effects. Antonie van Leeuwenhoek 2002, 82, 279–289. [Google Scholar]
- Matsuo, K.; Ota, H.; Akamatsu, T.; Sugiyama, A.; Katsuyama, T. Histochemistry of the surface mucous gel layer of the human colon. Gut 1997, 40, 782–789. [Google Scholar]
- Swidsinski, A.; Loening-Baucke, V.; Theissig, F.; Engelhardt, H.; Bengmark, S.; Koch, S.; Lochs, H.; Dörffel, Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 2007, 56, 343–350. [Google Scholar]
- Maassen, C.B.; van Holten-Neelen, C.; Balk, F.; den Bak-Glashouwer, M.J.; Leer, R.J.; Laman, J.D.; Boersma, W.J.; Claassen, E. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 2000, 18, 2613–2623. [Google Scholar]
- Deplancke, B.; Gaskins, H.R. Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001, 73, 1131S–1141S. [Google Scholar]
- Valeur, N.; Engel, P.; Carbajal, N.; Connolly, E.; Ladefoged, K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 2004, 70, 1176–1181. [Google Scholar]
- Weiss, G.; Jespersen, L. Transcriptional analysis of genes associated with stress and adhesion in Lactobacillus acidophilus NCFM during the passage through an in vitro gastrointestinal tract model. J. Mol. Microbiol. Biotechnol. 2010, 18, 206–214. [Google Scholar]
- Blum, S. Adhesion studies for probiotics: Need for validation and refinement. Trends Food Sci. Technol. 1999, 10, 405–410. [Google Scholar]
- Fuller, R. Probiotics in man and animals. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar]
- Laboisse, C.; Jarry, A.; Branka, J.E.; Merlin, D.; Bou-Hanna, C.; Vallette, G. Recent aspects of the regulation of intestinal mucus secretion. Proc. Nutr. Soc. 1996, 55, 259–264. [Google Scholar]
- Akiba, Y.; Guth, P.H.; Engel, E.; Nastaskin, I.; Kaunitz, J.D. Dynamic regulation of mucus gel thickness in rat duodenum. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G437–G447. [Google Scholar]
- Byrd, J.C.; Bresalier, R.S. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 77–99. [Google Scholar]
- Gendler, S.J. MUC1, the renaissance molecule. J. Mammary Gland Biol. Neoplasia 2001, 6, 339–353. [Google Scholar]
- Chambers, J.A.; Hollingsworth, M.A.; Trezise, A.E.; Harris, A. Developmental expression of mucin genes MUC1 and MUC2. J. Cell Sci. 1994, 107, 413–424. [Google Scholar]
- Allen, A.; Hutton, D.A.; Pearson, J.P. The MUC2 gene product: A human intestinal mucin. Int. J. Biochem. Cell Biol. 1998, 30, 797–801. [Google Scholar]
- Pratt, W.S.; Crawley, S.; Hicks, J.; Ho, J.; Nash, M.; Kim, Y.S.; Gum, J.R.; Swallow, D.M. Multiple transcripts of MUC3: Evidence for two genes, MUC3A and MUC3B. Biochem. Biophys. Res. Commun. 2000, 275, 916–923. [Google Scholar]
- Carraway, K.L.; Perez, A.; Idris, N.; Jepson, S.; Arango, M.; Komatsu, M.; Haq, B.; Price-Schiavi, S.A.; Zhang, J.; Carraway, C.A. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, in cancer and epithelia: To protect and to surviv. Prog. Nucleic Acid Res. Mol. Biol. 2002, 71, 149–185. [Google Scholar]
- Porchet, N.; Nguyen, V.C.; Dufosse, J.; Audie, J.P.; Guyonnet-Duperat, V.; Gross, M.S.; Denis, C.; Degand, P.; Bernheim, A.; Aubert, J.P. Molecular cloning and chromosomal localization of a novel human tracheo-bronchial mucin cDNA containing tandemly repeated sequences of 48 base Pairs. Biochem. Biophys. Res. Commun. 1991, 175, 414–422. [Google Scholar]
- Desseyn, J.L.; Guyonnet-Dupérat, V.; Porchet, N.; Aubert, J.P.; Laine, A. Human mucin gene MUC5B, the 10.7-Kb large central exon encodes various alternate subdomains resulting in a super-repeat. Structural evidence for a 11p15.5 gene family. J. Biol. Chem. 1997, 272, 3168–3178. [Google Scholar] [PubMed]
- Nielsen, P.A.; Bennett, E.P.; Wandall, H.H.; Therkildsen, M.H.; Hannibal, J.; Clausen, H. Identification of a major human high molecular weight salivary mucin (MG1) as tracheobronchial mucin MUC5B. Glycobiology 1997, 7, 413–419. [Google Scholar]
- Escande, F.; Aubert, J.P.; Porchet, N.; Buisine, M.P. Human mucin gene MUC5AC: Organization of its 5′-region and central repetitive region. Biochem. J. 2001, 358, 763–772. [Google Scholar]
- Nordman, H.; Davies, J.R.; Lindell, G.; de Bolós, C.; Real, F.; Carlstedt, I. Gastric MUC5AC and MUC6 are large oligomeric mucins that differ in size, glycosylation and tissue distribution. Biochem. J. 2002, 364, 191–200. [Google Scholar]
- Toribara, N.W.; Ho, S.B.; Gum, E.; Gum, J.R.; Lau, P.; Kim, Y.S. The carboxyl-terminal sequence of the human secretory mucin, MUC6. Analysis of the primary amino acid sequence. J. Biol. Chem. 1997, 272, 16398–16403. [Google Scholar] [PubMed]
- Bartman, A.E.; Buisine, M.P.; Aubert, J.P.; Niehans, G.A.; Toribara, N.W.; Kim, Y.S.; Kelly, E.J.; Crabtree, J.E.; Ho, S.B. The MUC6 secretory mucin gene is expressed in a wide variety of epithelial tissues. J. Pathol. 1998, 186, 398–405. [Google Scholar]
- Bobek, L.A.; Tsai, H.; Biesbrock, A.R.; Levine, M.J. Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7). J. Biol. Chem. 1993, 268, 20563–20569. [Google Scholar] [PubMed]
- Bobek, L.A.; Liu, J.; Sait, S.N.; Shows, T.B.; Bobek, Y.A.; Levine, M.J. Structure and chromosomal localization of the human salivary mucin gene, MUC7. Genomics 1996, 31, 277–282. [Google Scholar]
- Shankar, V.; Pichan, P.; Eddy, R.L.; Tonk, V.; Nowak, N.; Sait, S.N.; Shows, T.B.; Schultz, R.E.; Gotway, G.; Elkins, R.C.; et al. Chromosomal localization of a human mucin gene (MUC8) and cloning of the cDNA corresponding to the carboxy terminus. Am. J. Respir. Cell Mol. Biol. 1997, 16, 232–241. [Google Scholar]
- Williams, S.J.; McGuckin, M.A.; Gotley, D.C.; Eyre, H.J.; Sutherland, G.R.; Antalis, T.M. Two novel mucin genes down-regulated in colorectal cancer identified by differential display. Cancer Res. 1999, 59, 4083–4089. [Google Scholar]
- Williams, S.J.; Wreschner, D.H.; Tran, M.; Eyre, H.J.; Sutherland, G.R.; McGuckin, M.A. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J. Biol. Chem. 2001, 276, 18327–18336. [Google Scholar]
- Liu, C.; Shao, Z.M.; Zhang, L.; Beatty, P.; Sartippour, M.; Lane, T.; Livingston, E.; Nguyen, M. Human endomucin is an endothelial marker. Biochem. Biophys. Res. Commun. 2001, 288, 129–136. [Google Scholar]
- Pallesen, L.T.; Berglund, L.; Rasmussen, L.K.; Petersen, T.E.; Rasmussen, J.T. Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur. J. Biochem. 2002, 269, 2755–2763. [Google Scholar]
- Yin, B.W.T.; Dnistrian, A.; Lloyd, K.O. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int. J. Cancer 2002, 98, 737–740. [Google Scholar]
- McLemore, M.R.; Aouizerat, B. Introducing the MUC16 gene: Implications for prevention and early detection in epithelial ovarian cancer. Biol. Res. Nurs. 2005, 6, 262–267. [Google Scholar]
- Gum, J.R.; Crawley, S.C.; Hicks, J.W.; Szymkowski, D.E.; Kim, Y.S. MUC17, a novel membrane-tethered mucin. Biochem. Biophys. Res. Commun. 2002, 291, 466–475. [Google Scholar]
- Moniaux, N.; Junker, W.M.; Singh, A.P.; Jones, A.M.; Batra, S.K. Characterization of human mucin muc17. Complete coding sequence and organization. J. Biol. Chem. 2006, 281, 23676–23685. [Google Scholar]
- Lehmann, J.M.; Riethmüller, G.; Johnson, J.P. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. USA 1989, 86, 9891–9895. [Google Scholar]
- Johnson, J.P.; Rothbächer, U.; Sers, C. The progression associated antigen MUC18: A unique member of the immunoglobulin supergene family. Melanoma Res. 1993, 3, 337–340. [Google Scholar]
- Chen, Y.; Zhao, Y.H.; Kalaslavadi, T.B.; Hamati, E.; Nehrke, K.; Le, A.D.; Ann, D.K.; Wu, R. Genome-wide search and identification of a novel gel-forming mucin muc19/muc19 in glandular tissues. Am. J. Respir. Cell Mol. Biol. 2004, 30, 155–165. [Google Scholar]
- Higuchi, T.; Orita, T.; Nakanishi, S.; Katsuya, K.; Watanabe, H.; Yamasaki, Y.; Waga, I.; Nanayama, T.; Yamamoto, Y.; Munger, W.; et al. Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J. Biol. Chem. 2004, 279, 1968–1979. [Google Scholar] [PubMed]
- Yi, Y.; Kamata-Sakurai, M.; Denda-Nagai, K.; Itoh, T.; Okada, K.; Ishii-Schrade, K.; Iguchi, A.; Sugiura, D.; Irimura, T. Mucin 21/epiglycanin modulates cell adhesion. J. Biol. Chem. 2010, 285, 21233–21240. [Google Scholar]
- Itoh, Y.; Kamata-Sakurai, M.; Denda-Nagai, K.; Nagai, S.; Tsuiji, M.; Ishii-Schrade, K.; Okada, K.; Goto, A.; Fukayama, M.; Irimura, T. Identification and expression of human epiglycanin/muc21: A novel transmembrane mucin. Glycobiology 2008, 18, 74–83. [Google Scholar]
- Kurosawa, N.; Kanemitsu, Y.; Matsui, T.; Shimada, K.; Ishihama, H.; Muramatsu, T. Genomic analysis of a murine cell-surface sialomucin, MGC-24/CD164. Eur. J. Biochem. 1999, 265, 466–472. [Google Scholar]
- National Center for Biotechnology Information. HomoloGene Home Page. Available online: http://www.ncbi.nlm.nih.gov/homologene (accessed on 30 October 2010).
- Université René Descartes-Paris. GENATLAS. Available online: http://genatlas.medecine.univ-paris5.fr (accessed on 30 October 2010).
- Genomics Intstitute of the Nevartis Research Foundation. BioGPS. Available online: http://biogps.gnf.org (accessed on 30 October 2010).
- Hanisch, F.G. O-Glycosylation of the mucin type. Biol. Chem. 2001, 382, 143–149. [Google Scholar]
- Dekker, J.; Rossen, J.W.A.; Büller, H.A.; Einerhand, A.W.C. The MUC family: An obituary. Trends Biochem. Sci. 2002, 27, 126–131. [Google Scholar]
- Allen, A.; Pearson, J.P. Mucus glycoproteins of the normal gastrointestinal tract. Eur. J. Gastroenterol. Hepatol. 1993, 5, 193–199. [Google Scholar]
- Moncada, D.M.; Kammanadiminti, S.J.; Chadee, K. Mucin and toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003, 19, 305–311. [Google Scholar]
- Liévin-Le Moal, V.; Servin, A.L.; Coconnier-Polter, M. The increase in mucin exocytosis and the upregulation of MUC genes encoding for membrane-bound mucins induced by the thiol-activated exotoxin listeriolysin O is a host cell defence response that inhibits the cell-entry of Listeria monocytogenes. Cell. Microbiol. 2005, 7, 1035–1048. [Google Scholar]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two MUC2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci.USA 2008, 105, 15064–15069. [Google Scholar]
- Strous, G.J.; Dekker, J. Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol. 1992, 27, 57–92. [Google Scholar]
- Dharmani, P.; Srivastava, V.; Kissoon-Singh, V.; Chadee, K. Role of intestinal mucins in innate host defense mechanisms against pathogens. J. Innate Immun. 2009, 1, 123–135. [Google Scholar]
- Marcaurelle, L.A.; Bertozzi, C.R. Recent advances in the chemical synthesis of mucin-like glycoproteins. Glycobiology 2002, 12, 69R–77R. [Google Scholar]
- Brockhausen, I.; Schutzbach, J.; Kuhns, W. Glycoproteins and their relationship to human disease. Acta Anat. (Basel) 1998, 161, 36–78. [Google Scholar] [CrossRef] [PubMed]
- Slomiany, A.; Zdebska, E.; Slomiany, B.L. Structures of the neutral oligosaccharides isolated from A-active human gastric mucin. J. Biol. Chem. 1984, 259, 14743–14749. [Google Scholar]
- Corfield, A.P.; Myerscough, N.; Gough, M.; Brockhausen, I.; Schauer, R.; Paraskeva, C. Glycosylation patterns of mucins in colonic disease. Biochem. Soc. Trans. 1995, 23, 840–845. [Google Scholar]
- Sheahan, D.G.; Jervis, H.R. Comparative histochemistry of gastrointestinal mucosubstances. Am. J. Anat. 1976, 146, 103–131. [Google Scholar]
- Chance, D.L.; Mawhinney, T.P. Carbohydrate sulfation effects on growth of pseudomonas aeruginosa. Microbiology 2000, 146, 1717–1725. [Google Scholar]
- Fontaine, N.; Meslin, J.C.; Doré, J. Selective in vitro degradation of the sialylated fraction of germ-free rat mucins by the caecal flora of the rat. Reprod. Nutr. Dev. 1998, 38, 289–296. [Google Scholar]
- Roberto, A.M.; Wright, D.P. Bacterial glycosulphatases and sulphomucin degradation. Can. J. Gastroenterol. 1997, 11, 361–366. [Google Scholar]
- McCool, D.J.; Forstner, J.F.; Forstner, G.G. Synthesis and secretion of mucin by the human colonic tumour cell line LS180. Biochem. J. 1994, 302, 111–118. [Google Scholar]
- Sonnenburg, J.L.; Angenent, L.T.; Gordon, J.I. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004, 5, 569–573. [Google Scholar] [PubMed]
- Carrington, S.D.; Clyne, M.; Reid, C.J.; FitzPatrick, E.; Corfield, A.P. Microbial Interaction with Mucus and Mucins. In Microbial Glycobiology: Structures, Relevance and Applications; Moran, A., Holst, O., Brennan, P., von Itzstein, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 655–671. [Google Scholar]
- Varki, A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 1993, 3, 97–130. [Google Scholar]
- Gagneux, P.; Varki, A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 1999, 9, 747–755. [Google Scholar]
- Patsos, G.; Corfield, A. Management of the human mucosal defensive barrier: Evidence for glycan legislation. Biol. Chem. 2009, 390, 581–590. [Google Scholar]
- Robbe, C.; Capon, C.; Maes, E.; Rousset, M.; Zweibaum, A.; Zanetta, J.P.; Michalski, J.C. Evidence of regio-specific glycosylation in human intestinal mucins: Presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 2003, 278, 46337–46348. [Google Scholar]
- Brockhausen, I. Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta 1999, 1473, 67–95. [Google Scholar]
- Kelly, R.J.; Rouquier, S.; Giorgi, D.; Lennon, G.G.; Lowe, J.B. Sequence and expression of a candidate for the human secretor blood group Alpha(1,2)Fucosyltransferase gene (FUT2). homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J. Biol. Chem. 1995, 270, 4640–4649. [Google Scholar] [PubMed]
- Hoskins, L.C.; Boulding, E.T. Degradation of blood group antigens in human colon ecosystems. II. A gene interaction in man that affects the fecal population density of certain enteric bacteria. J. Clin. Invest. 1976, 57, 74–82. [Google Scholar] [PubMed]
- Salyers, A.A.; Pajeau, M.; McCarthy, R.E. Importance of mucopolysaccharides as substrates for Bacteroides thetaiotaomicron growing in intestinal tracts of exgermfree mice. Appl. Environ. Microbiol. 1988, 54, 1970–1976. [Google Scholar]
- Hwa, V.; Salyers, A.A. Analysis of two chondroitin sulfate utilization mutants of Bacteroides thetaiotaomicron that differ in their abilities to compete with the wild type in the gastrointestinal tracts of germfree mice. Appl. Environ. Microbiol. 1992, 58, 869–876. [Google Scholar]
- Corfield, A.P.; Wagner, S.A.; Clamp, J.R.; Kriaris, M.S.; Hoskins, L.C. Mucin degradation in the human colon: Production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 1992, 60, 3971–3978. [Google Scholar] [PubMed]
- Katayama, T.; Fujita, K.; Yamamoto, K. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J. Biosci. Bioeng. 2005, 99, 457–465. [Google Scholar]
- Ruiz-Palacios, G.M.; Cervantes, L.E.; Ramos, P.; Chavez-Munguia, B.; Newburg, D.S. Campylobacter jejuni binds intestinal H(O) antigen (Fuc Alpha 1, 2Gal Beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003, 278, 14112–14120. [Google Scholar] [PubMed]
- Oakey, H.J.; Harty, D.W.; Knox, K.W. Enzyme production by lactobacilli and the potential link with infective endocarditis. J. Appl. Bacteriol. 1995, 78, 142–148. [Google Scholar]
- Ashida, H.; Miyake, A.; Kiyohara, M.; Wada, J.; Yoshida, E.; Kumagai, H.; Katayama, T.; Yamamoto, K. Two distinct Alpha-l-Fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 2009, 19, 1010–1017. [Google Scholar]
- Lindén, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar]
- Hashimoto, K.; Goto, S.; Kawano, S.; Aoki-Kinoshita, K.F.; Ueda, N.; Hamajima, M.; Kawasaki, T.; Kanehisa, M. KEGG as a Glycome Informatics Resource. Glycobiology 2006, 16, 63R–70R. [Google Scholar]
- Raman, R.; Venkataraman, M.; Ramakrishnan, S.; Lang, W.; Raguram, S.; Sasisekharan, R. Advancing glycomics: Implementation strategies at the consortium for functional glycomics. Glycobiology 2006, 16, 82R–90R. [Google Scholar]
- Tannock, G.W.; Munro, K.; Harmsen, H.J.; Welling, G.W.; Smart, J.; Gopal, P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillusrhamnosus DR20. Appl. Environ. Microbiol. 2000, 66, 2578–2588. [Google Scholar]
- Garrido, D.; Suau, A.; Pochart, P.; Cruchet, S.; Gotteland, M. Modulation of the fecal microbiota by the intake of a Lactobacillus johnsonii La1-Containing product in human volunteers. FEMS Microbiol. Lett. 2005, 248, 249–256. [Google Scholar]
- Wadström, T.; Andersson, K.; Sydow, M.; Axelsson, L.; Lindgren, S.; Gullmar, B. Surface properties of lactobacilli isolated from the small intestine of pigs. J. Appl. Bacteriol. 1987, 62, 513–520. [Google Scholar]
- Lichtenberger, L.M. The hydrophobic barrier properties of gastrointestinal mucus. Annu. Rev. Physiol. 1995, 57, 565–583. [Google Scholar]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and Aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar]
- Ouwehand, A.C.; Kirjavainen, P.V.; Grönlund, M.M.; Isolauri, E.; Salminen, S.J. Adhesion of probiotic micro-organisms to intestinal mucus. Int. Dairy J. 1999, 9, 623–630. [Google Scholar]
- Muñoz-Provencio, D.; Llopis, M.; Antolín, M.; de Torres, I.; Guarner, F.; Pérez-Martínez, G.; Monedero, V. Adhesion properties of Lactobacillus casei strains to resected intestinal fragments and components of the extracellular matrix. Arch. Microbiol. 2009, 191, 153–161. [Google Scholar]
- Jacobsen, C.N.; Rosenfeldt, N.V.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Paerregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [PubMed]
- Jonsson, H.; Ström, E.; Roos, S. Addition of mucin to the growth medium triggers mucus-binding activity in different strains of Lactobacillus reuteri in vitro. FEMS Microbiol. Lett. 2001, 204, 19–22. [Google Scholar]
- MacKenzie, D.A.; Tailford, L.E.; Hemmings, A.M.; Juge, N. Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J. Biol. Chem. 2009, 284, 32444–32453. [Google Scholar]
- Dam, T.K.; Brewer, C.F. Multivalent lectin-carbohydrate interactions energetics and mechanisms of binding. Adv. Carbohydr. Chem. Biochem. 2010, 63, 139–164. [Google Scholar]
- Roos, S.; Jonsson, H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 2002, 148, 433–442. [Google Scholar]
- Boekhorst, J.; Helmer, Q.; Kleerebezem, M.; Siezen, R.J. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 2006, 152, 273–280. [Google Scholar]
- Altermann, E.; Russell, W.M.; Azcarate-Peril, M.A.; Barrangou, R.; Buck, B.L.; McAuliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M.; et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus Ncfm. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar]
- Mackenzie, D.A.; Jeffers, F.; Parker, M.L.; Vibert-Vallet, A.; Bongaerts, R.J.; Roos, S.; Walter, J.; Juge, N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 2010, 156, 3368–3378. [Google Scholar]
- Pridmore, R.D.; Berger, B.; Desiere, F.; Vilanova, D.; Barretto, C.; Pittet, A.C.; Zwahlen, M.C.; Rouvet, M.; Altermann, E.; Barrangou, R.; et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 2004, 101, 2512–2517. [Google Scholar]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar]
- Connell, I.; Agace, W.; Klemm, P.; Schembri, M.; Mărild, S.; Svanborg, C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 1996, 93, 9827–9832. [Google Scholar]
- Macías-Rodríguez, M.E.; Zagorec, M.; Ascencio, F.; Vázquez-Juárez, R.; Rojas, M. Lactobacillus fermentum BCS87 expresses mucus- and mucin-binding proteins on the cell surface. J. Appl. Microbiol. 2009, 107, 1866–1874. [Google Scholar]
- Buck, B.L.; Altermann, E.; Svingerud, T.; Klaenhammer, T.R. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005, 71, 8344–8351. [Google Scholar]
- Pretzer, G.; Snel, J.; Molenaar, D.; Wiersma, A.; Bron, P.A.; Lambert, J.; de Vos, W.M.; van der Meer, R.; Smits, M.A.; Kleerebezem, M. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 2005, 187, 6128–6136. [Google Scholar]
- Rojas, M.; Ascencio, F.; Conway, P.L. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 2002, 68, 2330–2336. [Google Scholar]
- Miyoshi, Y.; Okada, S.; Uchimura, T.; Satoh, E. A mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cell. Biosci. Biotechnol. Biochem. 2006, 70, 1622–1628. [Google Scholar]
- Jacobson, G.R.; Rosenbusch, J.P. Abundance and membrane association of elongation factor Tu in E. coli. Nature 1976, 261, 23–26. [Google Scholar] [PubMed]
- Dallo, S.F.; Kannan, T.R.; Blaylock, M.W.; Baseman, J.B. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 2002, 46, 1041–1051. [Google Scholar]
- Granato, D.; Bergonzelli, G.E.; Pridmore, R.D.; Marvin, L.; Rouvet, M.; Corthésy-Theulaz, I.E. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 2004, 72, 2160–2169. [Google Scholar]
- Nakamura, J.; Ito, D.; Nagai, K.; Umehara, Y.; Hamachi, M.; Kumagai, C. Rapid and sensitive detection of hiochi bacteria by amplification of hiochi bacterial common antigen gene by PCR method and characterization of the antigen. J. Ferment. Bioeng. 1997, 83, 161–167. [Google Scholar]
- Ramiah, K.; van Reenen, C.A.; Dicks, L.M.T. Expression of the mucus adhesion genes mub and MapA, adhesion-like factor EF-Tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PC. Int. J. Food Microbiol. 2007, 116, 405–409. [Google Scholar]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunesekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar]
- The UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38, D142–D148. [CrossRef] [PubMed]
- Sleytr, U.B.; Bayley, H.; Sára, M.; Breitwieser, A.; Küpcü, S.; Mader, C.; Weigert, S.; Unger, F.M.; Messner, P.; Jahn-Schmid, B. Applications of s-layers. FEMS Microbiol. Rev. 1997, 20, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, J.; Shuai, J.; Chen, J.; Zhang, Z.; Fang, W. The s-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int. J. Food Microbiol. 2007, 115, 307–312. [Google Scholar]
- Sánchez, B.; Arias, S.; Chaignepain, S.; Denayrolles, M.; Schmitter, J.M.; Bressollier, P.; Urdaci, M.C. Identification of surface proteins involved in the adhesion of a probiotic Bacillus cereus strain to mucin and fibronectin. Microbiology 2009, 155, 1708–1716. [Google Scholar]
- Zhang, Y.C.; Zhang, L.W.; Tuo, Y.F.; Guo, C.F.; Yi, H.X.; Li, J.Y.; Han, X.; Du, M. Inhibition of Shigella sonnei adherence to HT-29 cells by lactobacilli from Chinese fermented food and preliminary characterization of s-layer protein involvement. Res. Microbiol. 2010, 161, 667–672. [Google Scholar]
- Bøhle, L.A.; Brede, D.A.; Diep, D.B.; Holo, H.; Nes, I.F. The mucus adhesion promoting protein (MapA) of Lactobacillus reuteri is specifically degraded to an antimicrobial peptide. Appl. Environ. Microbiol. 2010, 76, 7306–7309. [Google Scholar]
- Ouwehand, A.C.; Salminen, S. In vitro adhesion assays for probiotics and their in vivo relevance: A review. Microb. Ecol. Health Dis. 2003, 15, 175–184. [Google Scholar]
- Ouwehand, A.C.; Tuomola, E.M.; Tölkkö, S.; Salminen, S. Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int. J. Food Microbiol. 2001, 64, 119–126. [Google Scholar]
- Kankaanpää, P.E.; Salminen, S.J.; Isolauri, E.; Lee, Y.K. The influence of polyunsaturated fatty acids on probiotic growth and adhesion. FEMS Microbiol. Lett. 2001, 194, 149–153. [Google Scholar]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar]
- Juntunen, M.; Kirjavainen, P.V.; Ouwehand, A.C.; Salminen, S.J.; Isolauri, E. Adherence of probiotic bacteria to human intestinal mucus in healthy infants and during rotavirus infection. Clin. Diagn. Lab. Immunol. 2001, 8, 293–296. [Google Scholar]
- Schiffrin, E.J.; Brassart, D.; Servin, A.L.; Rochat, F.; Donnet-Hughes, A. Immune modulation of blood leukocytes in humans by lactic acid bacteria: Criteria for strain selection. Am. J. Clin. Nutr. 1997, 66, 515S–520S. [Google Scholar]
- Vélez, M.P.; Petrova, M.I.; Lebeer, S.; Verhoeven, T.L.A.; Claes, I.; Lambrichts, I.; Tynkkynen, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol. Med. Microbiol. 2010, 59, 386–398. [Google Scholar]
- Lee, Y.; Puong, K. Competition for adhesion between probiotics and human gastrointestinal pathogens in the presence of carbohydrate. Br. J. Nutr. 2002, 88, S101–S108. [Google Scholar]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar]
- Gueimonde, M.; Jalonen, L.; He, F.; Hiramatsu, M.; Salminen, S. Adhesion and competitive inhibition and displacement of human enteropathogens by selected lactobacilli. Food Res. Int. 2006, 39, 467–471. [Google Scholar]
- Majamaa, H.; Isolauri, E.; Saxelin, M.; Vesikari, T. Lactic acid bacteria in the treatment of acute rotavirus gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 1995, 20, 333–338. [Google Scholar]
- O’Halloran, S.; Feeney, M.; Morrissey, D.; Murphy, L.; Thornton, G.; Shanahan, F.; O’Sullivan, G.C.; Collins, J.K. Adhesion of potential probiotic bacteria to human epithelial cell lines. Int. Dairy J. 1998, 8, 596. [Google Scholar]
- Laparra, J.M.; Sanz, Y. Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett. Appl. Microbiol. 2009, 49, 695–701. [Google Scholar]
- Tallon, R.; Arias, S.; Bressollier, P.; Urdaci, M.C. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 2007, 102, 442–451. [Google Scholar]
- Li, X.J.; Yue, L.Y.; Guan, X.F.; Qiao, S.Y. The adhesion of putative probiotic lactobacilli to cultured epithelial cells and porcine intestinal mucus. J. Appl. Microbiol. 2008, 104, 1082–1091. [Google Scholar]
- Pinto, M.; Robine-Leon, S.; Appay, M.D. Enterocyte-like differentiation and polarization of the human colon Carcinoma Cell Line Caco-2 in culture. Biol. Cell 1983, 47, 323–330. [Google Scholar]
- Rousset, M. The human colon carcinoma cell lines HT-29 and Caco-2: Two in vitro models for the study of intestinal differentiation. Biochimie 1986, 68, 1035–1040. [Google Scholar]
- Lenaerts, K.; Bouwman, F.G.; Lamers, W.H.; Renes, J.; Mariman, E.C. Comparative proteomic analysis of cell lines and scrapings of the human intestinal epithelium. BMC Genomics 2007, 8, 91. [Google Scholar]
- Crociani, J.; Grill, J.P.; Huppert, M.; Ballongue, J. Adhesion of different Bifidobacteria strains to human enterocyte-like Caco-2 cells and comparison with in vivo study. Lett. Appl. Microbiol. 1995, 21, 146–148. [Google Scholar]
- Lesuffleur, T.; Barbat, A.; Dussaulx, E.; Zweibaum, A. Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 1990, 50, 6334–6343. [Google Scholar]
- Leteurtre, E.; Gouyer, V.; Rousseau, K.; Moreau, O.; Barbat, A.; Swallow, D.; Huet, G.; Lesuffleur, T. Differential mucin expression in colon carcinoma HT-29 clones with variable resistance to 5-fluorouracil and methotrexate. Biol. Cell 2004, 96, 145–151. [Google Scholar]
- Bernet, M.F.; Brassart, D.; Neeser, J.R.; Servin, A.L. Lactobacillus acidophilus LA1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 1994, 35, 483–489. [Google Scholar]
- Gopal, P.K.; Prasad, J.; Smart, J.; Gill, H.S. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichiacoli. Int. J. Food Microbiol. 2001, 67, 207–216. [Google Scholar]
- Lesuffleur, T.; Porchet, N.; Aubert, J.P.; Swallow, D.; Gum, J.R.; Kim, Y.S.; Real, F.X.; Zweibaum, A. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J. Cell Sci. 1993, 106, 771–783. [Google Scholar]
- Kerneis, S.; Bernet, M.F.; Coconnier, M.H.; Servin, A.L. Adhesion of human enterotoxigenic Escherichia coli to human mucus secreting HT-29 cell subpopulations in culture. Gut 1994, 35, 1449–1454. [Google Scholar]
- Nutten, S.; Sansonetti, P.; Huet, G.; Bourdon-Bisiaux, C.; Meresse, B.; Colombel, J.F.; Desreumaux, P. Epithelial inflammation response induced by Shigella flexneri depends on mucin gene expression. Microbes Infect. 2002, 4, 1121–1124. [Google Scholar]
- Walter, E.; Janich, S.; Roessler, B.J.; Hilfinger, J.M.; Amidon, G.L. HT29-MTX/Caco-2 cocultures as an in vitro model for the intestinal epithelium: In vitro-in vivo correlation with permeability data from rats and humans. J. Pharm. Sci. 1996, 85, 1070–1076. [Google Scholar]
- Pontier, C.; Pachot, J.; Botham, R.; Lenfant, B.; Arnaud, P. HT29-MTX and Caco-2/TC7 monolayers as predictive models for human intestinal absorption: Role of the mucus layer. J. Pharm. Sci. 2001, 90, 1608–1619. [Google Scholar]
- Chen, X.M.; Elisia, I.; Kitts, D.D. Defining conditions for the co-culture of Caco-2 and HT29-MTX cells using taguchi design. J. Pharmacol. Toxicol. Methods 2010, 61, 334–342. [Google Scholar]
- Henriksson, A.; Szewzyk, R.; Conway, P.L. Characteristics of the adhesive determinants of Lactobacillus fermentum 104. Appl. Environ. Microbiol. 1991, 57, 499–502. [Google Scholar]
- Vesterlund, S.; Paltta, J.; Karp, M.; Ouwehand, A. Adhesion of bacteria to resected human colonic tissue: Quantitative analysis of bacterial adhesion and viability. Res. Microbiol. 2005, 156, 238–244. [Google Scholar]
- Phillips, A.D.; Navabpour, S.; Hicks, S.; Dougan, G.; Wallis, T.; Frankel, G. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer’s patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 2000, 47, 377–381. [Google Scholar]
- Girard, F.; Dziva, F.; van Diemen, P.; Phillips, A.D.; Stevens, M.P.; Frankel, G. Adherence of enterohemorrhagic Escherichia coli O157, O26, and O111 strains to bovine intestinal explants ex viv. Appl. Environ. Microbiol. 2007, 73, 3084–3090. [Google Scholar]
- Day, C.J.; Tiralongo, J.; Hartnell, R.D.; Logue, C.; Wilson, J.C.; von Itzstein, M.; Korolik, V. Differential carbohydrate recognition by Campylobacter jejuni strain 11168: Influences of temperature and growth conditions. PLoS One 2009, 4, 781–790. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).