Improving Personalized Meal Planning with Large Language Models: Identifying and Decomposing Compound Ingredients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Data Analysis Tools and Techniques
2.3. Statistical Analysis
2.4. Large Language Models
2.5. Methodology
2.5.1. Generation of Meal Plans:
| Prompt 1 Meal generation (Prompt) |
| Create a detailed one-day <INPUT_meal_type> plan. Ensure the meal plan includes precise portion sizes, specific quantities or weights of individual ingredients, and diverse cooking methods. Include compound ingredients like Chicken Cacciatore, Vegetable Ratatouille, or similar. However, avoid including ingredients included in the following list: <INPUT_list>, Structure the results in a formatted HTML format similar to the provided example: <INPUT_html_format>. |
2.5.2. Decomposition of Compound Ingredients:
| Prompt 2 Identification and decomposition of the compound ingredients. (Prompt) |
| Analyze the meal plan provided in an HTML table (enclosed within <table> and </table> tags): <INPUT_meal_plan>. Identify compound ingredients listed under the ‘Ingredient’ column that consist of multiple basic ingredients. Use the ‘Ingredient Details’ column for additional information. For each identified compound ingredient, report the names of these items along with their specific ingredients and estimated quantities based on the ‘Portion Size (g, dL)’ column, formatted in a dictionary of dictionaries. Ensure all ingredients are specific (e.g., ‘carrots’, ‘spinach’, etc., instead of ‘vegetables’). If items can be further broken down, do it. Example format: ‘Complex Food Item 1’: ‘Ingredient 1’: ‘50 g’, ‘Ingredient 2’: ‘100 g’, “Complex Food Item 2”: ‘Ingredient 1’: ‘200 g’, ‘Ingredient 2’: ‘150 g’. Return only the formatted dictionary encapsulated by dollar signs ($Content$). Dictionary should not contain any comments. If no complex food items meeting the criteria are identified, return an empty dictionary. Ensure that the quantities of each ingredient accurately reflect the specified portion size within each compound ingredient as detailed in the meal plan. For ingredients where the exact quantity cannot be determined, provide a reasonable estimate instead of leaving it unspecified. |
2.5.3. USDA FoodData Subset Creation
2.5.4. Mapping to USDA FoodData
| Prompt 3 Mapping the ingredient to FoodData Central’s list of ingredients (Prompt) |
| You are tasked with matching a given ingredient <food_item> to the most appropriate one from the following list of ingredients: <list_of_food_items>. Each ingredient possesses unique characteristics that influence its compatibility with others. Your objective is to identify which ingredient from the list best complements the given one, considering factors such as flavor profile, cooking methods, and culinary traditions. Return only the ‘fdcId’ value of the best match encapsulated in dollar signs (e.g., $21341$). Do not include any comments. |
2.5.5. Nutrient Composition Calculation
2.5.6. Aggregation of Nutrient Data
2.5.7. Evaluation Process
- Identification by Nutritionists: Nutritionists review 15 meal plans each to identify compound ingredients. These identifications serve as ground truth positives.
- Comparison with Model Predictions: The compound ingredients identified by the nutritionists are compared with those predicted by the model. This comparison is used to calculate accuracy and F1-score. Accuracy measures the proportion of correctly identified compound ingredients out of the total number of ingredients. F1-score provides a balance between precision and recall, offering a comprehensive measure especially useful in cases of class imbalance.
- Quality control: The nutritionists assess the decomposed basic ingredients and their quantities to confirm that they are realistic and consistent with the compound ingredients.
3. Results
3.1. Summary of Compound Ingredients in the Meal Plans
3.2. Identification of Compound Ingredients
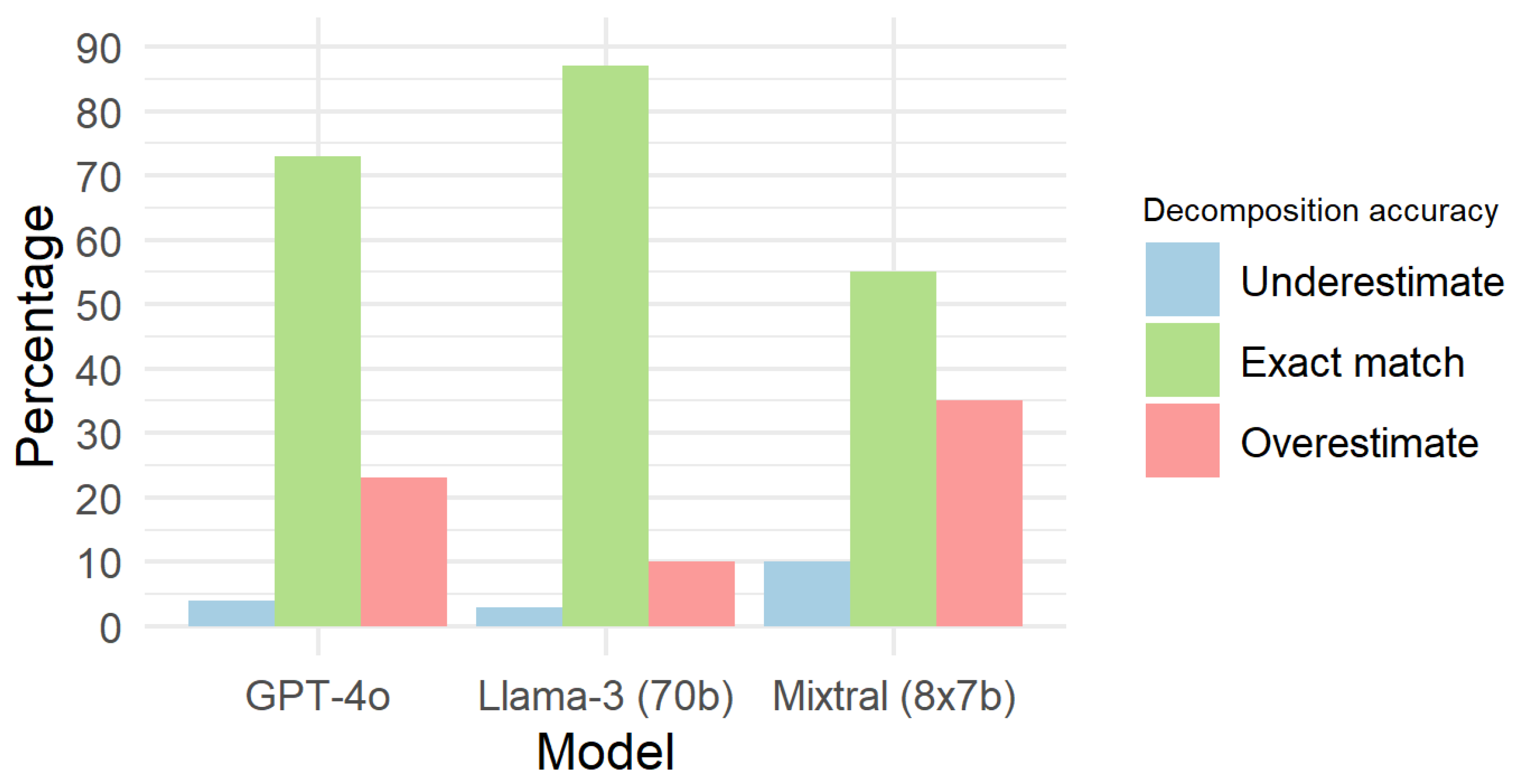
3.3. Decomposition of Compound Ingredients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACC | accuracy |
| AI | artificial intelligence |
| CI | confidence interval |
| CNN | convolutional neural network |
| FNDDS | food and nutrient database for dietary studies |
| GPT | generative pre-trained transformer |
| HTML | hypertext markup language |
| IP | Internet Protocol |
| IQR | interquartile range |
| LLM | large language model |
| SMoE | sparse mixture of experts |
| USDA | U.S. Department of Agriculture |
References
- Theodore, A.T.P.; Nfor, K.A.; Kim, J.-I.; Kim, H.-C. Applications of artificial intelligence, machine learning, and deep learning in nutrition: A systematic review. Nutrients 2024, 16, 1073. [Google Scholar] [CrossRef] [PubMed]
- Santhuja, P.; Reddy, E.G.; Choudri, S.R.; Muthulekshmi, M.; Balaji, S. Intelligent Personalized Nutrition Guidance System Using IoT and Machine Learning Algorithm. In Proceedings of the 2023 Second International Conference on Smart Technologies for Smart Nation (SmartTechCon), Singapore, 18–19 August 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 250–254. [Google Scholar]
- Maurya, A.; Wable, R.; Shinde, R.; John, S.; Jadhav, R.; Dakshayani, R. Chronic kidney disease prediction and recommendation of suitable diet plan by using machine learning. In Proceedings of the 2019 International Conference on Nascent Technologies in Engineering (ICNTE), Navi Mumbai, India, 4–5 January 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Li, T.; Wei, W.; Xing, S.; Min, W.; Zhang, C.; Jiang, S. Deep learning-based near-infrared hyperspectral imaging for food nutrition estimation. Foods 2023, 12, 3145. [Google Scholar] [CrossRef] [PubMed]
- Sripada, N.K.; Challa, S.C.; Kothakonda, S. AI-Driven Nutritional Assessment Improving Diets with Machine Learning and Deep Learning for Food Image Classification. In Proceedings of the 2023 International Conference on Self Sustainable Artificial Intelligence Systems (ICSSAS), Erode, India, 18–20 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 138–144. [Google Scholar]
- Kim, H.; Lim, D.H.; Kim, Y. Classification and prediction on the effects of nutritional intake on overweight/obesity, dyslipidemia, hypertension and type 2 diabetes mellitus using deep learning model: 4–7th Korea national health and nutrition examination survey. Int. J. Environ. Res. Public Health 2021, 18, 5597. [Google Scholar] [CrossRef] [PubMed]
- Alrige, M.A.; Chatterjee, S.; Medina, E.; Nuval, J. Applying the concept of nutrient-profiling to promote healthy eating and raise individuals’ awareness of the nutritional quality of their food. In Proceedings of the AMIA Annual Symposium Proceedings. American Medical Informatics Association, Washington, DC, USA, 4–8 November 2017; Volume 2017, p. 393. [Google Scholar]
- Lu, Y.; Stathopoulou, T.; Vasiloglou, M.F.; Pinault, L.F.; Kiley, C.; Spanakis, E.K.; Mougiakakou, S. goFOODTM: An artificial intelligence system for dietary assessment. Sensors 2020, 20, 4283. [Google Scholar] [CrossRef] [PubMed]
- Phalle, A.; Gokhale, D. Navigating next-gen nutrition care using artificial intelligence-assisted dietary assessment tools—A scoping review of potential applications. Front. Nutr. 2025, 12, 1518466. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, J.; Shen, J.; An, R. Artificial Intelligence Applications to Measure Food and Nutrient Intakes: Scoping Review. J. Med. Internet Res. 2024, 26, e54557. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Xia, F.; Chi, E.; Le, Q.V.; Zhou, D. Chain-of-thought prompting elicits reasoning in large language models. Adv. Neural Inf. Process. Syst. 2022, 35, 24824–24837. [Google Scholar]
- Naveed, H.; Khan, A.U.; Qiu, S.; Saqib, M.; Anwar, S.; Usman, M.; Barnes, N.; Mian, A. A comprehensive overview of large language models. arXiv 2023, arXiv:2307.06435. [Google Scholar]
- Ma, P.; Tsai, S.; He, Y.; Jia, X.; Zhen, D.; Yu, N.; Wang, Q.; Ahuja, J.K.; Wei, C.I. Large Language Models in Food Science: Innovations, Applications, and Future. Trends Food Sci. Technol. 2024, 148, 104488. [Google Scholar] [CrossRef]
- Abdulla, W. Mask R-CNN for Object Detection and Instance Segmentation on Keras and TensorFlow. GitHub Repository. 2017. Available online: https://github.com/matterport/Mask_RCNN (accessed on 12 January 2025).
- Osowiecka, K.; Pokorna, N.; Skrypnik, D. Assessment of negative factors affecting the intestinal microbiota in people with excessive body mass compared to people with normal body mass. Proceedings 2020, 61, 5. [Google Scholar] [CrossRef]
- OpenAI. Hello GPT-4o|OpenAI. 2024. Available online: https://openai.com/index/hello-gpt-4o/ (accessed on 29 June 2024).
- Meta. Introducing Meta Llama 3: The Most Capable Openly Available LLM to Date. 2024. Available online: https://ai.meta.com/blog/meta-llama-3/ (accessed on 29 June 2024).
- Jiang, A.Q.; Sablayrolles, A.; Roux, A.; Mensch, A.; Savary, B.; Bamford, C.; Chaplot, D.S.; de las Casas, D.; Hanna, E.B.; Bressand, F.; et al. Mixtral of Experts. arXiv 2024, arXiv:2401.04088. [Google Scholar]
- OpenAI. OpenAI API. 2024. Available online: https://openai.com/index/openai-api/ (accessed on 3 July 2024).
- Perplexity AI. Perplexity API. 2024. Available online: https://www.perplexity.ai/settings/api (accessed on 3 July 2024).
- USDA. U.S. Department of Agriculture, Agricultural Research Service FoodData Central. 2019. Available online: https://fdc.nal.usda.gov/ (accessed on 29 June 2024).
- Hicks, S.A.; Strümke, I.; Thambawita, V.; Hammou, M.; Riegler, M.A.; Halvorsen, P.; Parasa, S. On evaluation metrics for medical applications of artificial intelligence. Sci. Rep. 2022, 12, 5979. [Google Scholar] [CrossRef] [PubMed]
- Hieronimus, B.; Hammann, S.; Podszun, M.C. Can the AI tools ChatGPT and Bard generate energy, macro-and micro-nutrient sufficient meal plans for different dietary patterns? Nutr. Res. 2024, 128, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Park, J.S.; Sharma, K.; Velazquez, A.; Li, L.; Ostrominski, J.W.; Tran, T.; Seitter Pérez, R.H.; Shin, J.-H. Qualitative evaluation of artificial intelligence-generated weight management diet plans. Front. Nutr. 2024, 11, 1374834. [Google Scholar] [CrossRef] [PubMed]
- Paineau, D.; Beaufils, F.; Boulier, A.; Cassuto, D.; Chwalow, J.; Combris, P.; Couet, C.; Jouret, B.; Lafay, L.; Laville, M.; et al. The cumulative effect of small dietary changes may significantly improve nutritional intakes in free-living children and adults. Eur. J. Clin. Nutr. 2010, 64, 782–791. [Google Scholar] [CrossRef] [PubMed][Green Version]
- USGovernment. Developer Manual. Available online: https://fdc.nal.usda.gov/ (accessed on 4 September 2024).

| Evaluator | GPT-4o | Llama-3 (70B) | Mixtral (8x7B) | p-Value (GPT-4o vs. Llama-3) | p-Value (GPT-4o vs. Mixtral) | p-Value (Llama-3 vs. Mixtral) |
|---|---|---|---|---|---|---|
| 1 | 0.838 (0.73–0.95) | 0.885 (0.79–0.98) | 0.566 (0.39–0.74) | 0.48 | <0.05 | <0.01 |
| 2 | 0.806 (0.71–0.91) | 0.889 (0.83–0.95) | 0.729 (0.62–0.84) | 0.19 | 0.1 | <0.05 |
| 3 | 0.862 (0.77–0.95) | 0.906 (0.86–0.96) | 0.701 (0.6–0.8) | 0.62 | <0.01 | <0.01 |
| Overall | 0.835 (0.78–0.89) | 0.893 (0.85–0.94) | 0.666 (0.59–0.75) | 0.12 | <0.05 | <0.05 |
| Evaluator | GPT-4o | Llama-3 (70B) | Mixtral (8x7B) | p-Value (GPT-4o vs. Llama-3) | p-Value (GPT-4o vs. Mixtral) | p-Value (Llama-3 vs. Mixtral) |
|---|---|---|---|---|---|---|
| 1 | 0.842 (0.75–0.94) | 0.875 (0.74–1.01) | 0.612 (0.48–0.74) | 0.36 | <0.01 | <0.01 |
| 2 | 0.824 (0.74–0.91) | 0.902 (0.85–0.96) | 0.751 (0.64–0.86) | 0.17 | 0.17 | <0.05 |
| 3 | 0.861 (0.78–0.94) | 0.904 (0.88–0.93) | 0.708 (0.65–0.76) | 0.36 | <0.01 | <0.01 |
| Overall | 0.842 (0.79–0.89) | 0.894 (0.84–0.95) | 0.690 (0.62–0.76) | 0.14 | <0.05 | <0.05 |
| Model | Compound Ingredients |
|---|---|
| GPT-4o | ‘Scrambled Eggs’ ‘Oatmeal with Fresh Fruit’ ‘Grilled Vegetable Frittata’ ‘Whole Grain Pancakes’ ‘Steel-Cut Oatmeal’ |
| Llama-3 (70B) | ‘Oatmeal with Fresh Fruit’ ‘Grilled Vegetable Frittata’ ‘Whole Grain Pancakes’ |
| Mixtral (8x7B) | ‘Oatmeal with Fresh Fruit’ ‘Grilled Vegetable Frittata’ ‘Whole Grain Pancakes’ ‘Whole Wheat Bagel with Light Cream Cheese’ ‘Chicken Marsala’ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopitar, L.; Bedrač, L.; Strath, L.J.; Bian, J.; Stiglic, G. Improving Personalized Meal Planning with Large Language Models: Identifying and Decomposing Compound Ingredients. Nutrients 2025, 17, 1492. https://doi.org/10.3390/nu17091492
Kopitar L, Bedrač L, Strath LJ, Bian J, Stiglic G. Improving Personalized Meal Planning with Large Language Models: Identifying and Decomposing Compound Ingredients. Nutrients. 2025; 17(9):1492. https://doi.org/10.3390/nu17091492
Chicago/Turabian StyleKopitar, Leon, Leon Bedrač, Larissa J. Strath, Jiang Bian, and Gregor Stiglic. 2025. "Improving Personalized Meal Planning with Large Language Models: Identifying and Decomposing Compound Ingredients" Nutrients 17, no. 9: 1492. https://doi.org/10.3390/nu17091492
APA StyleKopitar, L., Bedrač, L., Strath, L. J., Bian, J., & Stiglic, G. (2025). Improving Personalized Meal Planning with Large Language Models: Identifying and Decomposing Compound Ingredients. Nutrients, 17(9), 1492. https://doi.org/10.3390/nu17091492






