A Genome-Wide Association Study of Circulating Serum Choline, Betaine, Dimethylglycine, and Their Ratios
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sample
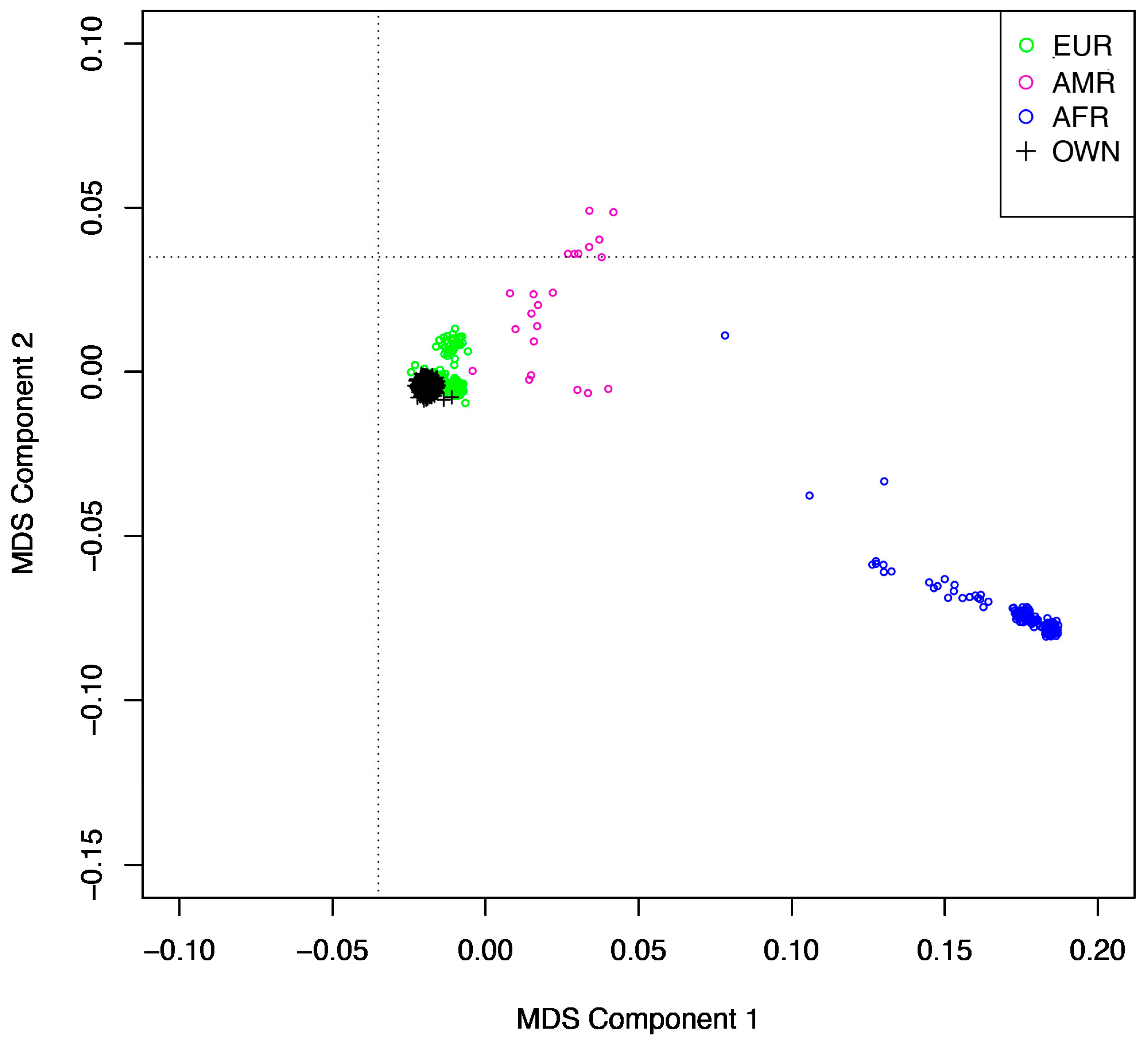
2.2. Genetic Quality Control
2.3. Genome-Wide Association and Statistical Analysis
3. Results
3.1. Participant Characteristics
3.2. Metabolite Distributions
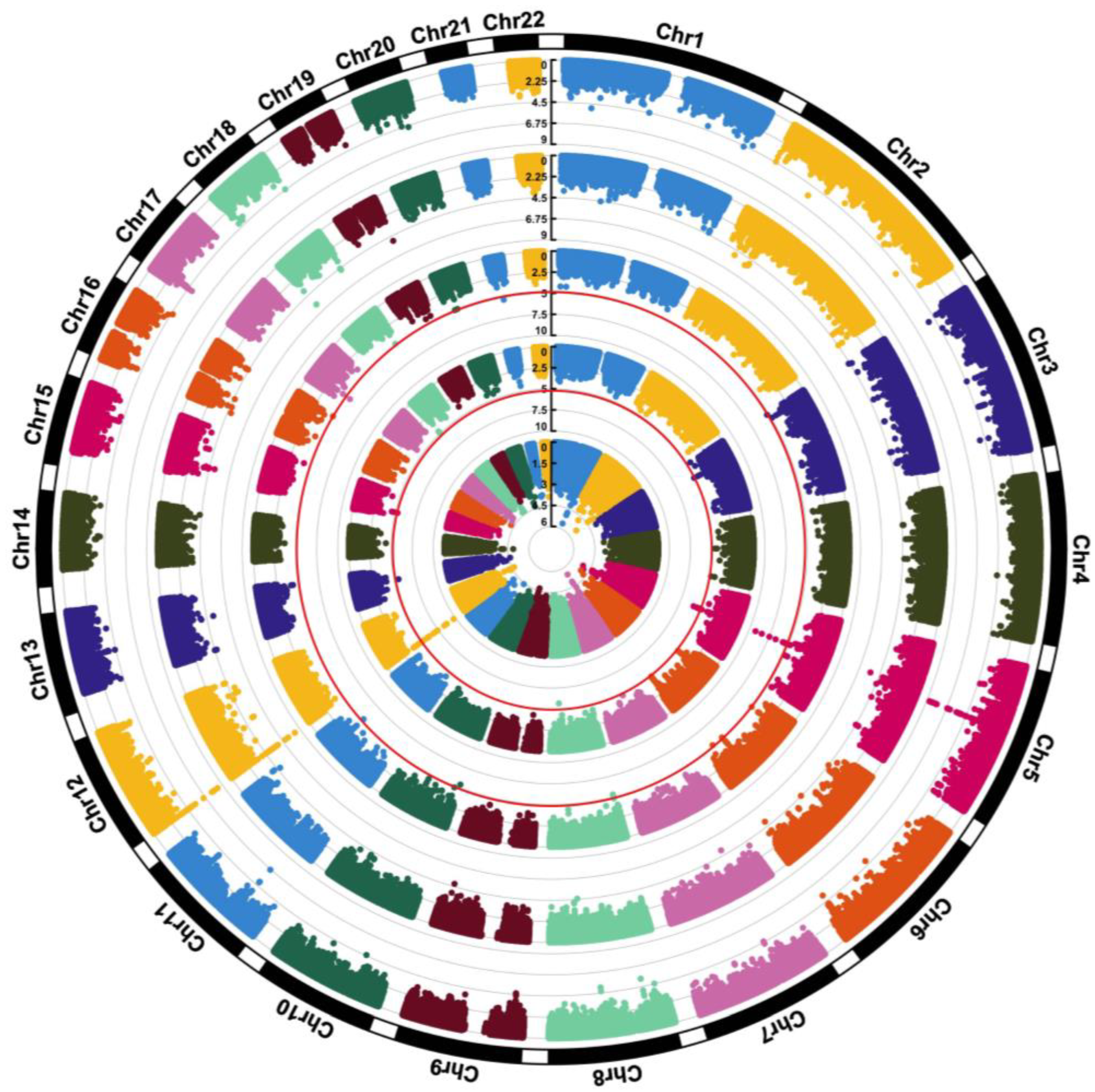
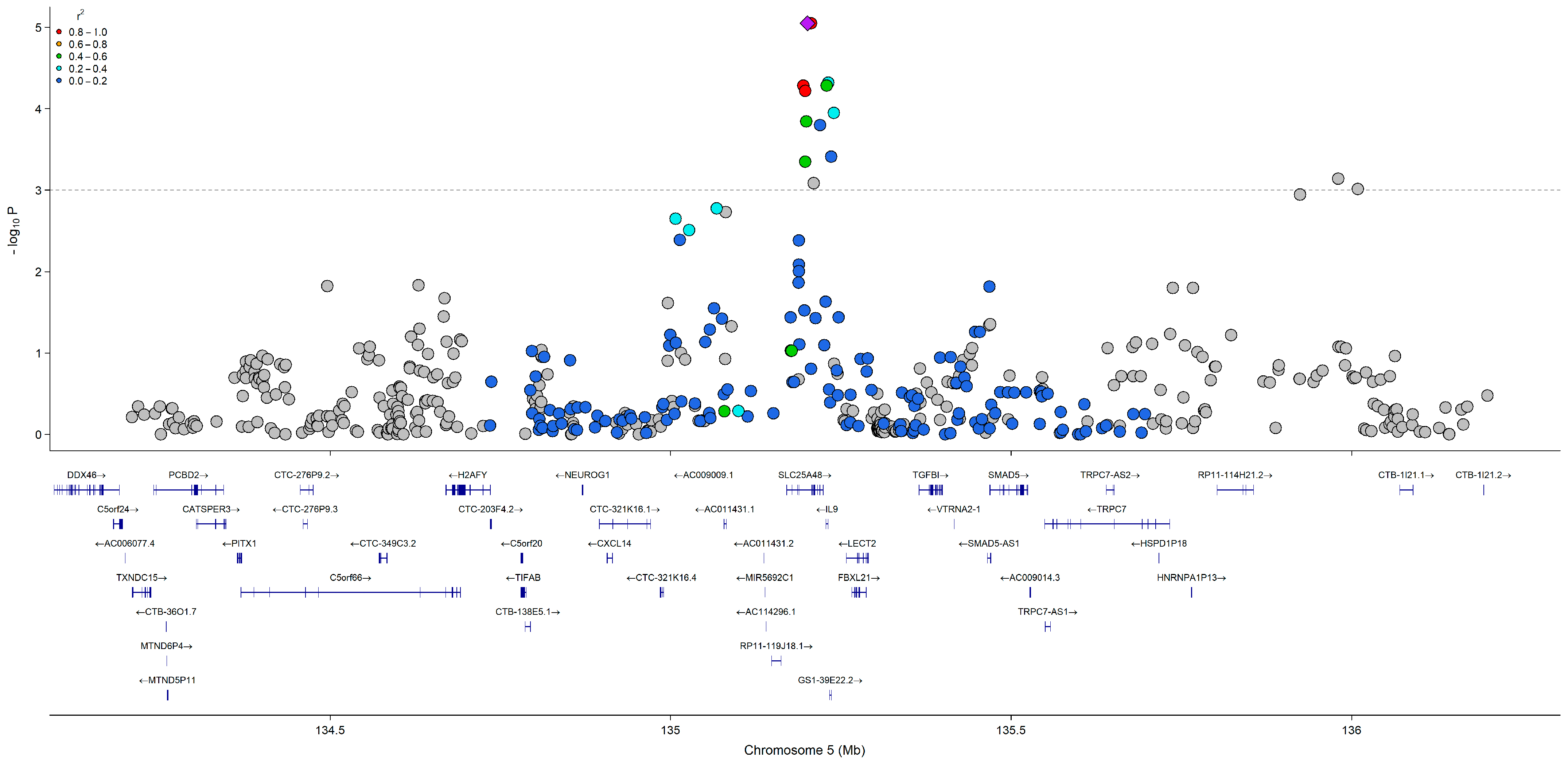
3.3. GWAS of Serum Choline, Betaine, and DMG
3.4. GWAS of Serum Choline-to-Betaine and Betaine-to-DMG Ratios
3.5. SNP Literature Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BHMT | betaine-homocysteine methyltransferase |
| CSGNM | Collaborative Study of Genes, Nutrients and Metabolites |
| dbGaP | database of Genotypes and Phenotypes |
| dbSNP | database of Single Nucleotide Polymorphisms |
| DMG | dimethylglycine |
| DMGDH | dimethylglycine dehydrogenase |
| FDR | false discovery rate |
| GTEx | Genotype-Tissue Expression |
| GWAS | genome-wide association studies |
| HoloTC | holotranscobalamin |
| IBS | identity-by-state |
| LDAH | lipid droplet-associated phosphatidylserine hydrolase |
| MAF | minor allele frequency |
| MASLD | metabolic dysfunction-associated steatotic liver disease |
| MDS | multidimensional scaling |
| PC | principal component |
| SLC | solute carrier |
| SNP | single nucleotide polymorphism |
| TMAO | trimethylamine N-oxide |
References
- Wallace, T.C.; Blusztajn, J.K.; Caudill, M.A.; Klatt, K.C.; Natker, E.; Zeisel, S.H.; Zelman, K.M. Choline: The underconsumed and underappreciated essential nutrient. Nutr. Today 2018, 53, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline. Available online: https://www.ncbi.nlm.nih.gov/books/NBK114310/ (accessed on 1 October 2024).
- Holm, P.I.; Ueland, P.M.; Kvalheim, G.; Lien, E.A. Determination of choline, betaine, and dimethylglycine in plasma by a high-throughput method based on normal-phase chromatography-tandem mass spectrometry. Clin. Chem. 2003, 49, 286–294. [Google Scholar] [CrossRef] [PubMed]
- de Zwart, F.J.; Slow, S.; Payne, R.J.; Lever, M.; George, P.M.; Gerrard, J.A.; Chambers, S.T. Glycine betaine and glycine betaine analogues in common foods. Food Chem. 2003, 83, 197–204. [Google Scholar] [CrossRef]
- Ganz, A.B.; Klatt, K.C.; Caudill, M.A. Common genetic variants alter metabolism and influence dietary choline requirements. Nutrients 2017, 9, 837. [Google Scholar] [CrossRef] [PubMed]
- Kenny, T.C.; Khan, A.; Son, Y.; Yue, L.; Heissel, S.; Sharma, A.; Pasolli, H.A.; Liu, Y.; Gamazon, E.R.; Alwaseem, H.; et al. Integrative genetic analysis identifies FLVCR1 as a plasma-membrane choline transporter in mammals. Cell Metab. 2023, 35, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Louck, L.E.; Cara, K.C.; Klatt, K.; Wallace, T.C.; Chung, M. The relationship of circulating choline and choline-related metabolite levels with health outcomes: A scoping review of genome-wide association studies and Mendelian randomization studies. Adv. Nutr. 2024, 15, 100164. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.L.; Carter, T.C.; Scott, J.M.; Troendle, J.F.; Gibney, E.R.; Shane, B.; Kirke, P.N.; Ueland, P.M.; Brody, L.C.; Molloy, A.M. Do high blood folate concentrations exacerbate metabolic abnormalities in people with low vitamin B-12 status? Am. J. Clin. Nutr. 2011, 94, 495–500. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. dbGaP (Database of Genotypes and Phenotypes): Accession phs000789.v1.p1. Available online: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000789.v1.p1 (accessed on 1 October 2024).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. dbSNP (Database of Single Nucleotide Polymorphisms). Available online: https://www.ncbi.nlm.nih.gov/snp/ (accessed on 1 October 2024).
- Boughton, A.P.; Welch, R.P.; Flickinger, M.; VandeHaar, P.; Taliun, D.; Abecasis, G.R.; Boehnke, M. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics 2021, 37, 3017–3018. [Google Scholar] [CrossRef]
- Genotype-Tissue Expression (GTEx) Project. GTEx Portal. Available online: https://gtexportal.org/home/ (accessed on 1 June 2024).
- Ganz, A.B.; Cohen, V.V.; Swersky, C.C.; Stover, J.; Vitiello, G.A.; Lovesky, J.; Chuang, J.C.; Shields, K.; Fomin, V.G.; Lopez, Y.S.; et al. Genetic variation in choline-metabolizing enzymes alters choline metabolism in young women consuming choline intakes meeting current recommendations. Int. J. Mol. Sci. 2017, 18, 252. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Unlu, G.; Lin, P.; Liu, Y.; Kilic, E.; Kenny, T.C.; Birsoy, K.; Gamazon, E.R. Metabolic gene function discovery platform GeneMAP identifies SLC25A48 as necessary for mitochondrial choline import. Nat. Genet. 2024, 56, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Verkerke, A.R.P.; Shi, X.; Li, M.; Higuchi, Y.; Yamamuro, T.; Katoh, D.; Nishida, H.; Auger, C.; Abe, I.; Gerszten, R.E.; et al. SLC25A48 controls mitochondrial choline import and metabolism. Cell Metab. 2024, 36, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Randall, J.C.; Winkler, T.W.; Kutalik, Z.; Berndt, S.I.; Jackson, A.U.; Monda, K.L.; Kilpeläinen, T.O.; Esko, T.; Mägi, R.; Li, S.; et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013, 9, e1003500. [Google Scholar] [CrossRef] [PubMed]
- Speliotes, E.K.; Yerges-Armstrong, L.M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D.; Gudnason, V.; Eiriksdottir, G.; Garcia, M.E.; Launer, L.J.; et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7, e1001324. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, A.; Belardinilli, F.; Bailetti, D.; Sponziello, M.; D’Erasmo, L.; Polimeni, L.; Baratta, F.; Pastori, D.; Ceci, F.; Montali, A.; et al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (NAFLD) by a candidate genes resequencing strategy. Sci. Rep. 2018, 8, 3702. [Google Scholar] [CrossRef] [PubMed]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Heid, I.M.; Jackson, A.U.; Randall, J.C.; Winkler, T.W.; Qi, L.; Steinthorsdottir, V.; Thorleifsson, G.; Zillikens, M.C.; Speliotes, E.K.; Mägi, R.; et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010, 42, 949–960. [Google Scholar] [CrossRef]
- Saliba-Gustafsson, P.; Justesen, J.M.; Ranta, A.; Sharma, D.; Bielczyk-Maczynska, E.; Li, J.; Najmi, L.A.; Apodaka, M.; Aspichueta, P.; Björck, H.M.; et al. A functional genomic framework to elucidate novel causal metabolic dysfunction-associated fatty liver disease genes. Hepatology 2024. published ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Vohnoutka, R.B.; Kuppa, A.; Hegde, Y.; Chen, Y.; Pant, A.; Tohme, M.E.; Choi, E.K.; McCarty, S.M.; Bagchi, D.P.; Du, X.; et al. Knockout of murine Lyplal1 confers sex-specific protection against diet-induced obesity. J. Mol. Endocrinol. 2023, 70, e220131. [Google Scholar] [CrossRef]
| Variable | Value (n = 2220) |
|---|---|
| Age (y), mean ± SD (min, max) | 22.4 ± 1.69 (19.0, 28.0) |
| Sex, No. (%) | |
| Male | 918 (41.4) |
| Female | 1302 (58.6) |
| Serum choline (µmol/L), mean ± SD (min, max) | 7.65 ± 1.81 (3.30, 19.7) |
| SNP | Log (β) | SE | p-Value | Gene |
|---|---|---|---|---|
| rs10779349 | 0.0141 | 0.00327 | 1.63 × 10−5 | LYPLAL1: intron variant |
| rs4846551 | 0.0165 | 0.00373 | 1.08 × 10−5 | LYPLAL1: intron variant |
| rs6671991 | 0.0182 | 0.00385 | 2.40 × 10−6 | LYPLAL1: intron variant |
| rs10205941 | −0.0140 | 0.00322 | 1.57 × 10−5 | None |
| rs6890039 | −0.0250 | 0.00561 | 8.96 × 10−6 | SLC25A48: intron variant |
| rs6596270 | −0.0250 | 0.00561 | 8.96 × 10−6 | SLC25A48: synonymous variant |
| rs4272127 | −0.0169 | 0.00415 | 4.79 × 10−5 | SLC25A48: intron variant |
| rs6601167 | 0.0228 | 0.00522 | 1.26 × 10−5 | None |
| rs226915 | −0.0229 | 0.00517 | 9.97 × 10−6 | LOC100507336: intron variant |
| rs2261033 | 0.0171 | 0.00399 | 2.00 × 10−5 | PRRC2A: intron variant |
| rs7812117 | 0.0170 | 0.00417 | 4.57 × 10−5 | NXPH1: intron variant |
| rs1050290 | −0.0162 | 0.00371 | 1.40 × 10−5 | MEOX2: 5′-UTR variant LINC02587: 2KB upstream variant |
| rs740566 | −0.0167 | 0.00360 | 3.57 × 10−6 | LINC02587: 2KB upstream variant MEOX2: 2KB upstream variant |
| rs12669299 | −0.0152 | 0.00356 | 2.03 × 10−5 | None |
| rs2155939 | −0.0189 | 0.00464 | 4.81 × 10−5 | NCALD: intron variant |
| rs10819341 | 0.0264 | 0.00620 | 2.11 × 10−5 | None |
| rs882895 | 0.0251 | 0.00606 | 3.55 × 10−5 | None |
| rs7944388 | −0.0522 | 0.0126 | 3.82 × 10−5 | None |
| rs1178704 | 0.0455 | 0.0111 | 4.31 × 10−5 | PTPRQ: intron variant LOC105369867: intron variant |
| rs12813355 | −0.0165 | 0.00388 | 2.21 × 10−5 | LOC105369867: intron variant |
| rs149583 | 0.0247 | 0.00584 | 2.48 × 10−5 | ATXN8OS: intron variant |
| rs4904895 | 0.0167 | 0.00383 | 1.30 × 10−5 | LRFN5: intron variant |
| rs1227005 | −0.0207 | 0.00449 | 4.08 × 10−6 | PID1: intron variant |
| rs1362243 | −0.0161 | 0.00392 | 4.21 × 10−5 | PID1: intron variant |
| SNP | Log (β) | SE | p-Value | Gene |
|---|---|---|---|---|
| rs10001408 | −0.0321737 | 0.00786715 | 4.48 × 10−5 | TMEM150C: intron variant |
| rs10016201 | −0.0336097 | 0.00796971 | 2.57 × 10−5 | TMEM150C: intron variant |
| rs10208223 | −0.0248463 | 0.00603189 | 3.94 × 10−5 | LOC730100: intron variant |
| rs1051104 | −0.0300326 | 0.00604692 | 7.34 × 10−7 | SLC6A12: intron variant |
| rs10944 | −0.0250323 | 0.00544036 | 4.44 × 10−6 | BHMT2: 3′-UTR variant |
| rs11062011 | −0.0331439 | 0.00561343 | 4.10 × 10−9 | SLC6A12 |
| rs12208357 | −0.0795363 | 0.01839576 | 1.60 × 10−5 | SLC22A1: missense variant LOC124901452: 2KB upstream variant |
| rs12566456 | −0.0891763 | 0.02174221 | 4.25 × 10−5 | KCNH1: intron variant |
| rs12827 | 0.08231739 | 0.02003506 | 4.13 × 10−5 | IQSEC3-AS1: intron variant IQSEC3: 3′-UTR variant |
| rs13088151 | −0.0340056 | 0.00806313 | 2.57 × 10−5 | CNTN4: intron variant |
| rs141112 | 0.03233318 | 0.00772166 | 2.93 × 10−5 | HLA-DOA HLA-DPA1 |
| rs1644004 | −0.0248609 | 0.00551393 | 6.87 × 10−6 | DMGDH: intron variant |
| rs16965026 | −0.0507124 | 0.01204763 | 2.66 × 10−5 | LOC105370772: intron variant |
| rs17692962 | −0.0316486 | 0.00573103 | 3.74 × 10−8 | IQSEC3: intron variant IQSEC3-AS1: intron variant |
| rs183355 | −0.0250172 | 0.00568667 | 1.14 × 10−5 | SLC6A12: 3′-UTR variant IQSEC3-AS1: 2KB upstream variant |
| rs185077 | −0.0238111 | 0.0054365 | 1.24 × 10−5 | DMGDH: intron variant |
| rs1902623 | −0.0534341 | 0.01237439 | 1.64 × 10−5 | RPS1 |
| rs1903530 | −0.0332367 | 0.00770002 | 1.66 × 10−5 | RNA5SP173 |
| rs1986680 | −0.0394049 | 0.00961203 | 4.29 × 10−5 | SYT16 |
| rs2063345 | 0.02946126 | 0.00698375 | 2.56 × 10−5 | SLC22A3: intron variant |
| rs2075228 | −0.0305374 | 0.00629958 | 1.34 × 10−6 | SLC6A12: intron variant |
| rs216244 | 0.02198786 | 0.0052052 | 2.50 × 10−5 | SLC6A12: intron variant |
| rs2174914 | 0.02927708 | 0.00702309 | 3.18 × 10−5 | SLC22A3: intron variant |
| rs2284329 | 0.02429431 | 0.00544388 | 8.50 × 10−6 | SLC6A12: intron variant |
| rs2284330 | 0.02460784 | 0.00530129 | 3.66 × 10−6 | SLC6A12: intron variant |
| rs2291922 | −0.0435999 | 0.00680552 | 1.82 × 10−10 | IQSEC3-AS1: noncoding transcript variant IQSEC3: intron variant |
| rs233162 | −0.0269973 | 0.00663008 | 4.83 × 10−5 | ZNF385D: intron variant |
| rs2457575 | 0.02982937 | 0.00698443 | 2.03 × 10−5 | SLC22A3: intron variant |
| rs2568131 | −0.0590011 | 0.01437736 | 4.21 × 10−5 | NAV2: intron variant LOC124902644: 2KB upstream variant LOC124902644: 2KB upstream variant |
| rs2661834 | 0.02939491 | 0.00699923 | 2.78 × 10−5 | SLC22A3: intron variant |
| rs2702450 | −0.0315948 | 0.00772003 | 4.42 × 10−5 | RNA5SP173 |
| rs2834932 | 0.02879038 | 0.00705378 | 4.63 × 10−5 | LOC100506403: intron variant |
| rs2858458 | 0.03220939 | 0.00772612 | 3.18 × 10−5 | HLA-DOA HLA-DPA1 |
| rs2876711 | −0.0270062 | 0.0057624 | 2.95 × 10−6 | LOC105370265 KCTD12 |
| rs293711 | 0.02357265 | 0.00575914 | 4.41 × 10−5 | BPIFB9P |
| rs3128924 | 0.02731147 | 0.00636548 | 1.86 × 10−5 | HLA-DPA2 COL11A2P1 |
| rs3135021 | 0.03408974 | 0.00817301 | 3.15 × 10−5 | HLA-DPA1: intron variant HLA-DPB1: intron variant |
| rs377551 | 0.02871405 | 0.00694405 | 3.68 × 10−5 | SLC22A3: intron variant |
| rs379569 | 0.03232336 | 0.00772832 | 3.00 × 10−5 | HLA-DOA HLA-DPA1 |
| rs380468 | 0.03220939 | 0.00772612 | 3.18 × 10−5 | HLA-DOA HLA-DPA1 |
| rs382198 | 0.03233488 | 0.00772772 | 2.97 × 10−5 | HLA-DOA HLA-DPA1 |
| rs394487 | 0.02899207 | 0.00694306 | 3.09 × 10−5 | SLC22A3: intron variant |
| rs412065 | 0.03222353 | 0.00772846 | 3.17 × 10−5 | HLA-DOA HLA-DPA1 |
| rs426523 | 0.03220939 | 0.00772612 | 3.18 × 10−5 | HLA-DOA HLA-DPA1 |
| rs439852 | 0.03220939 | 0.00772612 | 3.18 × 10−5 | HLA-DOA HLA-DPA1 |
| rs444821 | 0.03220939 | 0.00772612 | 3.18 × 10−5 | HLA-DOA HLA-DPA1 |
| rs495360 | −0.0277466 | 0.00527505 | 1.58 × 10−7 | SLC6A12 |
| rs4974157 | 0.02405487 | 0.00537685 | 8.08 × 10−6 | ERC2: intron variant |
| rs505387 | −0.0300213 | 0.00613914 | 1.08 × 10−6 | SLC6A12: 3′-UTR variant |
| rs510714 | −0.0257809 | 0.00595824 | 1.58 × 10−5 | SLC6A12: intron variant |
| rs542736 | −0.0291588 | 0.0058391 | 6.39 × 10−7 | SLC6A12: intron variant |
| rs557881 | −0.0281208 | 0.00517767 | 6.22 × 10−8 | SLC6A12: missense variant |
| rs562487 | −0.0247541 | 0.00535511 | 4.01 × 10−6 | BHMT: 2KB upstream variant |
| rs569919 | 0.02760991 | 0.0067914 | 4.96 × 10−5 | SLC22A3 |
| rs570680 | −0.0338119 | 0.00559611 | 1.79 × 10−9 | SLC6A13: intron variant |
| rs579292 | −0.0226518 | 0.00530637 | 2.05 × 10−5 | LOC105371956: intron variant |
| rs586199 | −0.0264621 | 0.00548411 | 1.49 × 10−6 | BHMT |
| rs614564 | 0.02621931 | 0.00624998 | 2.84 × 10−5 | LOC105378088: 2KB upstream variant |
| rs631151 | −0.0253154 | 0.00552072 | 4.78 × 10−6 | DMGDH: intron variant |
| rs633159 | −0.0289333 | 0.00590165 | 1.02 × 10−6 | SLC6A12: 3′-UTR variant |
| rs6457709 | 0.03100779 | 0.00739251 | 2.84 × 10−5 | HLA-DPA1 |
| rs663310 | 0.03222585 | 0.0077285 | 3.17 × 10−5 | HLA-DOA HLA-DPA1 |
| rs7015522 | 0.04962682 | 0.01045299 | 2.19 × 10−6 | CARSIP2 |
| rs7171449 | −0.0502894 | 0.01103103 | 5.43 × 10−6 | LOC105370772: intron variant |
| rs7599255 | −0.0301309 | 0.0072983 | 3.79 × 10−5 | ALK: intron variant |
| rs7613818 | 0.02124971 | 0.00522684 | 4.96 × 10−5 | ERC2: intron variant |
| rs7782195 | 0.02406508 | 0.0056471 | 2.12 × 10−5 | MAGI2: intron variant |
| rs7960096 | −0.025664 | 0.00575139 | 8.52 × 10−6 | IQSEC3-AS1: intron variant IQSEC3: 3′-UTR variant |
| rs7971499 | −0.0223223 | 0.00513152 | 1.42 × 10−5 | SLC6A12: 3′-UTR variant |
| rs8093227 | −0.031431 | 0.00719859 | 1.32 × 10−5 | LOC105371956: intron variant |
| rs9293761 | 0.02368349 | 0.00547998 | 1.62 × 10−5 | ARSB |
| rs9873480 | 0.02583322 | 0.00609977 | 2.38 × 10−5 | ERC2: intron variant |
| SNP | Log (β) | SE | p-Value | Gene |
|---|---|---|---|---|
| rs10043368 | −0.0163339 | 0.004018 | 4.97 × 10−5 | ZNF366: intron variant |
| rs1022954 | 0.02230921 | 0.00541474 | 3.93 × 10−5 | MYO16: intron variant |
| rs10514154 | −0.0362616 | 0.00824616 | 1.15 × 10−5 | DMGDH: intron variant |
| rs1055749 | 0.02127731 | 0.0048998 | 1.47 × 10−5 | MCPH1-AS1: intron variant MCPH1: 3′-UTR variant |
| rs1057091 | 0.02250277 | 0.00490949 | 4.83 × 10−6 | MCPH1: missense variant MCPH1-AS1: intron variant |
| rs11252395 | −0.0238639 | 0.00517109 | 4.16 × 10−6 | LOC105376368 |
| rs11996743 | −0.0157811 | 0.00385836 | 4.47 × 10−5 | ZFPM2-AS1: intron variant |
| rs12211424 | −0.0381197 | 0.00851944 | 8.05 × 10−6 | ZNF184: intron variant |
| rs1385961 | −0.0368556 | 0.00818561 | 7.07 × 10−6 | RNU6-214P |
| rs1385962 | −0.0360201 | 0.00813816 | 1.01 × 10−5 | RNU6-214P |
| rs1422096 | −0.026488 | 0.00628629 | 2.61 × 10−5 | LOC105379107: intron variant |
| rs16876302 | −0.0280152 | 0.00587175 | 1.95 × 10−6 | DMGDH: intron variant |
| rs17054397 | 0.04219231 | 0.01032575 | 4.54 × 10−5 | ITK: 3′-UTR variant |
| rs17082203 | −0.0343162 | 0.00814404 | 2.61 × 10−5 | RNU6-214P |
| rs17082229 | −0.0344952 | 0.00814271 | 2.37 × 10−5 | RNU6-214P |
| rs17155846 | −0.0274697 | 0.00628538 | 1.30 × 10−5 | LOC105379107: intron variant |
| rs17551120 | 0.03625213 | 0.00799345 | 6.06 × 10−6 | STXBP5-AS1: intron variant |
| rs1805072 | −0.0373494 | 0.00824437 | 6.21 × 10−6 | DMGDH: synonymous variant |
| rs1805074 | −0.0278176 | 0.0058738 | 2.32 × 10−6 | DMGDH: missense variant |
| rs1872624 | −0.0158555 | 0.00383831 | 3.75 × 10−5 | B4GALT7: intron variant |
| rs1998084 | 0.01772614 | 0.00411422 | 1.72 × 10−5 | LOC100505664: intron variant |
| rs2034900 | −0.0281137 | 0.00587331 | 1.81 × 10−6 | DMGDH: intron variant |
| rs2431332 | 0.03143612 | 0.00567521 | 3.40 × 10−8 | DMGDH: intron variant |
| rs2433149 | 0.02181202 | 0.00490801 | 9.27 × 10−6 | MCPH1-AS1: intron variant MCPH1: 3′-UTR variant |
| rs250513 | 0.02676484 | 0.00618679 | 1.59 × 10−5 | DMGDH: intron variant |
| rs28326 | 0.03198695 | 0.00733647 | 1.36 × 10−5 | DMGDH: intron variant |
| rs3758562 | 0.02019445 | 0.00488116 | 3.65 × 10−5 | PRF1: intron variant |
| rs3792849 | −0.0178398 | 0.00427434 | 3.11 × 10−5 | B4GALT7: intron variant |
| rs3892245 | 0.04203865 | 0.01032946 | 4.87 × 10−5 | ITK: 3′-UTR variant |
| rs4365962 | −0.0370252 | 0.00818995 | 6.49 × 10−6 | RNU6-214P |
| rs4421087 | 0.02065935 | 0.00451312 | 4.97 × 10−6 | SNP not documented in dbGaP |
| rs4458584 | −0.0161164 | 0.00384608 | 2.90 × 10−5 | B4GALT7 |
| rs4522 | −0.0175608 | 0.00395074 | 9.23 × 10−6 | HSBP1: 3′-UTR variant |
| rs4685654 | −0.0295262 | 0.00651226 | 6.10 × 10−6 | LRRN1 |
| rs4750458 | 0.03575054 | 0.00826126 | 1.58 × 10−5 | FRMD4A: intron variant |
| rs4891832 | −0.0223609 | 0.00518445 | 1.68 × 10−5 | LOC105372180 |
| rs506500 | 0.02136081 | 0.00460962 | 3.80 × 10−6 | BHMT: intron variant LOC124901012: intron variant |
| rs585800 | 0.02595743 | 0.00498842 | 2.14 × 10−7 | BHMT: 3′-UTR variant |
| rs6044680 | −0.0179242 | 0.0041538 | 1.67 × 10−5 | RNU1-131P |
| rs631305 | 0.04246729 | 0.00716996 | 3.66 × 10−9 | BHMT |
| rs642431 | 0.039844 | 0.00640411 | 5.88 × 10−10 | DMGDH: intron variant BHMT2: 2KB upstream variant |
| rs642934 | 0.039844 | 0.00640411 | 5.88 × 10−10 | DMGDH: intron variant BHMT2: 2KB upstream variant |
| rs6774615 | −0.0302061 | 0.0069094 | 1.29 × 10−5 | SLC6A6: intron variant |
| rs6796254 | 0.02049116 | 0.0050114 | 4.49 × 10−5 | LOC105376959 |
| rs694290 | 0.02072496 | 0.00461362 | 7.42 × 10−6 | BHMT: intron variant |
| rs7217560 | 0.02483543 | 0.00528401 | 2.76 × 10−6 | COPS3 |
| rs7220577 | 0.02080798 | 0.0049688 | 2.93 × 10−5 | COPS3 |
| rs731124 | −0.0275095 | 0.005856 | 2.79 × 10−6 | DMGDH: noncoding transcript variant |
| rs7934785 | −0.0188417 | 0.00412737 | 5.27 × 10−6 | CNTN5: intron variant |
| rs8050249 | −0.0162909 | 0.00400241 | 4.86 × 10−5 | HS3ST4: intron variant |
| rs8134775 | 0.0272707 | 0.00600288 | 5.85 × 10−6 | LINC00310 |
| rs9301307 | 0.01529968 | 0.00372773 | 4.20 × 10−5 | MYO16: intron variant |
| rs933684 | −0.0279284 | 0.00587222 | 2.10 × 10−6 | DMGDH: intron variant |
| rs9489944 | −0.0301432 | 0.00737781 | 4.55 × 10−5 | RNU6-214P |
| Gene | SNP | Log (β) | SE | p-Value |
|---|---|---|---|---|
| MTHFR | rs1801133 | −0.00587 | 0.00397 | 0.139 |
| rs1801131 | 0.00172 | 0.00415 | 0.679 | |
| MTHFD1 | rs2236225 | NA | NA | NA |
| RFC | rs1051266 | −0.00683 | 0.00344 | 0.0469 |
| MTR | rs1805087 | 0.00192 | 0.00560 | 0.731 |
| MTRR | rs1801394 | −0.00250 | 0.00338 | 0.460 |
| CHKA | rs10791957 | 0.00365 | 0.00340 | 0.284 |
| CHDH | rs9001 | NA | NA | NA |
| rs12676 | NA | NA | NA | |
| BHMT | rs3733890 | 0.00514 | 0.00469 | 0.274 |
| PEMT | rs12325817 | NA | NA | NA |
| rs4646343 | NA | NA | NA | |
| rs2266782 | −0.00102 | 0.00351 | 0.772 | |
| SLC44A1 | rs7873937 | NA | NA | NA |
| rs3199966 | 0.00182 | 0.00957 | 0.849 |
| SNP | β | SE | p-Value | Gene |
|---|---|---|---|---|
| rs6681578 | 0.00991093 | 0.00236933 | 2.99 × 10−5 | CACHD1: intron variant |
| rs2164720 | 0.01467305 | 0.00330247 | 9.31 × 10−6 | LDAH: intron variant |
| rs13026309 | 0.00674695 | 0.00146867 | 4.60 × 10−6 | THADA: intron variant |
| rs3136247 | 0.01103863 | 0.00244463 | 6.65 × 10−6 | MSH6: intron variant |
| rs1800932 | 0.01074649 | 0.00243654 | 1.08 × 10−5 | MSH6: synonymous variant |
| rs7711253 | 0.0165464 | 0.00354381 | 3.21 × 10−6 | BDP1: intron variant |
| rs7707956 | 0.01125355 | 0.00271287 | 3.48 × 10−5 | No documented gene consequence |
| rs7706073 | 0.01920656 | 0.00467906 | 4.20 × 10−5 | LINC01950: intron variant |
| rs2288068 | 0.01567417 | 0.00376542 | 3.27 × 10−5 | CYFIP2: intron variant |
| rs17307478 | 0.01523564 | 0.0036602 | 3.27 × 10−5 | KIAA0319: intron variant |
| rs13215160 | 0.01626786 | 0.00388485 | 2.93 × 10−5 | GABRR1: intron variant |
| rs12208357 | 0.02262707 | 0.00509501 | 9.40 × 10−6 | SLC22A1: missense variant LOC124901452: 2KB upstream variant |
| rs756513 | 0.0067078 | 0.0015018 | 8.35 × 10−6 | No documented gene consequence |
| rs17364251 | 0.01394374 | 0.00331615 | 2.72 × 10−5 | No documented gene consequence |
| rs10818754 | 0.01277969 | 0.00295672 | 1.61 × 10−5 | RC3H2: 3′-UTR variant |
| rs11222960 | 0.0269977 | 0.00623105 | 1.54 × 10−5 | NTM: intron variant |
| rs2291922 | 0.01111452 | 0.00188971 | 4.69 × 10−9 | IQSEC3-AS1: noncoding transcript variant IQSEC3: intron variant |
| rs17692962 | 0.00774825 | 0.00159149 | 1.20 × 10−6 | IQSEC3: intron variant IQSEC3-AS1: intron variant |
| rs7960096 | 0.00691708 | 0.0015951 | 1.51 × 10−5 | IQSEC3-AS1: intron variant IQSEC3: 3′-UTR variant |
| rs183355 | 0.00698278 | 0.00157733 | 1.00 × 10−5 | SLC6A12: 3′-UTR variant IQSEC3-AS1: 2KB upstream variant |
| rs633159 | 0.00810786 | 0.00163617 | 7.77 × 10−7 | SLC6A12: 3′-UTR variant |
| rs505387 | 0.00795451 | 0.00170302 | 3.18 × 10−6 | SLC6A12: 3′-UTR variant |
| rs559759 | 0.00648629 | 0.00156154 | 3.40 × 10−5 | SLC6A12: 3′-UTR variant |
| rs7971499 | 0.00660971 | 0.0014223 | 3.56 × 10−6 | SLC6A12: 3′-UTR variant |
| rs1051104 | 0.00772504 | 0.00167796 | 4.39 × 10−6 | SLC6A12: intron variant |
| rs542736 | 0.00768298 | 0.001621 | 2.28 × 10−6 | SLC6A12: intron variant |
| rs2075228 | 0.00782415 | 0.0017485 | 8.04 × 10−6 | SLC6A12: intron variant |
| rs2284329 | −0.0065039 | 0.0015107 | 1.74 × 10−5 | SLC6A12: intron variant |
| rs2284330 | −0.0065495 | 0.0014704 | 8.84 × 10−6 | SLC6A12: intron variant |
| rs557881 | 0.00668919 | 0.00143866 | 3.52 × 10−6 | SLC6A12: missense variant |
| rs495360 | 0.0071813 | 0.00146364 | 9.96 × 10−7 | SLC6A12 |
| rs11062011 | 0.00837156 | 0.00156026 | 8.93 × 10−8 | SLC6A12 |
| rs570680 | 0.00858784 | 0.00155116 | 3.46 × 10−8 | SLC6A13: intron variant |
| rs2220168 | 0.00789697 | 0.00192738 | 4.33 × 10−5 | ITPR2: intron variant |
| rs11174335 | 0.00889376 | 0.00189291 | 2.78 × 10−6 | TAFA2: intron variant |
| rs11174342 | 0.00902436 | 0.00195687 | 4.23 × 10−6 | TAFA2: intron variant LOC124902950: intron variant |
| rs12578778 | 0.01992362 | 0.0047782 | 3.17 × 10−5 | ANO4: intron variant |
| rs16934132 | 0.00691779 | 0.0015842 | 1.32 × 10−5 | ACACB: intron variant |
| rs7976552 | 0.00744949 | 0.00163517 | 5.51 × 10−6 | ACACB: intron variant |
| rs16947978 | 0.02028651 | 0.00453305 | 8.02 × 10−6 | KSR2: intron variant |
| rs1681688 | 0.00584176 | 0.00142875 | 4.49 × 10−5 | TMEM132D: intron variant |
| rs17078172 | 0.01505929 | 0.0035616 | 2.45 × 10−5 | HMGA1P6 |
| rs7335200 | 0.01471746 | 0.00353061 | 3.18 × 10−5 | HMGA1P6 |
| rs1984163 | 0.00950216 | 0.00221904 | 1.93 × 10−5 | HMGA1P6 |
| rs2876711 | 0.00724716 | 0.00159826 | 6.09 × 10−6 | LOC107984587 |
| rs10135123 | 0.01175498 | 0.00289155 | 4.97 × 10−5 | URK1 |
| rs7171449 | 0.01252207 | 0.00306117 | 4.46 × 10−5 | LOC105370772: intron variant |
| rs1902623 | 0.01417481 | 0.00343231 | 3.77 × 10−5 | RPS15P8 |
| rs2929644 | 0.02090622 | 0.00503746 | 3.45 × 10−5 | LOC102724253 |
| rs2929647 | 0.02416542 | 0.00541238 | 8.42 × 10−6 | LOC102724253 |
| rs11540961 | −0.0214329 | 0.0049166 | 1.37 × 10−5 | NUBP2: missense variant |
| rs4780333 | 0.01057356 | 0.00247552 | 2.03 × 10−5 | CIITA: intron variant |
| rs16973822 | 0.01613107 | 0.00380786 | 2.37 × 10−5 | RBBP6: intron variant |
| rs4243134 | 0.01381404 | 0.00335306 | 3.93 × 10−5 | MON1B SYCE1L |
| rs4569298 | 0.01372323 | 0.00335339 | 4.42 × 10−5 | MON1B SYCE1L |
| rs8093227 | 0.00961646 | 0.00199542 | 1.54 × 10−6 | LOC105371956: intron variant |
| rs1893459 | −0.007108 | 0.00166218 | 1.98 × 10−5 | NETO1: intron variant |
| rs12959392 | 0.00626331 | 0.00149733 | 2.99 × 10−5 | NETO1: intron variant |
| rs12150965 | 0.01503655 | 0.00338257 | 9.21 × 10−6 | CACNG8: 2KB upstream variant |
| rs13013933 | −0.022755 | 0.00549693 | 3.61 × 10−5 | GPC1: 3′-UTR variant |
| rs13088151 | 0.00945497 | 0.00223571 | 2.44 × 10−5 | CNTN4: intron variant |
| rs9883103 | 0.00739201 | 0.00179233 | 3.86 × 10−5 | VEPH1: intron variant LOC101928236: intron variant |
| rs6840205 | 0.00987881 | 0.00242079 | 4.65 × 10−5 | ATP10D: intron variant |
| rs16851681 | 0.00997056 | 0.00241252 | 3.72 × 10−5 | ATP10D: missense variant |
| rs11935423 | 0.0127963 | 0.00314413 | 4.87 × 10−5 | TRMT10A: intron variant |
| SNP | Log (β) | SE | p-Value | Gene |
|---|---|---|---|---|
| rs6425992 | −0.2338471 | 0.05303353 | 1.09 × 10−5 | FTLP18 |
| rs1277207 | 0.34910275 | 0.08397241 | 3.34 × 10−5 | AKNAD1: missense variant |
| rs7519217 | 0.32552111 | 0.07711205 | 2.53 × 10−5 | LOC343508 |
| rs7603744 | 0.67931996 | 0.1647434 | 3.87 × 10−5 | LINC01250: intron variant |
| rs722889 | 0.81880778 | 0.17393026 | 2.66 × 10−6 | RNA5SP94 |
| rs1841991 | 0.29815476 | 0.05141108 | 7.62 × 10−9 | ARSB: intron variant |
| rs1717570 | 0.29604386 | 0.05125945 | 8.77 × 10−9 | ARSB: intron variant |
| rs9293761 | 0.29800874 | 0.05053237 | 4.27 × 10−9 | ARSB: intron variant |
| rs185077 | −0.2600591 | 0.04999753 | 2.16 × 10−7 | DMGDH: intron variant |
| rs248383 | −0.2586772 | 0.0510865 | 4.46 × 10−7 | DMGDH: intron variant |
| rs248385 | −0.2648006 | 0.05126639 | 2.62 × 10−7 | DMGDH: synonymous variant |
| rs631151 | −0.2904679 | 0.05092897 | 1.33 × 10−8 | DMGDH: intron variant |
| rs1644004 | −0.2920737 | 0.05088538 | 1.08 × 10−8 | DMGDH: intron variant |
| rs642431 | −0.392191 | 0.08315844 | 2.55 × 10−6 | DMGDH: intron variant; BHMT2: 2KB upstream variant |
| rs642934 | −0.392191 | 0.08315844 | 2.55 × 10−6 | DMGDH: intron variant; BHMT2: 2KB upstream variant |
| rs10944 | −0.2970196 | 0.05019258 | 3.78 × 10−9 | BHMT2: 3′-UTR variant |
| rs586199 | −0.30439 | 0.05060727 | 2.11 × 10−9 | BHMY |
| rs631305 | −0.4153324 | 0.09307635 | 8.52 × 10−6 | BHMY |
| rs562487 | −0.2929906 | 0.04940669 | 3.51 × 10−9 | BHMT: 2KB upstream variant |
| rs506500 | −0.3581445 | 0.0594361 | 1.97 × 10−9 | BHMT: intron variant LOC124901012: intron variant |
| rs694290 | −0.3510983 | 0.05949219 | 4.16 × 10−9 | BHMT: intron variant |
| rs4421087 | −0.3334988 | 0.05816667 | 1.12 × 10−8 | SNP/gene not on file |
| rs585800 | −0.333203 | 0.06462924 | 2.76 × 10−7 | BHMT: 3′-UTR variant |
| rs1915706 | 0.23732281 | 0.0531033 | 8.26 × 10−6 | BHMT JMY |
| rs2364594 | 0.25671564 | 0.05616074 | 5.12 × 10−6 | BHMT |
| rs13158309 | 0.24662904 | 0.05341217 | 4.11 × 10−6 | JMY BHMT |
| rs2121107 | 0.25735015 | 0.05615016 | 4.84 × 10−6 | JMY BHMT |
| rs12189248 | 0.24216394 | 0.05412745 | 8.07 × 10−6 | LOC124901011: intron variant LOC124900205: 500B downstream variant |
| rs10078815 | 0.24999207 | 0.0554529 | 6.88 × 10−6 | LOC124901011: intron variant |
| rs2591392 | 0.22464765 | 0.05429781 | 3.65 × 10−5 | JMY: intron variant |
| rs2591387 | 0.23057766 | 0.05567505 | 3.58 × 10−5 | JMY: intron variant |
| rs13182512 | 0.22635436 | 0.0534241 | 2.36 × 10−5 | JMY: missense variant |
| rs7711939 | 0.23409743 | 0.05570361 | 2.75 × 10−5 | JMY: intron variant |
| rs10074882 | 0.23412197 | 0.05571608 | 2.75 × 10−5 | JMY: intron variant |
| rs1062326 | 0.22808771 | 0.05438296 | 2.85 × 10−5 | JMY: 3′-UTR variant LOC102724530: intron variant |
| rs2750473 | 0.33949293 | 0.0771035 | 1.12 × 10−5 | LOC107986623: intron variant |
| rs2297374 | 0.26371872 | 0.05811383 | 5.99 × 10−6 | SLC22A1: intron variant |
| rs11982432 | 0.37733778 | 0.0896363 | 2.66 × 10−5 | No documented gene consequence |
| rs763317 | −0.1991543 | 0.04890966 | 4.83 × 10−5 | EGFR: intron variant |
| rs12542609 | 0.63969567 | 0.1528459 | 2.96 × 10−5 | LOC100419761 |
| rs6986672 | 0.48012465 | 0.10450138 | 4.59 × 10−6 | KIF13B: missense variant |
| rs6981956 | 0.48095613 | 0.10448875 | 4.40 × 10−6 | KIF13B: intron variant |
| rs2461063 | −0.2420174 | 0.05667695 | 2.04 × 10−5 | LOC107986893 |
| rs1550734 | −0.2422015 | 0.05408795 | 7.92 × 10−6 | LOC107986893 |
| rs7040466 | −0.685511 | 0.16709438 | 4.24 × 10−5 | JKAMPP1 |
| rs7873941 | 0.47965638 | 0.11324424 | 2.37 × 10−5 | JKAMPP1 |
| rs16937205 | 0.7200178 | 0.17095181 | 2.64 × 10−5 | NAMPTP1 |
| rs586910 | 0.6629388 | 0.12728722 | 2.08 × 10−7 | PAMR1 FJX1 |
| rs659401 | 0.45442497 | 0.10393125 | 1.29 × 10−5 | PAMR1 FJX1 |
| rs10768154 | 0.45909896 | 0.10395536 | 1.05 × 10−5 | PAMR1 FJX1 |
| rs2291922 | −0.3251766 | 0.06319042 | 2.90 × 10−7 | IQSEC3-AS1: noncoding transcript variant IQSEC3: intron variant |
| rs17692962 | −0.2472614 | 0.05314565 | 3.47 × 10−6 | IQSEC3: intron variant IQSEC3-AS1: intron variant |
| rs633159 | −0.2239166 | 0.05470879 | 4.41 × 10−5 | SLC6A12: 3′-UTR variant |
| rs1051104 | −0.2449588 | 0.05603346 | 1.29 × 10−5 | SLC6A12: intron variant |
| rs542736 | −0.2330685 | 0.05413321 | 1.74 × 10−5 | SLC6A12: intron variant |
| rs2075228 | −0.2602225 | 0.05840137 | 8.78 × 10−6 | SLC6A12: intron variant |
| rs216244 | 0.20164584 | 0.04818403 | 2.97 × 10−5 | SLC6A12: intron variant |
| rs557881 | −0.2740004 | 0.04785595 | 1.17 × 10−8 | SLC6A12: missense variant |
| rs495360 | −0.2736733 | 0.04877753 | 2.27 × 10−8 | SLC6A12 |
| rs11062011 | −0.3062373 | 0.05191357 | 4.23 × 10−9 | SLC6A13 |
| rs570680 | −0.2973386 | 0.05191132 | 1.16 × 10−8 | SLC6A13: intron variant |
| rs901510 | −0.1992071 | 0.04841748 | 4.03 × 10−5 | LOC105370224 |
| rs7198724 | 0.58003513 | 0.13640424 | 2.20 × 10−5 | ZC3H18: intron variant |
| rs34790908 | 0.29965941 | 0.07283552 | 4.03 × 10−5 | TNFSF12: 2KB upstream variant TNFSF12-TNFSF13: 2KB upstream variant |
| rs4511593 | 0.24831263 | 0.05896752 | 2.65 × 10−5 | TNFSF12: intron variant TNFSF12-TNFSF13: intron variant |
| rs4227 | 0.28288183 | 0.06812976 | 3.42 × 10−5 | MPDU1: Noncoding transcript variant SOX15: 500B downstream variant |
| rs8073498 | 0.23498641 | 0.05318078 | 1.04 × 10−5 | ATP1B2 TP53 |
| rs7505206 | 0.20790143 | 0.05021527 | 3.60 × 10−5 | LOC107985171 |
| rs12479693 | −0.2026292 | 0.04944385 | 4.32 × 10−5 | PTPRT: intron variant LOC105372623: intron variant |
| rs7268202 | 0.50667127 | 0.11927352 | 2.25 × 10−5 | BCAS4 |
| rs870908 | 0.66192585 | 0.14697084 | 7.03 × 10−6 | RNA5SP117 |
| rs10048970 | 0.76015386 | 0.16784891 | 6.25 × 10−6 | TAMM41: intron variant |
| rs1529042 | 0.20293812 | 0.04929111 | 3.98 × 10−5 | GPR15 |
| rs901884 | −0.4497657 | 0.10136571 | 9.57 × 10−6 | NLGN1: intron variant |
| rs11940805 | 0.20149032 | 0.04789739 | 2.70 × 10−5 | LOC107986306: intron variant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Louck, L.E.; Klatt, K.C.; Wallace, T.C.; Ma, J.; Chung, M. A Genome-Wide Association Study of Circulating Serum Choline, Betaine, Dimethylglycine, and Their Ratios. Nutrients 2025, 17, 2630. https://doi.org/10.3390/nu17162630
Louck LE, Klatt KC, Wallace TC, Ma J, Chung M. A Genome-Wide Association Study of Circulating Serum Choline, Betaine, Dimethylglycine, and Their Ratios. Nutrients. 2025; 17(16):2630. https://doi.org/10.3390/nu17162630
Chicago/Turabian StyleLouck, Lauren E., Kevin C. Klatt, Taylor C. Wallace, Jiantao Ma, and Mei Chung. 2025. "A Genome-Wide Association Study of Circulating Serum Choline, Betaine, Dimethylglycine, and Their Ratios" Nutrients 17, no. 16: 2630. https://doi.org/10.3390/nu17162630
APA StyleLouck, L. E., Klatt, K. C., Wallace, T. C., Ma, J., & Chung, M. (2025). A Genome-Wide Association Study of Circulating Serum Choline, Betaine, Dimethylglycine, and Their Ratios. Nutrients, 17(16), 2630. https://doi.org/10.3390/nu17162630









