Influence of Dietary Polyunsaturated Fatty Acid Intake on Potential Lipid Metabolite Diagnostic Markers in Renal Cell Carcinoma: A Case-Control Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Plasma Sample Preparation
2.3. UPLC/Orbitrap MS and Metabolite Quantification Analysis
2.4. Metabolomic Data Processing
2.5. Identification of RCC Diagnostic Candidate Markers through Machine Learning
2.6. Receiver Operating Characteristic (ROC) Curve, Enrichment, and Pathway Analysis
2.7. Carnitine Palmitoyltransferase (CPT) Family Genes Expression Analysis and CPT1 Measurement
2.8. Dietary Fat and Nutrient Intake Analysis and Correlations with Metabolites
2.9. Statistical Analysis
3. Results
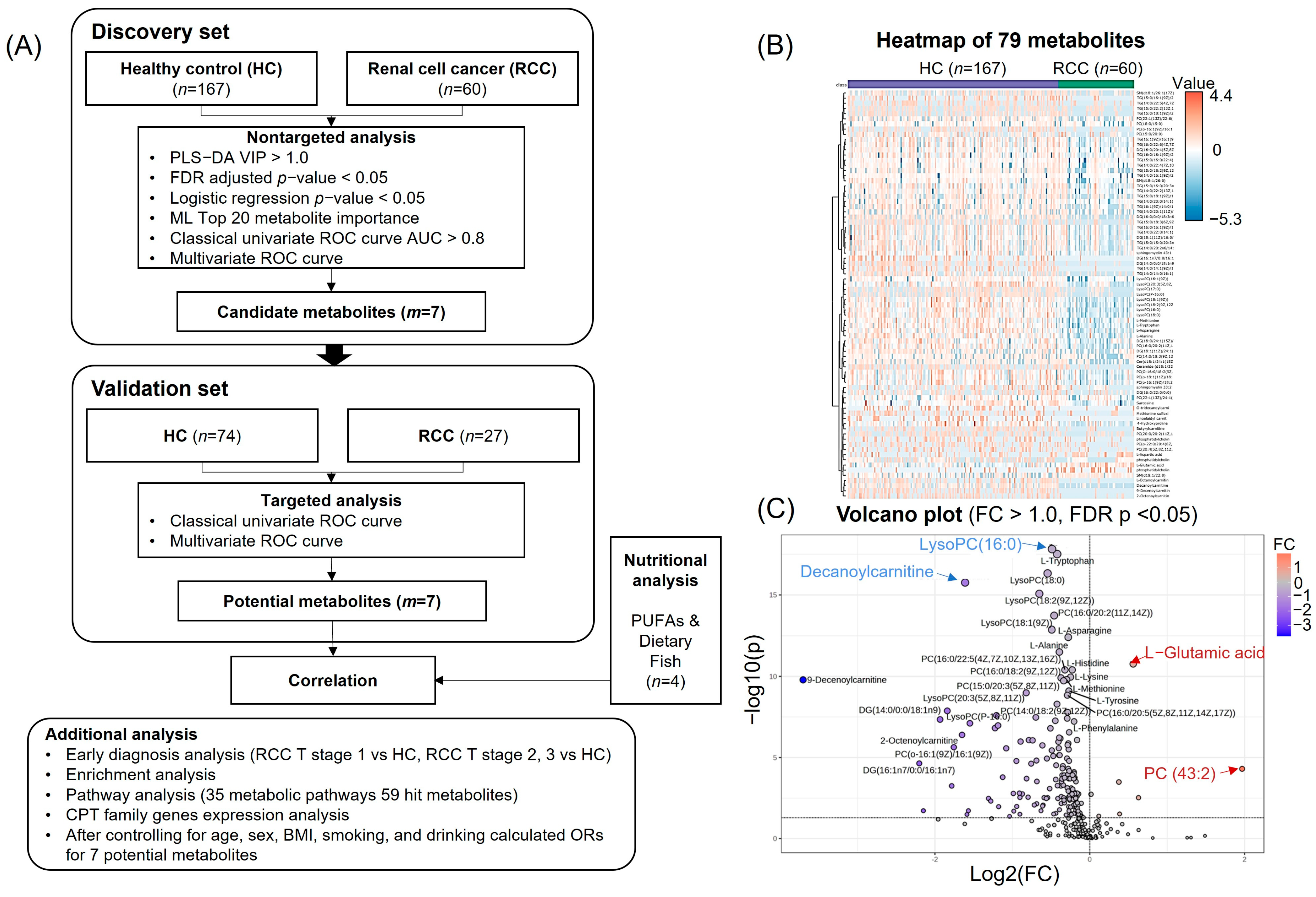
3.1. Clinical and Diagnostic Characteristics of HC and RCC Groups
3.2. Metabolomic Profiling for Identification of Candidate Metabolites
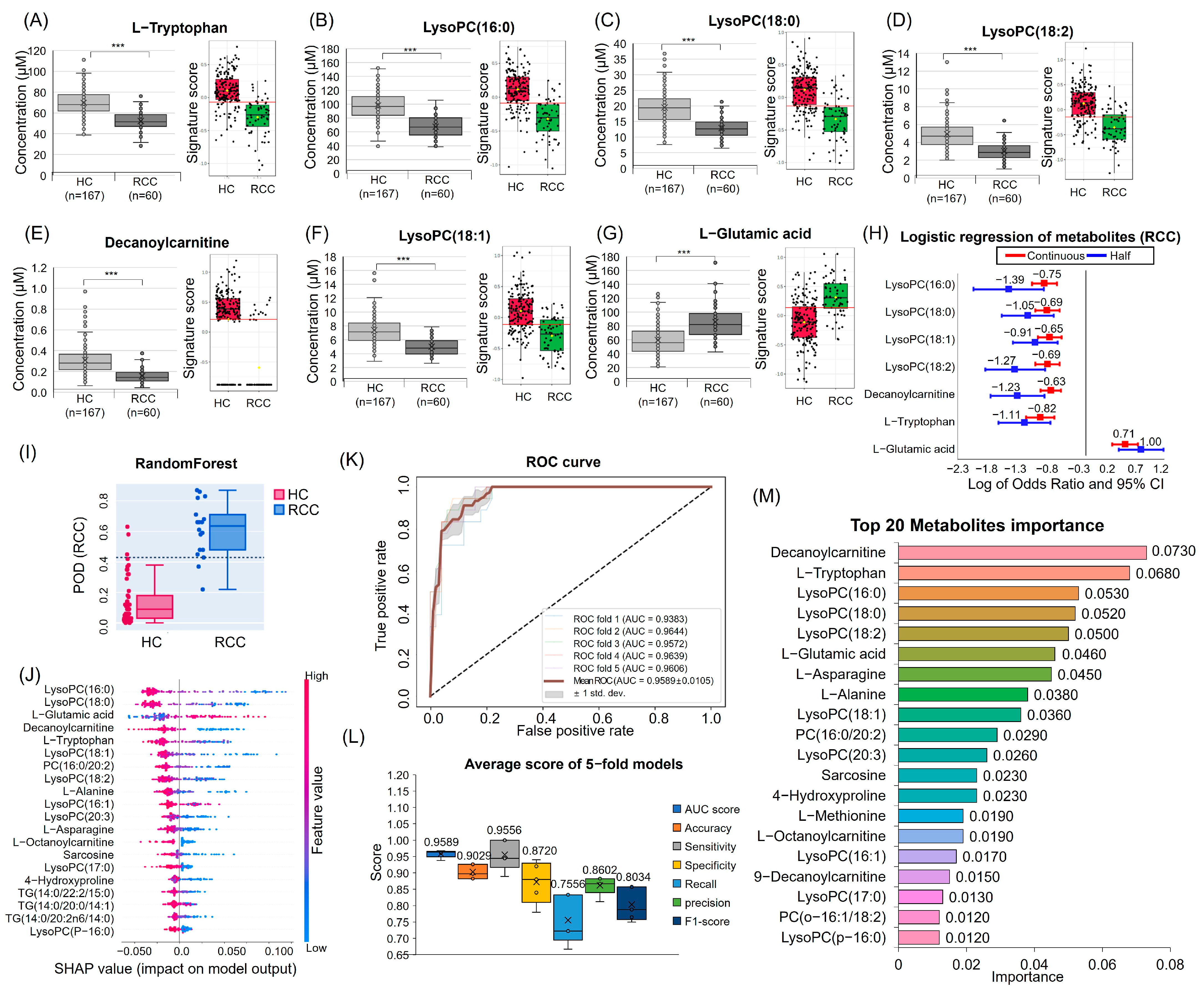
3.3. Quantitative Analysis and Multivariate Logistic Regression Analysis Results for Candidate Metabolites
3.4. ML for RCC Diagnostic Candidate Metabolites Prediction
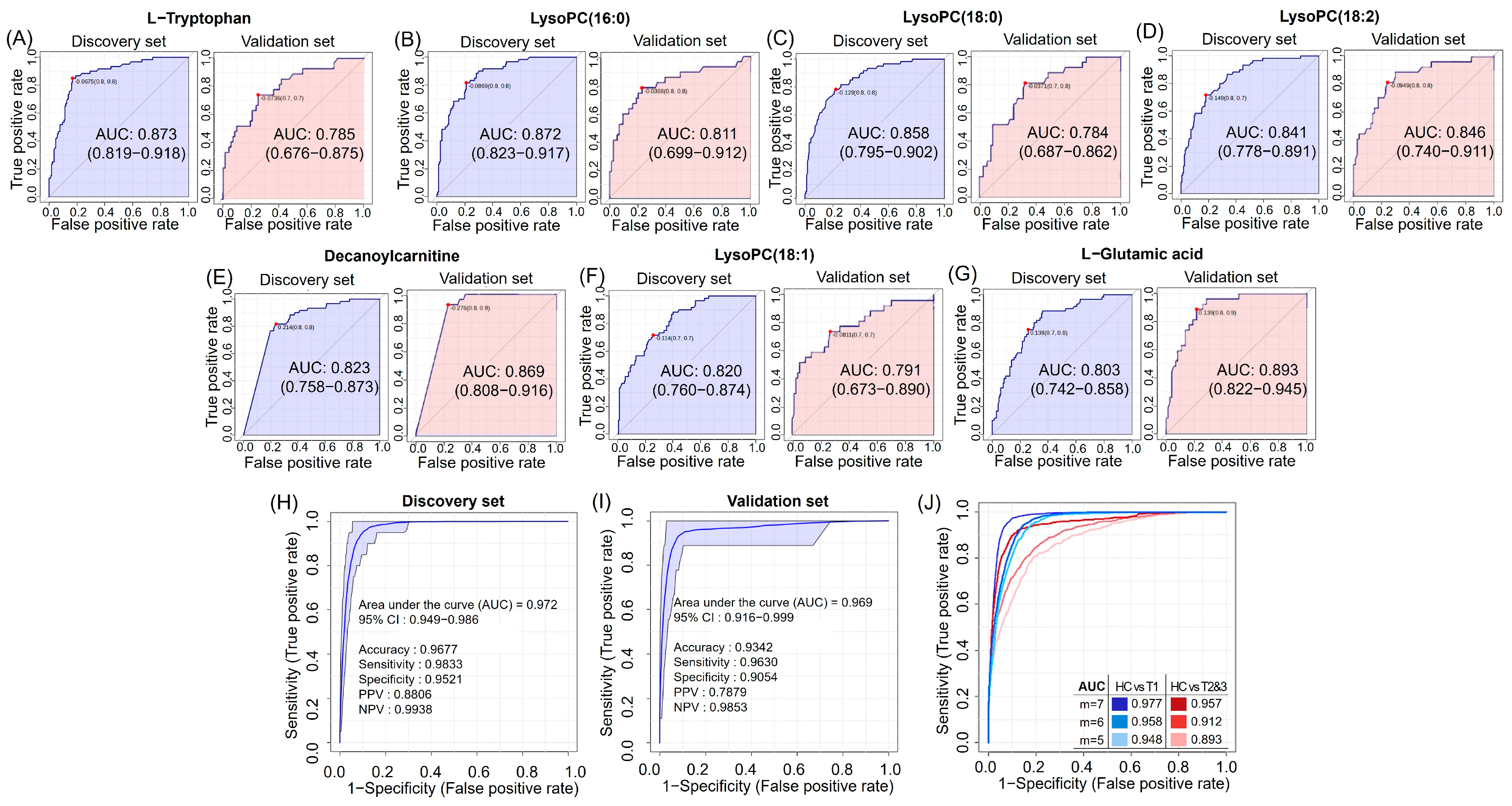
3.5. Discovery and Validation of Seven Potential RCC Diagnostic Markers
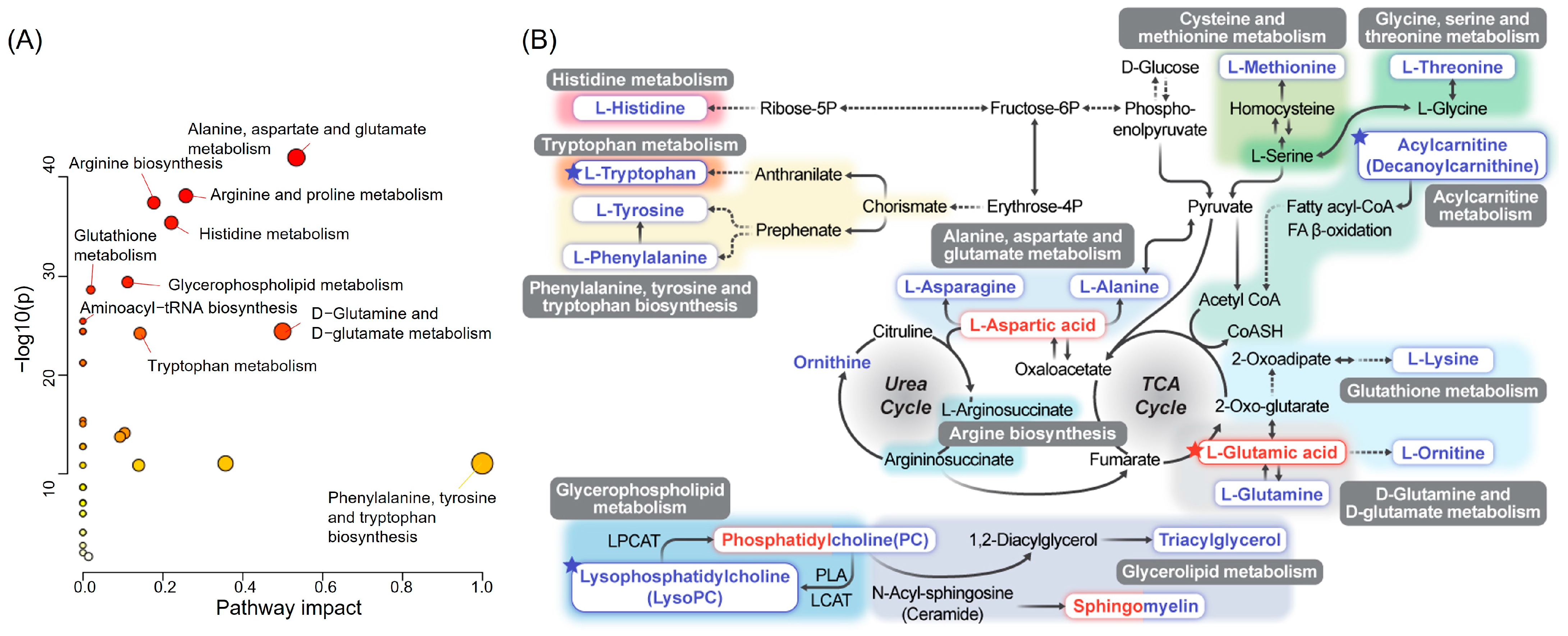
3.6. Enrichment and Pathway Analyses of HCs and RCC Differential Metabolites
3.7. Decanoylcarnitine Decrease in RCC Is Regulated by CPT1a Downregulation
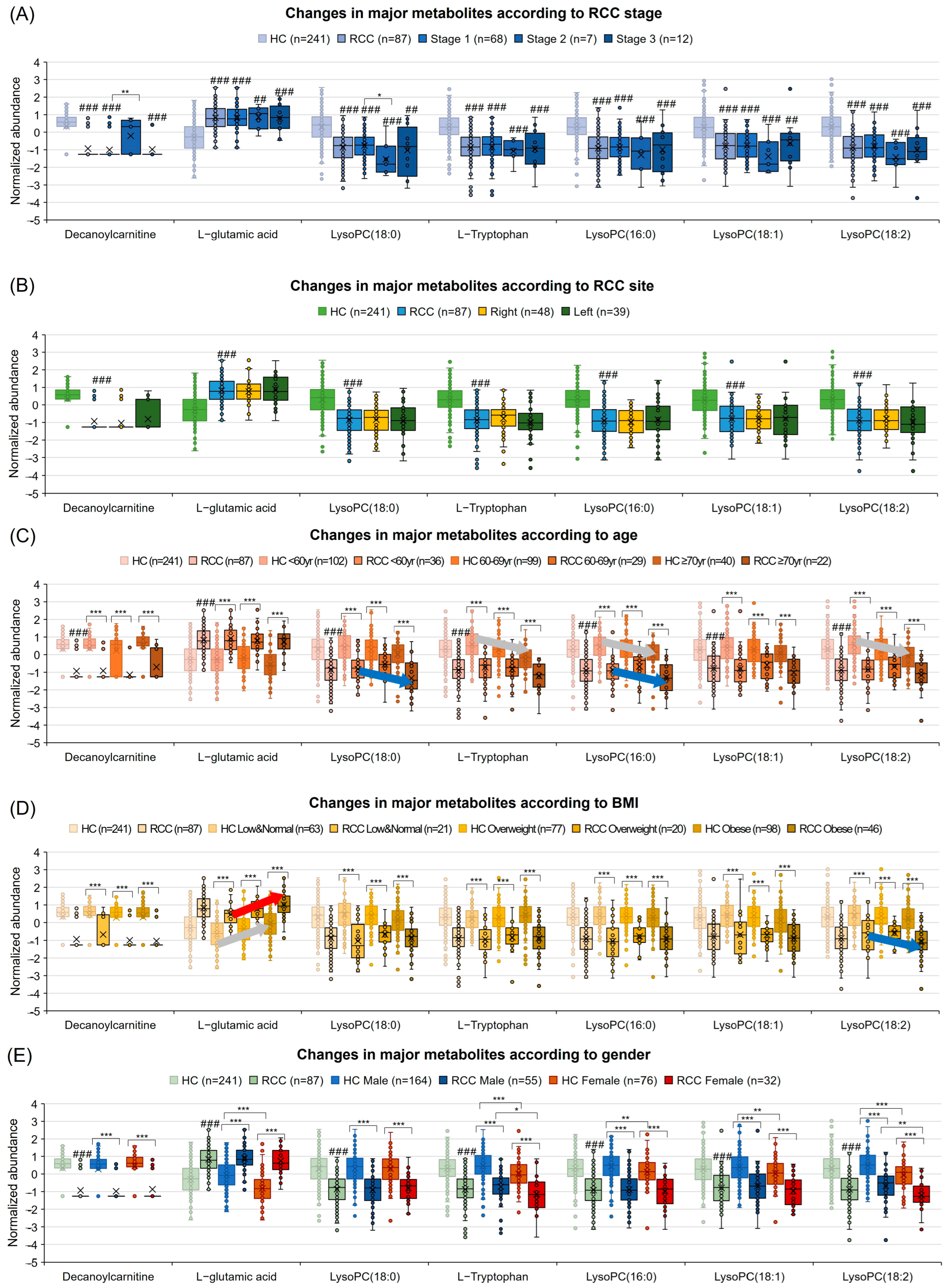
3.8. Diagnostic Potential Marker Levels Are Affected by Clinical Factors
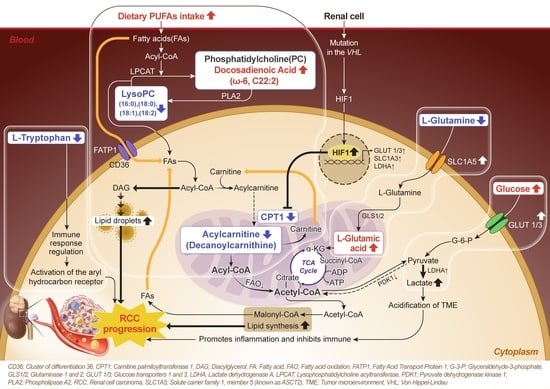
3.9. High PUFA Consumption Is Associated with an Increased Risk of RCC Due to Its Correlation with Blood Fatty Acids
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Li, F.; Aljahdali, I.A.; Zhang, R.; Nastiuk, K.L.; Krolewski, J.J.; Ling, X. Kidney cancer biomarkers and targets for therapeutics: Survivin (BIRC5), XIAP, MCL-1, HIF1α, HIF2α, NRF2, MDM2, MDM4, p53, KRAS and AKT in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 254. [Google Scholar] [CrossRef]
- Sanchez, D.J.; Simon, M.C. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim. Et Biophys. Acta (BBA) Rev. Cancer 2018, 1870, 23–31. [Google Scholar] [CrossRef]
- Perrotti, F.; Rosa, C.; Cicalini, I.; Sacchetta, P.; Del Boccio, P.; Genovesi, D.; Pieragostino, D. Advances in lipidomics for cancer biomarkers discovery. Int. J. Mol. Sci. 2016, 17, 1992. [Google Scholar] [CrossRef]
- Usher-Smith, J.; Simmons, R.K.; Rossi, S.H.; Stewart, G.D. Current evidence on screening for renal cancer. Nat. Rev. Urol. 2020, 17, 637–642. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, M.; Liu, X.; Sun, H.; Guo, Z.; Tang, X.; Wang, Z.; Li, J.; Li, H.; Sun, W. Urine metabolomics for renal cell carcinoma (RCC) prediction: Tryptophan metabolism as an important pathway in RCC. Front. Oncol. 2019, 9, 663. [Google Scholar] [CrossRef]
- Maslov, D.L.; Trifonova, O.P.; Lichtenberg, S.; Balashova, E.E.; Mamedli, Z.Z.; Alferov, A.A.; Stilidi, I.S.; Lokhov, P.G.; Kushlinskii, N.E.; Archakov, A.I. Blood Plasma Metabolome Profiling at Different Stages of Renal Cell Carcinoma. Cancers 2023, 15, 140. [Google Scholar] [CrossRef]
- Zira, A.N.; Theocharis, S.E.; Mitropoulos, D.; Migdalis, V.; Mikros, E. 1H NMR metabonomic analysis in renal cell carcinoma: A possible diagnostic tool. J. Proteome Res. 2010, 9, 4038–4044. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, M.; Cheng, X.; Liu, X.; Sun, H.; Guo, Z.; Li, J.; Tang, X.; Wang, Z.; Sun, W. LC-MS-based plasma metabolomics and lipidomics analyses for differential diagnosis of bladder cancer and renal cell carcinoma. Front. Oncol. 2020, 10, 717. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, M.; Liu, X.; Sun, H.; Guo, Z.; Tang, X.; Wang, Z.; Li, J.; He, L.; Zhang, W. Investigation of plasma metabolic and lipidomic characteristics of a Chinese cohort and a pilot study of renal cell carcinoma biomarker. Front. Oncol. 2020, 10, 1507. [Google Scholar] [CrossRef]
- Lee, H.-O.; Uzzo, R.G.; Kister, D.; Kruger, W.D. Combination of serum histidine and plasma tryptophan as a potential biomarker to detect clear cell renal cell carcinoma. J. Transl. Med. 2017, 15, 1–6. [Google Scholar] [CrossRef]
- Falegan, O.S.; Ball, M.W.; Shaykhutdinov, R.A.; Pieroraio, P.M.; Farshidfar, F.; Vogel, H.J.; Allaf, M.E.; Hyndman, M.E. Urine and serum metabolomics analyses may distinguish between stages of renal cell carcinoma. Metabolites 2017, 7, 6. [Google Scholar] [CrossRef]
- Zheng, H.; Ji, J.; Zhao, L.; Chen, M.; Shi, A.; Pan, L.; Huang, Y.; Zhang, H.; Dong, B.; Gao, H. Prediction and diagnosis of renal cell carcinoma using nuclear magnetic resonance-based serum metabolomics and self-organizing maps. Oncotarget 2016, 7, 59189. [Google Scholar] [CrossRef]
- Gao, H.; Dong, B.; Liu, X.; Xuan, H.; Huang, Y.; Lin, D. Metabonomic profiling of renal cell carcinoma: High-resolution proton nuclear magnetic resonance spectroscopy of human serum with multivariate data analysis. Anal. Chim. Acta 2008, 624, 269–277. [Google Scholar] [CrossRef]
- Nizioł, J.; Ossoliński, K.; Tripet, B.P.; Copié, V.; Arendowski, A.; Ruman, T. Nuclear magnetic resonance and surface-assisted laser desorption/ionization mass spectrometry-based serum metabolomics of kidney cancer. Anal. Bioanal. Chem. 2020, 412, 5827–5841. [Google Scholar] [CrossRef]
- Lin, L.; Yu, Q.; Yan, X.; Hang, W.; Zheng, J.; Xing, J.; Huang, B. Direct infusion mass spectrometry or liquid chromatography mass spectrometry for human metabonomics? A serum metabonomic study of kidney cancer. Analyst 2010, 135, 2970–2978. [Google Scholar] [CrossRef] [PubMed]
- Deja, S.; Litarski, A.; Mielko, K.A.; Pudełko-Malik, N.; Wojtowicz, W.; Zabek, A.; Szydełko, T.; Młynarz, P. Gender-specific metabolomics approach to kidney cancer. Metabolites 2021, 11, 767. [Google Scholar] [CrossRef]
- Lin, L.; Huang, Z.; Gao, Y.; Yan, X.; Xing, J.; Hang, W. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J. Proteome Res. 2011, 10, 1396–1405. [Google Scholar] [CrossRef]
- Manzi, M.; Palazzo, M.; Knott, M.E.; Beauseroy, P.; Yankilevich, P.; Giménez, M.I.; Monge, M.E. Coupled mass-spectrometry-based lipidomics machine learning approach for early detection of clear cell renal cell carcinoma. J. Proteome Res. 2020, 20, 841–857. [Google Scholar] [CrossRef]
- Hoerner, C.R.; Chen, V.J.; Fan, A.C. The ‘Achilles Heel’ of metabolism in renal cell carcinoma: Glutaminase inhibition as a rational treatment strategy. Kidney Cancer 2019, 3, 15–29. [Google Scholar] [CrossRef]
- Wettersten, H.I.; Aboud, O.A.; Lara Jr, P.N.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef]
- Heravi, G.; Yazdanpanah, O.; Podgorski, I.; Matherly, L.H.; Liu, W. Lipid metabolism reprogramming in renal cell carcinoma. Cancer Metastasis Rev. 2022, 41, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Deng, W.; Zhang, Z.; Zeng, Z.; Liu, Y.; Zhou, X.; Zhang, C.; Wang, G. Novel amino acid metabolism-related gene signature to predict prognosis in clear cell renal cell carcinoma. Front. Genet. 2022, 13, 982162. [Google Scholar] [CrossRef] [PubMed]
- Fritsche-Guenther, R.; Gloaguen, Y.; Eisenberger, A.; Kirwan, J.A. Analysis of adherent cell culture lysates with low metabolite concentrations using the Biocrates AbsoluteIDQ p400 HR kit. Sci. Rep. 2022, 12, 7933. [Google Scholar] [CrossRef] [PubMed]
- Jagga, Z.; Gupta, D. Machine learning for biomarker identification in cancer research–developments toward its clinical application. Per. Med. 2015, 12, 371–387. [Google Scholar] [CrossRef]
- Campi, R.; Rebez, G.; Klatte, T.; Roussel, E.; Ouizad, I.; Ingels, A.; Pavan, N.; Kara, O.; Erdem, S.; Bertolo, R. Effect of smoking, hypertension and lifestyle factors on kidney cancer—Perspectives for prevention and screening programmes. Nat. Rev. Urol. 2023, 20, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Spiegelman, D.; Hunter, D.J.; Albanes, D.; Bernstein, L.; Van Den Brandt, P.A.; Buring, J.E.; Cho, E.; English, D.R.; Freudenheim, J.L. Fat, protein, and meat consumption and renal cell cancer risk: A pooled analysis of 13 prospective studies. JNCI J. Natl. Cancer Inst. 2008, 100, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; La Vecchia, C.; DesMeules, M.; Negri, E.; Mery, L.; Group, C.C.R.E.R. Nutrient and fiber intake and risk of renal cell carcinoma. Nutr. Cancer 2008, 60, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, S.; Linseisen, J.; Overvad, K.; Lund Würtz, A.M.; Roswall, N.; Tjonneland, A.; Boutron-Ruault, M.C.; Racine, A.; Bastide, N.; Palli, D. Meat and fish consumption and the risk of renal cell carcinoma in the E uropean prospective investigation into cancer and nutrition. Int. J. Cancer 2015, 136, E423–E431. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Sobue, T.; Zha, L.; Kitamura, T.; Iwasaki, M.; Inoue, M.; Yamaji, T.; Tsugane, S.; Sawada, N. Dietary fish and n-3 polyunsaturated fatty acid consumption, and renal cell carcinoma risk in Japan: The Japan Public Health Center-based Prospective Study. Cancer Epidemiol. Biomark. Prev. 2023, 32, 1365–1372. [Google Scholar] [CrossRef]
- Du, W.; Zhang, L.; Brett-Morris, A.; Aguila, B.; Kerner, J.; Hoppel, C.L.; Puchowicz, M.; Serra, D.; Herrero, L.; Rini, B.I. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 2017, 8, 1769. [Google Scholar] [CrossRef]
- Du, Y.; Wang, Q.; Zhang, X.; Wang, X.; Qin, C.; Sheng, Z.; Yin, H.; Jiang, C.; Li, J.; Xu, T. Lysophosphatidylcholine acyltransferase 1 upregulation and concomitant phospholipid alterations in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 66. [Google Scholar] [CrossRef]
- Harayama, T.; Eto, M.; Shindou, H.; Kita, Y.; Otsubo, E.; Hishikawa, D.; Ishii, S.; Sakimura, K.; Mishina, M.; Shimizu, T. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 2014, 20, 295–305. [Google Scholar] [CrossRef]
- Lands, W.E. Metabolism of glycerolipides: A comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 1958, 231, 883–888. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. Lipids in cancer: A global view of the contribution of lipid pathways to metastatic formation and treatment resistance. Oncogenesis 2022, 11, 46. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, Y.; Liu, Q.; Wu, F.; Liu, X.; Qu, H.; Yuan, Y.; Ge, J.; Xu, Y.; Wang, H. The mRNA expression signature and prognostic analysis of multiple fatty acid metabolic enzymes in clear cell renal cell carcinoma. J. Cancer 2019, 10, 6599. [Google Scholar] [CrossRef]
- Christensen, E.; Hagve, T.-A.; Grønn, M.; Christophersen, B.O. β-oxidation of medium chain (C8C14) fatty acids studied in isolated liver cells. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1989, 1004, 187–195. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Mazza, T.; Pochini, L.; Galluccio, M.; Giangregorio, N.; Tonazzi, A.; Indiveri, C. Carnitine traffic in cells. Link with cancer. Front. Cell Dev. Biol. 2020, 8, 583850. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Reznik, E.; Lee, C.-H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, H.; Ren, Z.; Yi, X.; Zhang, Q.; Yang, Z.; Kuang, Y.; Zhu, Y. Overexpression CPT1A reduces lipid accumulation via PPARα/CD36 axis to suppress the cell proliferation in ccRCC: CPT1A inhibits ccRCC cell proliferation via PPARα/CD36 axis. Acta Biochim. Biophys. Sin. 2022, 54, 220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, X.; Lu, S.; Ou, K. Metabolic reprogramming of clear cell renal cell carcinoma. Front. Endocrinol. 2023, 14, 1195500. [Google Scholar] [CrossRef] [PubMed]
- Courtney, K.D.; Bezwada, D.; Mashimo, T.; Pichumani, K.; Vemireddy, V.; Funk, A.M.; Wimberly, J.; McNeil, S.S.; Kapur, P.; Lotan, Y. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 2018, 28, 793–800.e792. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.C.; Payne, L.B.; Rathmell, W.K. Hypoxia, angiogenesis, and metabolism in the hereditary kidney cancers. J. Clin. Investig. 2019, 129, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Du, G.; Li, L.; Peng, K. Cellular specificity of lactate metabolism and a novel lactate-related gene pair index for frontline treatment in clear cell renal cell carcinoma. Front. Oncol. 2023, 13, 1253783. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Opitz, C.A.; Somarribas Patterson, L.F.; Mohapatra, S.R.; Dewi, D.L.; Sadik, A.; Platten, M.; Trump, S. The therapeutic potential of targeting tryptophan catabolism in cancer. Br. J. Cancer 2020, 122, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Lanke, V.; Vinod, P. Network-based metabolic characterization of renal cell carcinoma. Sci. Rep. 2020, 10, 5955. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef] [PubMed]
- McClain, K.M.; Sampson, J.N.; Petrick, J.L.; Mazzilli, K.M.; Gerszten, R.E.; Clish, C.B.; Purdue, M.P.; Lipworth, L.; Moore, S.C. Metabolomic Analysis of Renal Cell Carcinoma in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites 2022, 12, 1189. [Google Scholar] [CrossRef]
- Adams, K.F.; Leitzmann, M.F.; Albanes, D.; Kipnis, V.; Moore, S.C.; Schatzkin, A.; Chow, W.-H. Body size and renal cell cancer incidence in a large US cohort study. Am. J. Epidemiol. 2008, 168, 268–277. [Google Scholar] [CrossRef]
- Semba, R.D.; Zhang, P.; Adelnia, F.; Sun, K.; Gonzalez-Freire, M.; Salem Jr, N.; Brennan, N.; Spencer, R.G.; Fishbein, K.; Khadeer, M. Low plasma lysophosphatidylcholines are associated with impaired mitochondrial oxidative capacity in adults in the Baltimore Longitudinal Study of Aging. Aging Cell 2019, 18, e12915. [Google Scholar] [CrossRef]
- Wolk, A.; Larsson, S.C.; Johansson, J.-E.; Ekman, P. Long-term fatty fish consumption and renal cell carcinoma incidence in women. JAMA 2006, 296, 1371–1376. [Google Scholar] [CrossRef]


| Characteristics | Discovery Set | Validation Set | ||||
|---|---|---|---|---|---|---|
| HC (n = 167) | RCC (n = 60) | p a | HC (n = 74) | RCC (n = 27) | p a | |
| Sex (Male) | 121 (72.5) | 35 (58.3) | 0.052 | 44 (59.5) | 20 (74.1) | 0.244 |
| Age (year) | 60.0 (8.84) | 62.5 (11.0) | 0.109 | 60.8 (9.90) | 60.2 (10.9) | 0.759 |
| BMI (kg/m2) | 24.6 (2.78) | 25.4 (3.99) | 0.170 | 24.3 (3.62) | 25.6 (2.41) | 0.024 |
| Low (<18.5) | 3 (1.80) | 2 (3.30) | 0.482 | 3 (4.10) | 0 (0.00) | 0.172 |
| Normal (18.5–22.9) | 41 (24.6) | 15 (25.0) | 19 (25.7) | 4 (14.8) | ||
| Overweight (23.0–24.9) | 51 (30.5) | 13 (21.7) | 26 (35.1) | 7 (25.9) | ||
| Obese (≥25) | 72 (42.1) | 30 (50.0) | 26 (35.1) | 16 (59.3) | ||
| Experiences of smoking | 86 (53.8) | 26 (43.3) | 0.177 | 40 (55.6) | 16 (59.3) | 0.822 |
| Experiences of drinking | 123 (79.9) | 35 (58.3) | 0.002 | 61 (83.6) | 17 (63.0) | 0.034 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-H.; Chung, J.-S.; Lee, H.-H.; Park, J.-H.; Kim, M.-K. Influence of Dietary Polyunsaturated Fatty Acid Intake on Potential Lipid Metabolite Diagnostic Markers in Renal Cell Carcinoma: A Case-Control Study. Nutrients 2024, 16, 1265. https://doi.org/10.3390/nu16091265
Kim Y-H, Chung J-S, Lee H-H, Park J-H, Kim M-K. Influence of Dietary Polyunsaturated Fatty Acid Intake on Potential Lipid Metabolite Diagnostic Markers in Renal Cell Carcinoma: A Case-Control Study. Nutrients. 2024; 16(9):1265. https://doi.org/10.3390/nu16091265
Chicago/Turabian StyleKim, Yeon-Hee, Jin-Soo Chung, Hyung-Ho Lee, Jin-Hee Park, and Mi-Kyung Kim. 2024. "Influence of Dietary Polyunsaturated Fatty Acid Intake on Potential Lipid Metabolite Diagnostic Markers in Renal Cell Carcinoma: A Case-Control Study" Nutrients 16, no. 9: 1265. https://doi.org/10.3390/nu16091265
APA StyleKim, Y.-H., Chung, J.-S., Lee, H.-H., Park, J.-H., & Kim, M.-K. (2024). Influence of Dietary Polyunsaturated Fatty Acid Intake on Potential Lipid Metabolite Diagnostic Markers in Renal Cell Carcinoma: A Case-Control Study. Nutrients, 16(9), 1265. https://doi.org/10.3390/nu16091265