Application of Computational Data Modeling to a Large-Scale Population Cohort Assists the Discovery of Inositol as a Strain-Specific Substrate for Faecalibacterium prausnitzii
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Modeling to Predict the Abundance of F. prausnitzii Using Nutrient Intake Data
2.3. Culture Conditions for Testing Selected Nutrients
2.4. Batch Fermentation
2.5. Bacterial DNA Extraction
2.6. Quantification of Total Bacteria and F. prausnitzii by Real Time PCR
2.7. 1H-Nuclear Magnetic Resonance (NMR) Metabolomics
2.8. General Statistical Analysis
3. Results
3.1. Characteristics of the Study Subjects
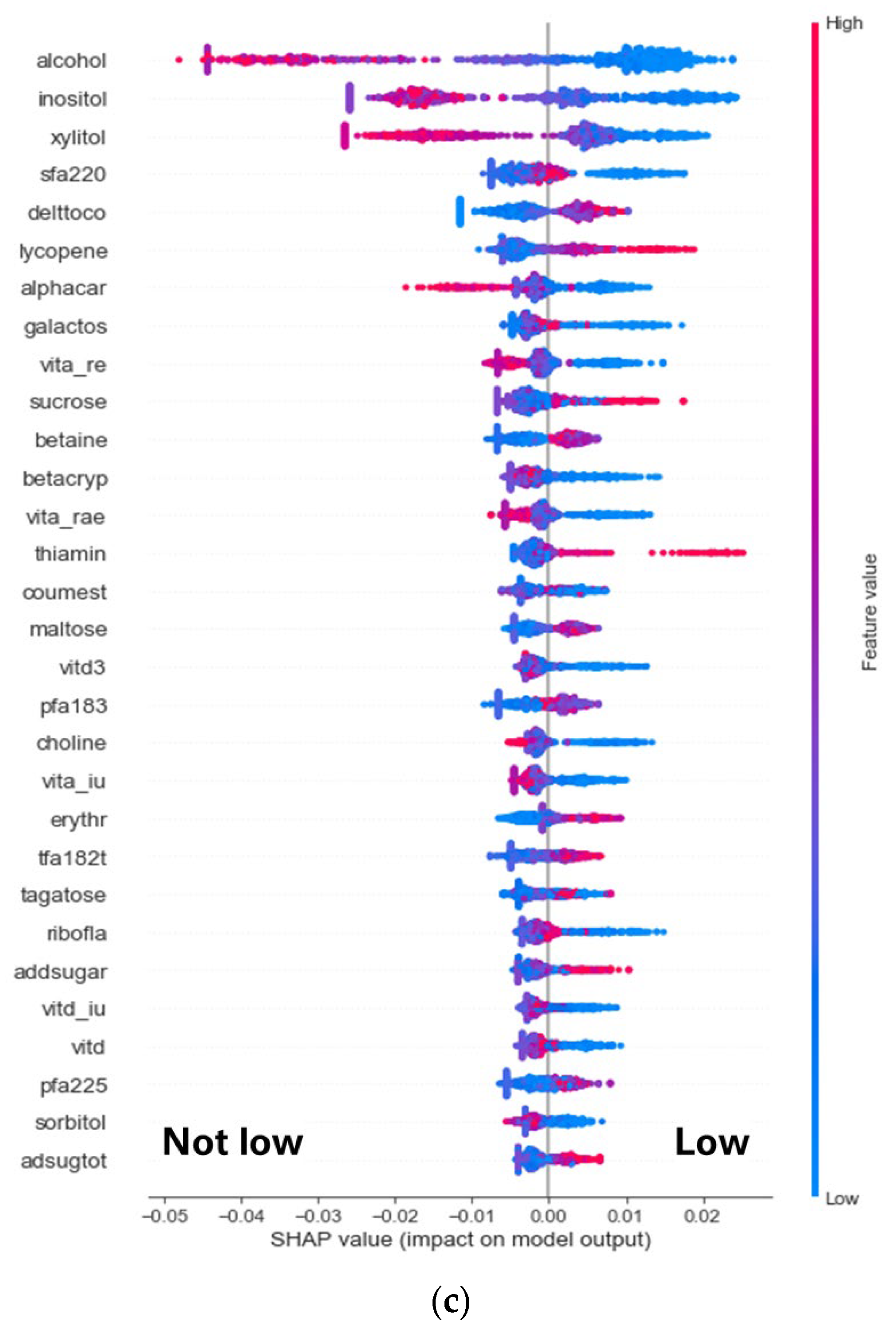
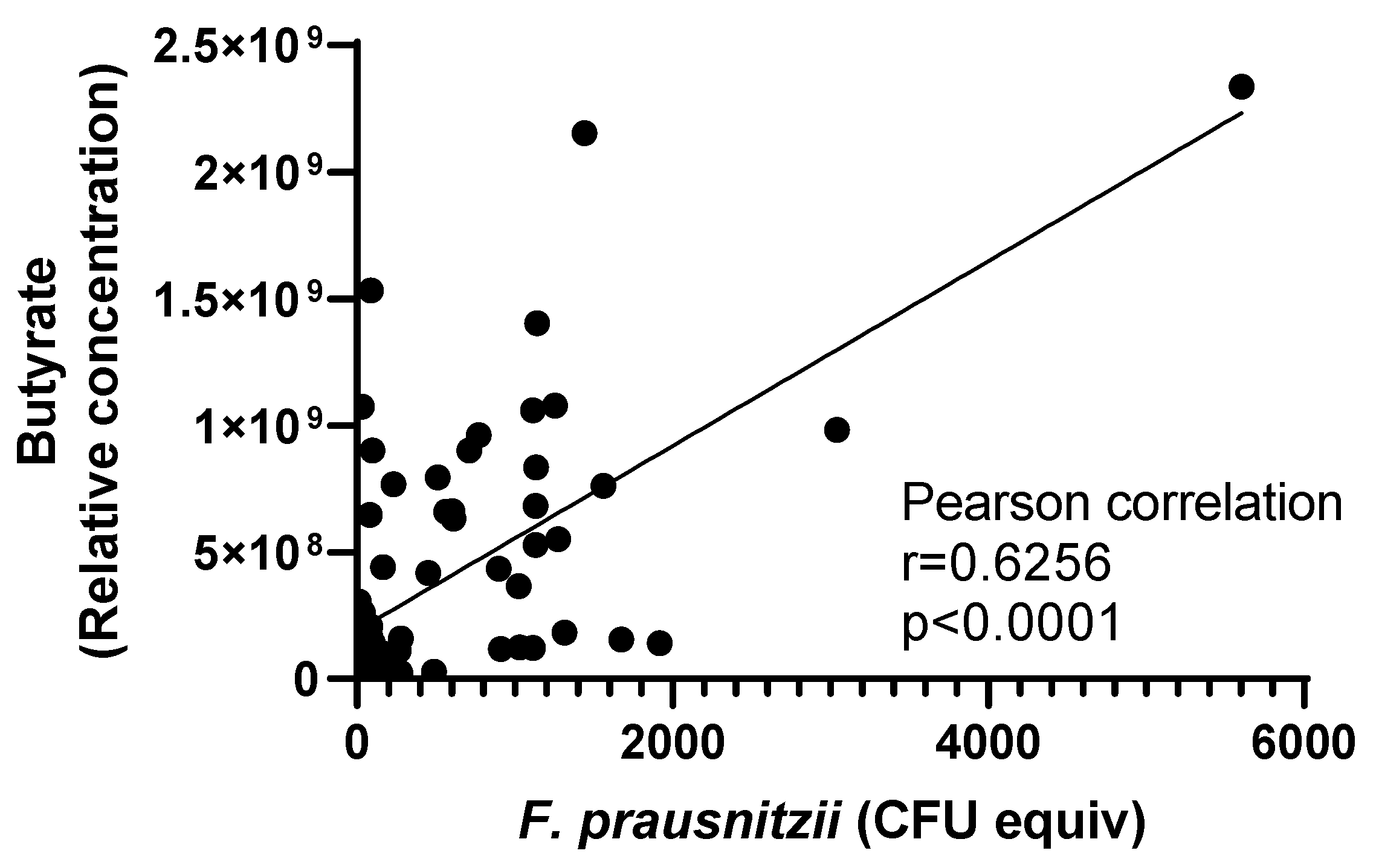
3.2. Discovery of Nutrients Associated with the Abundance of F. prausnitzii
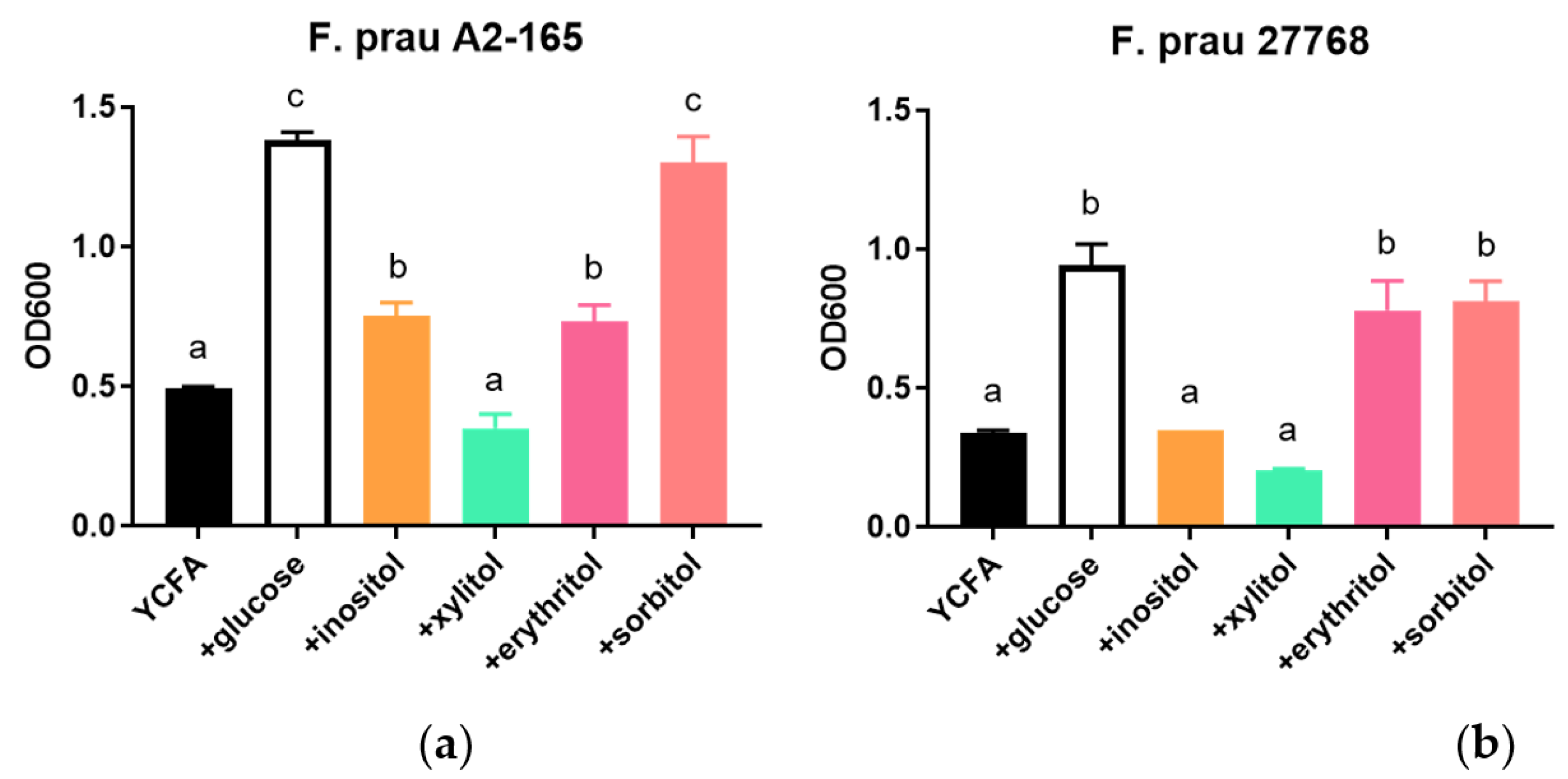
3.3. Growth of F. prausnitzii on Inositol-Based Media Is Strain Dependent
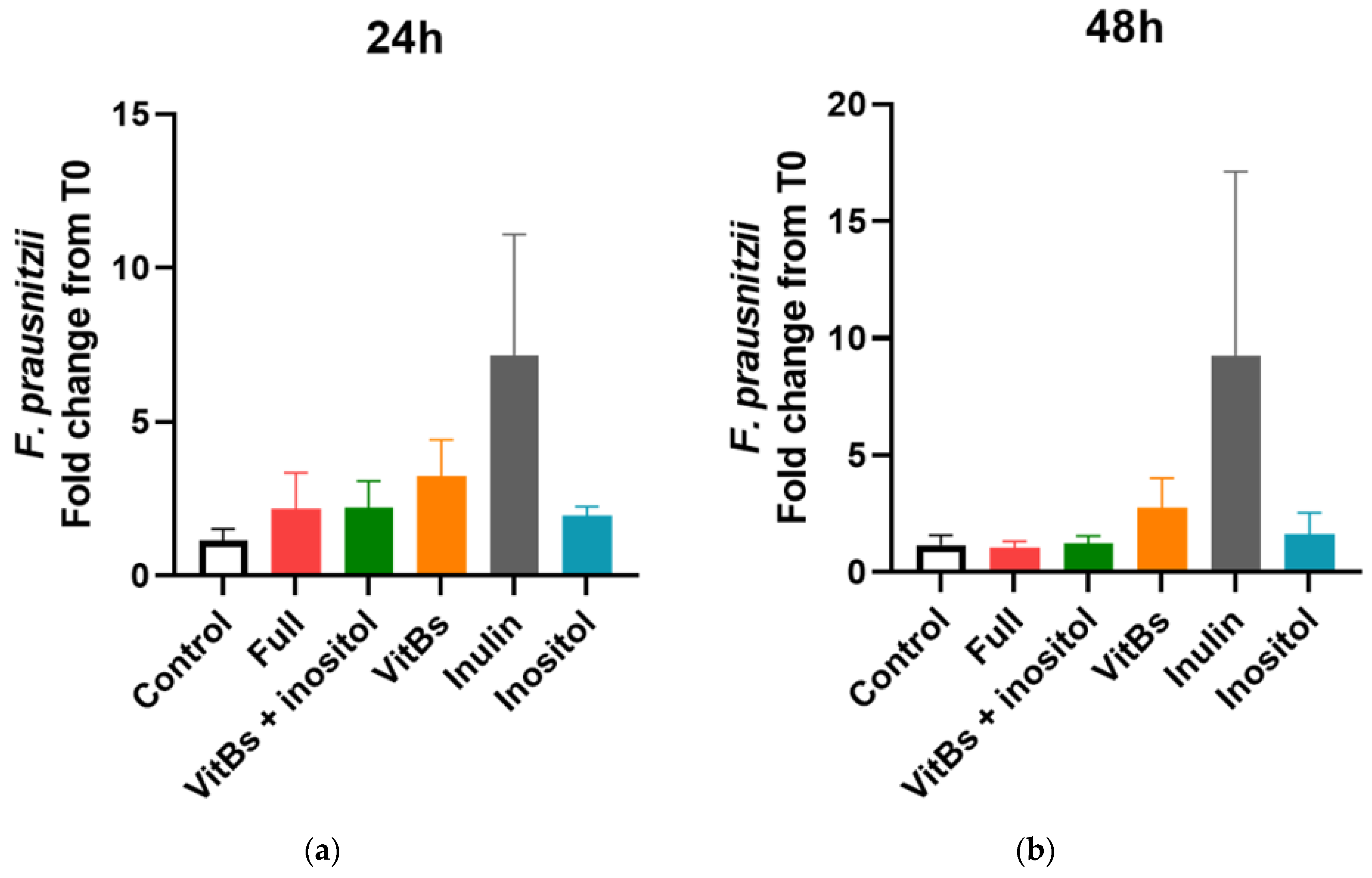
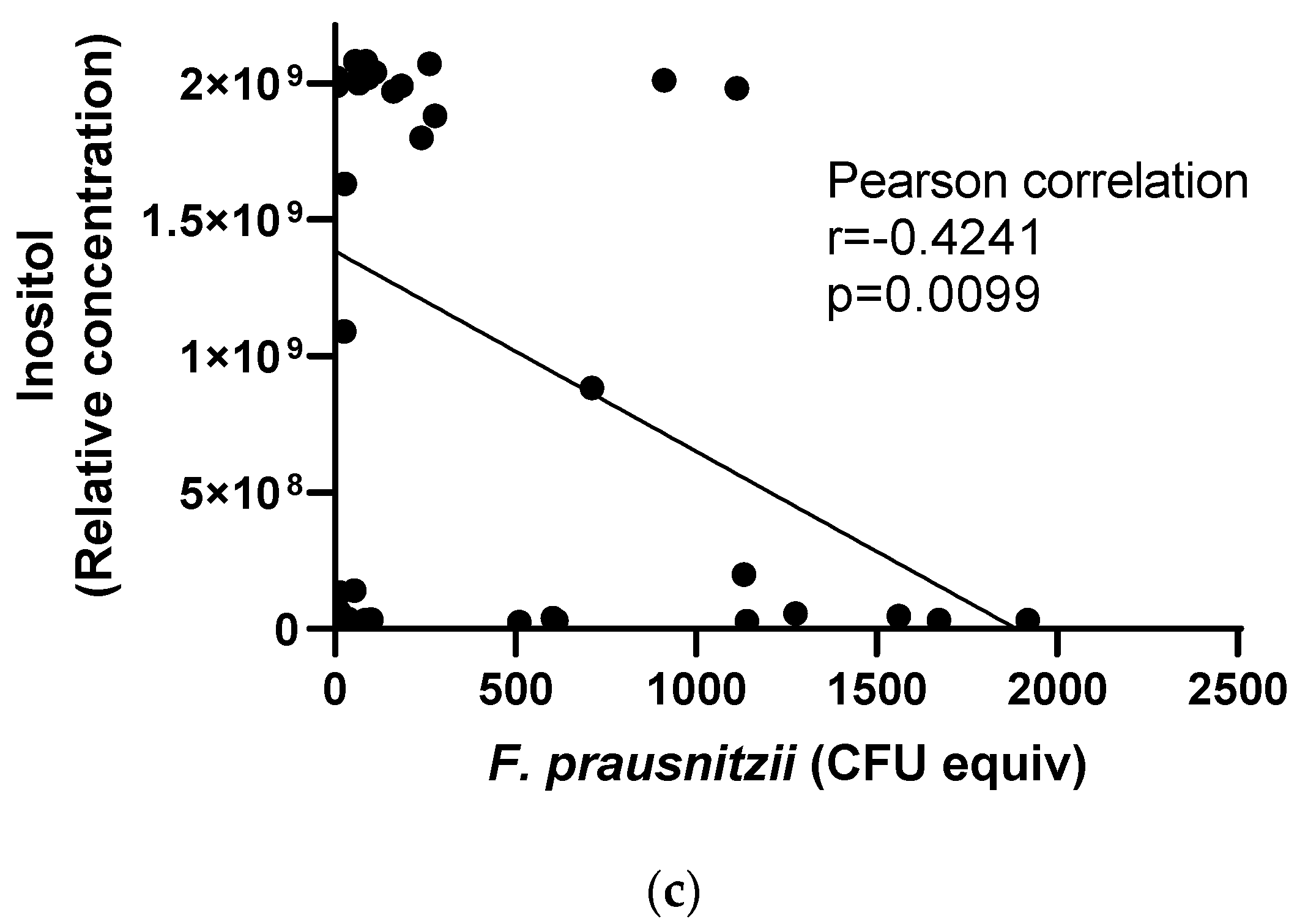
3.4. Responses of F. prausnitzii to Nutrients in a Mixed Community
4. Discussion
5. Conclusions
6. Patents
- (1)
- Systems and methods for estimating, from food frequency questionnaire-based nutrients intake data, the relative amounts of Faecalibacterium prausnitzii (Fprau) in the gut microbiome ecosystem and associated recommendations to improve Faecalibacterium prausnitzii [59].
- (2)
- Compositions and methods using at least one inositol or sorbitol to enhance the growth of Faecalibacterium prausnitzii [60].
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, H.; Chen, S.; He, J.; Zhou, Y.; Nie, Y. Systematic review and meta-analysis of the role of Faecalibacterium prausnitzii alteration in inflammatory bowel disease. J. Gastroenterol. Hepatol. 2021, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Furet, J.P.; Kong, L.C.; Tap, J.; Poitou, C.; Basdevant, A.; Bouillot, J.L.; Mariat, D.; Corthier, G.; Dore, J.; Henegar, C.; et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: Links with metabolic and low-grade inflammation markers. Diabetes 2010, 59, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef]
- Da Silva, H.E.; Teterina, A.; Comelli, E.M.; Taibi, A.; Arendt, B.M.; Fischer, S.E.; Lou, W.; Allard, J.P. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci. Rep. 2018, 8, 1466. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Martinez-Medina, M.; Surís-Valls, R.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.H.; Flint, H.J.; Garcia-Gil, L.J. Changes in the Abundance of Faecalibacterium prausnitzii Phylogroups I and II in the Intestinal Mucosa of Inflammatory Bowel Disease and Patients with Colorectal Cancer. Inflamm. Bowel Dis. 2015, 22, 28–41. [Google Scholar] [CrossRef]
- Martín, R.; Bermúdez-Humarán, L.G.; Langella, P. Searching for the Bacterial Effector: The Example of the Multi-Skilled Commensal Bacterium Faecalibacterium prausnitzii. Front. Microbiol. 2018, 9, 346. [Google Scholar] [CrossRef]
- Miquel, S.; Leclerc, M.; Martin, R.; Chain, F.; Lenoir, M.; Raguideau, S.; Hudault, S.; Bridonneau, C.; Northen, T.; Bowen, B.; et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio 2015, 6, e00300-15. [Google Scholar] [CrossRef]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef]
- Martín, R.; Miquel, S.; Chain, F.; Natividad, J.M.; Jury, J.; Lu, J.; Sokol, H.; Theodorou, V.; Bercik, P.; Verdu, E.F.; et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015, 15, 67. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Wu, H.; Bjornson, E.; Zhang, C.; Hakkarainen, A.; Räsänen, S.M.; Lee, S.; Mancina, R.M.; Bergentall, M.; Pietiläinen, K.H.; et al. An Integrated Understanding of the Rapid Metabolic Benefits of a Carbohydrate-Restricted Diet on Hepatic Steatosis in Humans. Cell Metab. 2018, 27, 559–571.e5. [Google Scholar] [CrossRef]
- Ruiz-Saavedra, S.; Salazar, N.; Suárez, A.; de Los Reyes-Gavilán, C.G.; Gueimonde, M.; González, S. Comparison of Different Dietary Indices as Predictors of Inflammation, Oxidative Stress and Intestinal Microbiota in Middle-Aged and Elderly Subjects. Nutrients 2020, 12, 3828. [Google Scholar] [CrossRef]
- Dewulf, E.M.; Cani, P.D.; Claus, S.P.; Fuentes, S.; Puylaert, P.G.; Neyrinck, A.M.; Bindels, L.B.; de Vos, W.M.; Gibson, G.R.; Thissen, J.P.; et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013, 62, 1112–1121. [Google Scholar] [CrossRef]
- Hustoft, T.N.; Hausken, T.; Ystad, S.O.; Valeur, J.; Brokstad, K.; Hatlebakk, J.G.; Lied, G.A. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2017, 29, e12969. [Google Scholar] [CrossRef]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef]
- Fernando, W.M.; Hill, J.E.; Zello, G.A.; Tyler, R.T.; Dahl, W.J.; Van Kessel, A.G. Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults. Benef. Microbes 2010, 1, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Hooda, S.; Boler, B.M.; Serao, M.C.; Brulc, J.M.; Staeger, M.A.; Boileau, T.W.; Dowd, S.E.; Fahey, G.C., Jr.; Swanson, K.S. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J. Nutr. 2012, 142, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Hyde, E.; Debelius, J.W.; Morton, J.T.; Gonzalez, A.; Ackermann, G.; Aksenov, A.A.; Behsaz, B.; Brennan, C.; Chen, Y.; et al. American Gut: An Open Platform for Citizen Science Microbiome Research. mSystems 2018, 3, e00031-18. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Navas-Molina, J.A.; Kosciolek, T.; McDonald, D.; Vázquez-Baeza, Y.; Ackermann, G.; DeReus, J.; Janssen, S.; Swafford, A.D.; Orchanian, S.B.; et al. Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods 2018, 15, 796–798. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.M.; Stewart, C.S.; Flint, H.J. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov. comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52 Pt 6, 2141–2146. [Google Scholar]
- Van den Abbeele, P.; Taminiau, B.; Pinheiro, I.; Duysburgh, C.; Jacobs, H.; Pijls, L.; Marzorati, M. Arabinoxylo-Oligosaccharides and Inulin Impact Inter-Individual Variation on Microbial Metabolism and Composition, Which Immunomodulates Human Cells. J. Agric. Food Chem. 2018, 66, 1121–1130. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Molino, S.; Navajas-Porras, B.; Valverde-Moya, Á.J.; Hinojosa-Nogueira, D.; López-Maldonado, A.; Pastoriza, S.; Rufián-Henares, J.Á. An in vitro batch fermentation protocol for studying the contribution of food to gut microbiota composition and functionality. Nat. Protoc. 2021, 16, 3186–3209. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148 Pt 1, 257–266. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Martinez-Medina, M.; Busquets, D.; Sabat-Mir, M.; Duncan, S.H.; Flint, H.J.; Aldeguer, X.; Garcia-Gil, L.J. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes. Int. J. Med. Microbiol. 2014, 304, 464–475. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Madrid-Gambin, F.; Oller-Moreno, S.; Fernandez, L.; Bartova, S.; Giner, M.P.; Joyce, C.; Ferraro, F.; Montoliu, I.; Moco, S.; Marco, S. AlpsNMR: An R package for signal processing of fully untargeted NMR-based metabolomics. Bioinformatics 2020, 36, 2943–2945. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Khan, T.M.; Duncan, S.H.; Harmsen, H.J.M.; Garcia-Gil, L.J.; Flint, H.J. Cultured Representatives of Two Major Phylogroups of Human Colonic Faecalibacterium prausnitzii Can Utilize Pectin, Uronic Acids, and Host-Derived Substrates for Growth. Appl. Environ. Microbiol. 2012, 78, 420–428. [Google Scholar] [CrossRef]
- Fitzgerald, C.B.; Shkoporov, A.N.; Sutton, T.D.S.; Chaplin, A.V.; Velayudhan, V.; Ross, R.P.; Hill, C. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genom. 2018, 19, 931. [Google Scholar] [CrossRef]
- Ruiz-Ojeda, F.J.; Plaza-Díaz, J.; Sáez-Lara, M.J.; Gil, A. Effects of Sweeteners on the Gut Microbiota: A Review of Experimental Studies and Clinical Trials. Adv. Nutr. 2019, 10 (Suppl. S1), S31–S48. [Google Scholar] [CrossRef]
- Fernández-Bañares, F. Carbohydrate Maldigestion and Intolerance. Nutrients 2022, 14, 1923. [Google Scholar] [CrossRef]
- Soto-Martin, E.C.; Warnke, I.; Farquharson, F.M.; Christodoulou, M.; Horgan, G.; Derrien, M.; Faurie, J.-M.; Flint, H.J.; Duncan, S.H.; Louis, P. Vitamin Biosynthesis by Human Gut Butyrate-Producing Bacteria and Cross-Feeding in Synthetic Microbial Communities. mBio 2020, 11, e00886-20. [Google Scholar] [CrossRef]
- Vital, M.; Karch, A.; Pieper, D.H. Colonic Butyrate-Producing Communities in Humans: An Overview Using Omics Data. mSystems 2017, 2, e00130-17. [Google Scholar] [CrossRef] [PubMed]
- Cotillard, A.; Cartier-Meheust, A.; Litwin, N.S.; Chaumont, S.; Saccareau, M.; Lejzerowicz, F.; Tap, J.; Koutnikova, H.; Lopez, D.G.; McDonald, D.; et al. A posteriori dietary patterns better explain variations of the gut microbiome than individual markers in the American Gut Project. Am. J. Clin. Nutr. 2021, 115, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Zuluaga, J.d.l.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, e00261-19. [Google Scholar] [CrossRef]
- Taylor, B.C.; Lejzerowicz, F.; Poirel, M.; Shaffer, J.P.; Jiang, L.; Aksenov, A.; Litwin, N.; Humphrey, G.; Martino, C.; Miller-Montgomery, S.; et al. Consumption of Fermented Foods Is Associated with Systematic Differences in the Gut Microbiome and Metabolome. mSystems 2020, 5, e00901-19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wang, X.; Li, J.; Jiang, R.; Chen, H.; Chen, T.; Yang, Y. Determine independent gut microbiota-diseases association by eliminating the effects of human lifestyle factors. BMC Microbiol. 2022, 22, 4. [Google Scholar] [CrossRef]
- Ullmann, T.; Peschel, S.; Finger, P.; Müller, C.L.; Boulesteix, A.-L. Over-optimism in unsupervised microbiome analysis: Insights from network learning and clustering. PLoS Comput. Biol. 2023, 19, e1010820. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; Pérez-Martínez, P.; Andrés-Lacueva, C.; Cardona, F.; Tinahones, F.; Queipo-Ortuño, M.I. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. 2016, 7, 1775–1787. [Google Scholar] [CrossRef]
- Clements, R.S., Jr.; Darnell, B. Myo-inositol content of common foods: Development of a high-myo-inositol diet. Am. J. Clin. Nutr. 1980, 33, 1954–1967. [Google Scholar] [CrossRef]
- Jiang, Z.; Sun, T.-y.; He, Y.; Gou, W.; Zuo, L.-s.-y.; Fu, Y.; Miao, Z.; Shuai, M.; Xu, F.; Xiao, C.; et al. Dietary fruit and vegetable intake, gut microbiota, and type 2 diabetes: Results from two large human cohort studies. BMC Med. 2020, 18, 371. [Google Scholar] [CrossRef]
- van Soest, A.P.M.; Hermes, G.D.A.; Berendsen, A.A.M.; van de Rest, O.; Zoetendal, E.G.; Fuentes, S.; Santoro, A.; Franceschi, C.; de Groot, L.C.P.G.M.; de Vos, W.M. Associations between Pro- and Anti-Inflammatory Gastro-Intestinal Microbiota, Diet, and Cognitive Functioning in Dutch Healthy Older Adults: The NU-AGE Study. Nutrients 2020, 12, 3471. [Google Scholar] [CrossRef]
- Sakamoto, M.; Sakurai, N.; Tanno, H.; Iino, T.; Ohkuma, M.; Endo, A. Genome-based, phenotypic and chemotaxonomic classification of Faecalibacterium strains: Proposal of three novel species Faecalibacterium duncaniae sp. nov. Faecalibacterium hattorii sp. nov. and Faecalibacterium gallinarum sp. nov. Int. J. Syst. Evol. Microbiol. 2022, 72, 5379. [Google Scholar] [CrossRef] [PubMed]
- Koecher, K.J.; Noack, J.A.; Timm, D.A.; Klosterbuer, A.S.; Thomas, W.; Slavin, J.L. Estimation and Interpretation of Fermentation in the Gut: Coupling Results from a 24 h Batch in Vitro System with Fecal Measurements from a Human Intervention Feeding Study Using Fructo-oligosaccharides, Inulin, Gum Acacia, and Pea Fiber. J. Agric. Food Chem. 2014, 62, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Scalera, V.; Natuzzi, D.; Prezioso, G. myo-inositol transport in rat intestinal brush border membrane vesicles, and its inhibition by D-glucose. Biochim. Biophys. Acta 1991, 1062, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, S.; Zolghadri, S.; Stanek, A. Beneficial Effects of Anti-Inflammatory Diet in Modulating Gut Microbiota and Controlling Obesity. Nutrients 2022, 14, 3985. [Google Scholar] [CrossRef]
- Kim, H.; Jeong, Y.; Kang, S.; You, H.J.; Ji, G.E. Co-Culture with Bifidobacterium catenulatum Improves the Growth, Gut Colonization, and Butyrate Production of Faecalibacterium prausnitzii: In Vitro and In Vivo Studies. Microorganisms 2020, 8, 788. [Google Scholar] [CrossRef]
- Costabile, A.; Kolida, S.; Klinder, A.; Gietl, E.; Bäuerlein, M.; Frohberg, C.; Landschütze, V.; Gibson, G.R. A double-blind, placebo-controlled, cross-over study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br. J. Nutr. 2010, 104, 1007–1017. [Google Scholar] [CrossRef]
- Ramnani, P.; Gaudier, E.; Bingham, M.; van Bruggen, P.; Tuohy, K.M.; Gibson, G.R. Prebiotic effect of fruit and vegetable shots containing Jerusalem artichoke inulin: A human intervention study. Br. J. Nutr. 2010, 104, 233–240. [Google Scholar] [CrossRef]
- Dogra, S. Systems and Methods for Estimating, from Food Frequency Questionnaire Based Nutrients Intake Data, The Relative Amounts of Faecalibacterium Prausnitzii (Fprau) in the Gut Microbiome Ecosystem and Associated Recommendations to Improve Faecalibacterium Prausnitzii. WO2022233924A1, 10 November 2022. Available online: https://worldwide.espacenet.com/patent/search/family/075825630/publication/WO2022233924A1?q=pn%3DWO2022233924A1 (accessed on 11 January 2023).
- Chou, C.; Dogra, S.; Dardinier, A. Compositions and Methods Using at least One of Inositol, Erythritol or Sorbitol to Enhance Growth of Faecalibacterium Prausnitzii. WO2022233922A1, 10 November 2022. Available online: https://worldwide.espacenet.com/patent/search/family/081941092/publication/WO2022233922A1?q=pn%3DWO2022233922A1 (accessed on 11 January 2023).




| Data Source | AGP | NHANES 2011–2012 | ||||
|---|---|---|---|---|---|---|
| HEI-2010 dietary component (max score) | Children 2–17 years (n = 68) | Adults 18–64 years (n = 2686) | Older adults ≥ 65 years (n = 852) | Children 2–17 years (n = 2857) | Adults 18–64 years (n = 4044) | Older Adults ≥ 65 years (n = 1032) |
| Dairy (10) | 5.33 (0.47) | 5.03 (0.06) | 5.73 (0.09) | 9.03 (0.22) | 5.78 (0.13) | 5.99 (0.16) |
| EmptyCalories (20) | 16.48 (0.49) | 17.59 (0.07) | 16.91 (0.11) | 11.50 (0.28) | 12.53 (0.28) | 14.99 (0.44) |
| FattyAcids (10) | 5.64 (0.44) | 6.01 (0.07) | 4.91 (0.12) | 3.29 (0.18) | 4.92 (0.19) | 5.60 (0.36) |
| GreensAndBeans (5) | 3.14 (0.25) | 4.39 (0.02) | 4.44 (0.04) | 0.70 (0.09) | 3.63 (0.16) | 3.58 (0.47) |
| RefinedGrains (10) | 8.04 (0.37) | 9.18 (0.04) | 9.46 (0.06) | 4.91 (0.16) | 6.36 (0.17) | 7.34 (0.31) |
| SeafoodAndPlantProteins (5) | 3.26 (0.24) | 4.47 (0.02) | 4.67 (0.03) | 3.05 (0.17) | 3.98 (0.22) | 4.91 (0.18) |
| Sodium (10) | 3.37 (0.34) | 2.43 (0.05) | 3.43 (0.09) | 4.85 (0.25) | 4.04 (0.08) | 3.66 (0.26) |
| TotalFruit (5) | 4.38 (0.15) | 3.67 (0.03) | 4.18 (0.05) | 3.91 (0.18) | 2.61 (0.11) | 3.84 (0.22) |
| TotalVegetables (5) | 3.94 (0.17) | 4.69 (0.01) | 4.69 (0.03) | 2.10 (0.09) | 3.54 (0.09) | 4.16 (0.19) |
| WholeGrains (10) | 3.93 (0.41) | 4.43 (0.07) | 3.96 (0.13) | 2.50 (0.10) | 2.75 (0.16 | 4.23 (0.34) |
| TotalScore (100) | 66.34 (1.38) | 70.73 (0.2) | 71.54 (0.32) | 55.07 (0.72) | 58.27 (0.98) | 68.29 (1.76) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dogra, S.K.; Dardinier, A.; Mainardi, F.; Siegwald, L.; Bartova, S.; Le Roy, C.; Chou, C.J. Application of Computational Data Modeling to a Large-Scale Population Cohort Assists the Discovery of Inositol as a Strain-Specific Substrate for Faecalibacterium prausnitzii. Nutrients 2023, 15, 1311. https://doi.org/10.3390/nu15061311
Dogra SK, Dardinier A, Mainardi F, Siegwald L, Bartova S, Le Roy C, Chou CJ. Application of Computational Data Modeling to a Large-Scale Population Cohort Assists the Discovery of Inositol as a Strain-Specific Substrate for Faecalibacterium prausnitzii. Nutrients. 2023; 15(6):1311. https://doi.org/10.3390/nu15061311
Chicago/Turabian StyleDogra, Shaillay Kumar, Adrien Dardinier, Fabio Mainardi, Léa Siegwald, Simona Bartova, Caroline Le Roy, and Chieh Jason Chou. 2023. "Application of Computational Data Modeling to a Large-Scale Population Cohort Assists the Discovery of Inositol as a Strain-Specific Substrate for Faecalibacterium prausnitzii" Nutrients 15, no. 6: 1311. https://doi.org/10.3390/nu15061311
APA StyleDogra, S. K., Dardinier, A., Mainardi, F., Siegwald, L., Bartova, S., Le Roy, C., & Chou, C. J. (2023). Application of Computational Data Modeling to a Large-Scale Population Cohort Assists the Discovery of Inositol as a Strain-Specific Substrate for Faecalibacterium prausnitzii. Nutrients, 15(6), 1311. https://doi.org/10.3390/nu15061311





