Identification and Selection of Prospective Probiotics for Enhancing Gastrointestinal Digestion: Application in Pharmaceutical Preparations and Dietary Supplements
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures and Growth Conditions
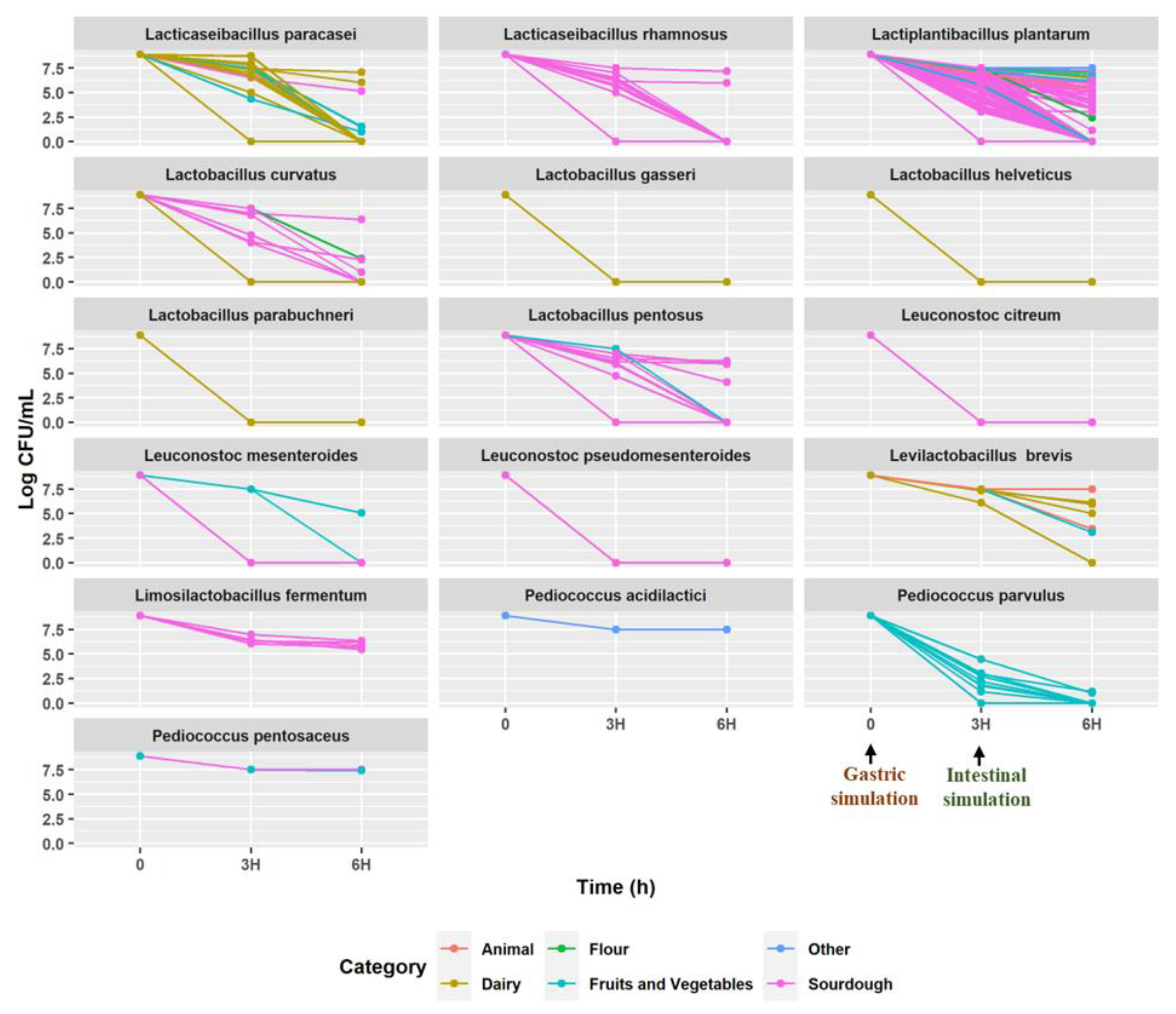
2.2. Resistance to Simulated Gastric and Intestinal Fluids under In Vitro Conditions
2.3. Raffinose Hydrolysis
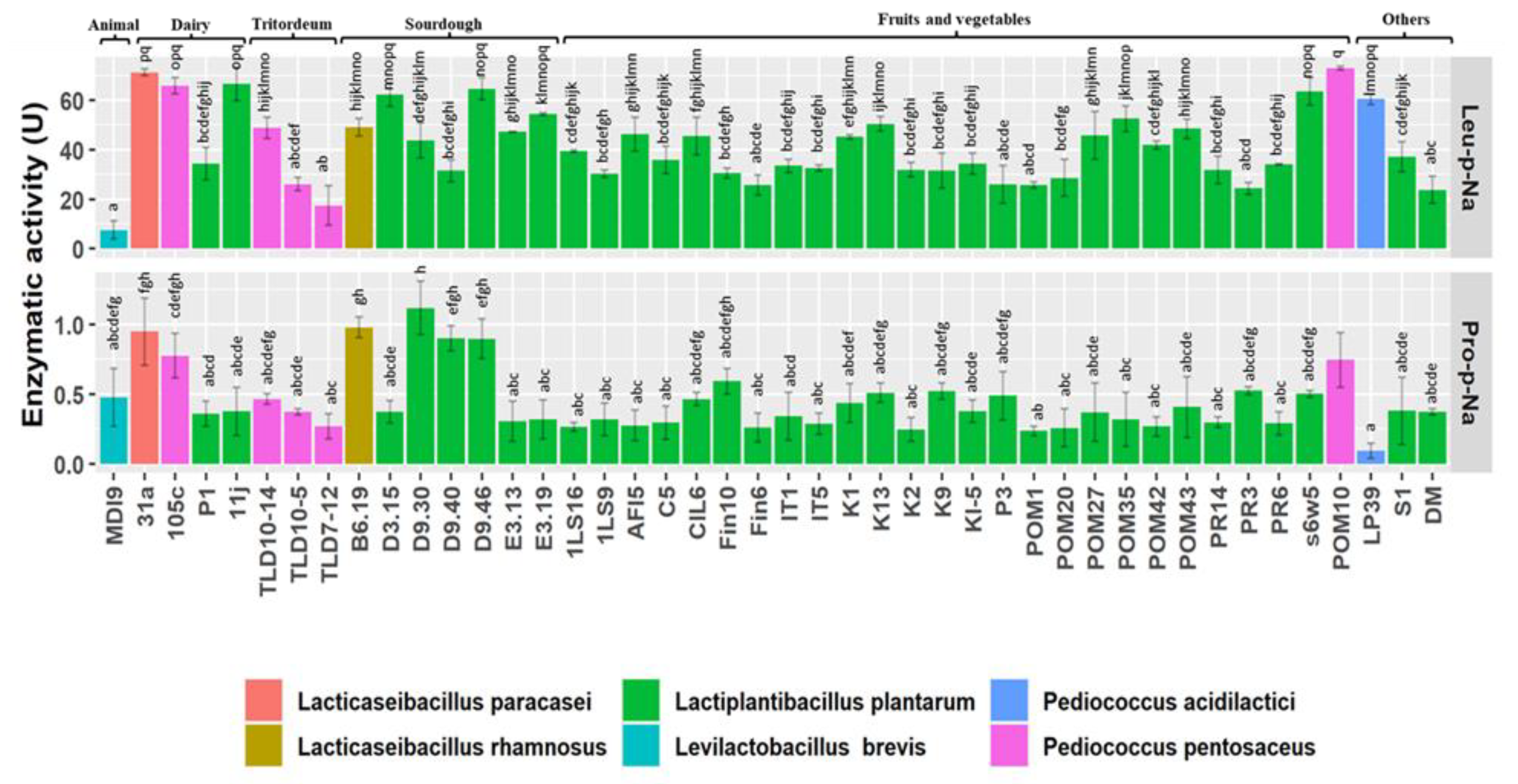
2.4. Peptidase Activity towards Leu-p-Na and Pro-p-Na Synthetic Substrates
2.5. Simulation of Digestion of Targeted Food Matrices
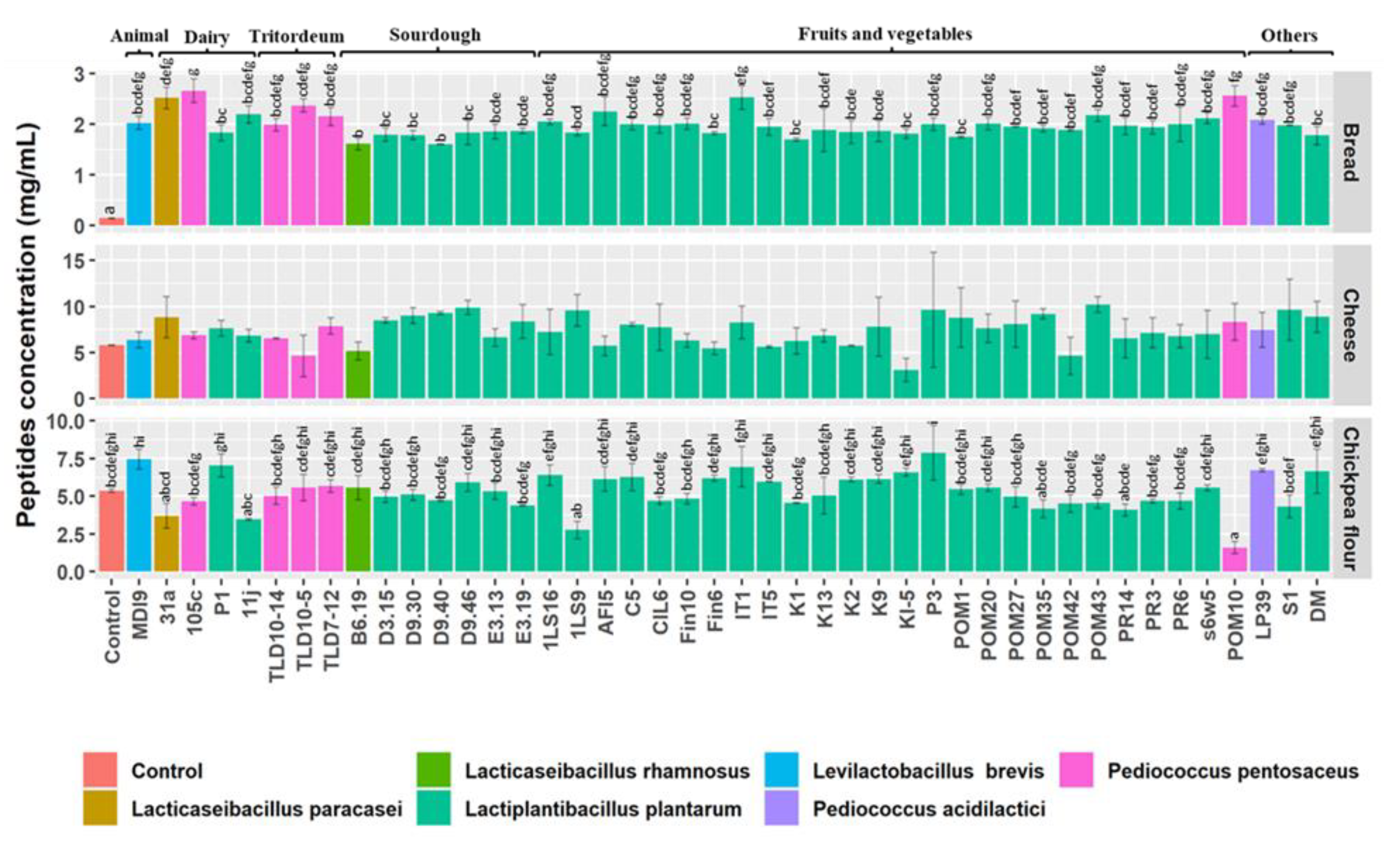
2.6. Determination of Total Peptides
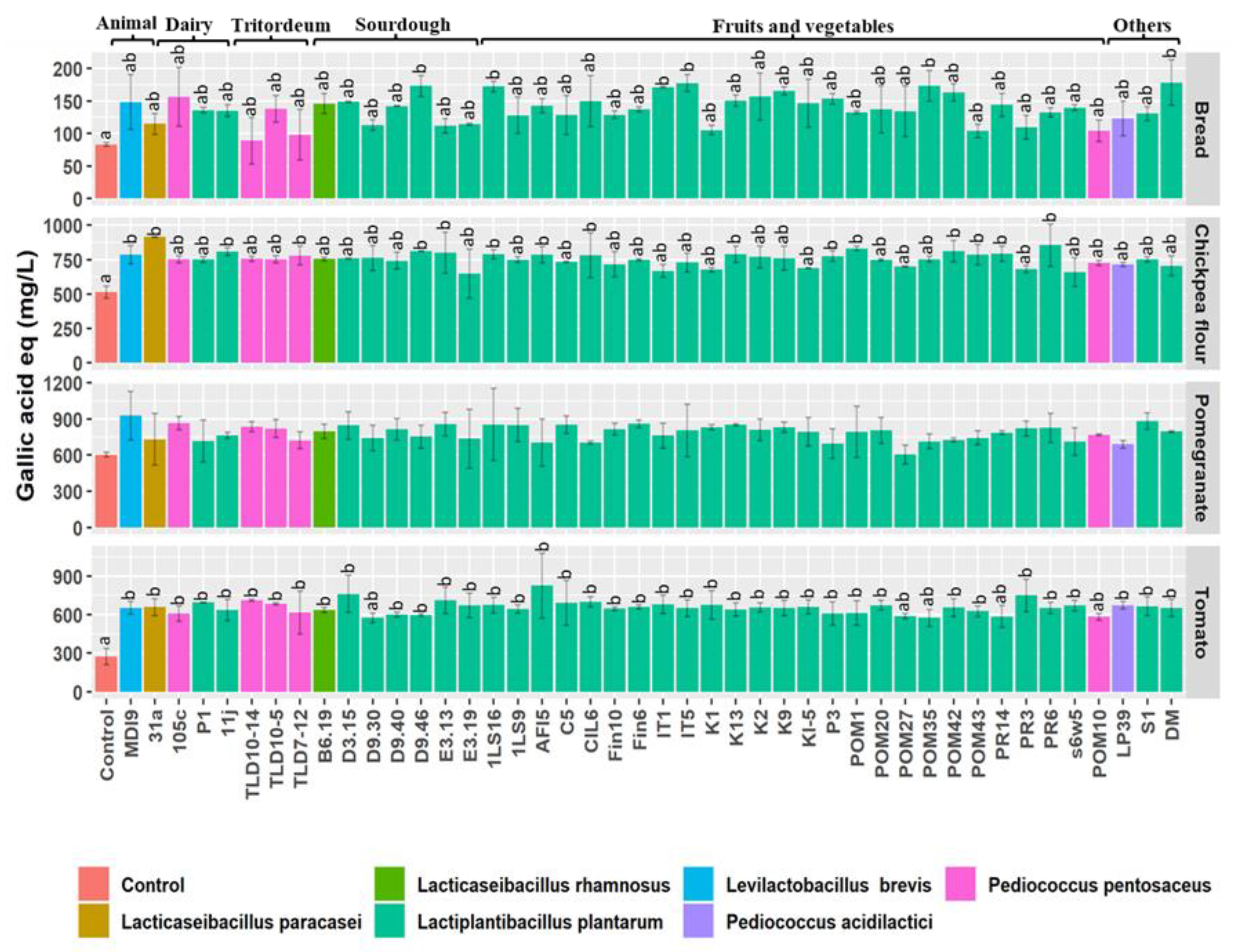
2.7. Determination of Total Free Phenolic Compounds
2.8. Scoring Procedure for the Selection of the Most Promising Probiotic Candidates
2.9. Statistical Analysis
3. Results
3.1. Selection of High-Resistant Strains
3.2. Raffinose Hydrolysis
3.3. Peptidase Activity
3.4. Total Concentration of Peptides
3.5. Total Free Phenolic Compounds
3.6. Final Selection of the Most Promising Probiotic Candidates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ghoshal, U.C.; Shukla, R.; Ghoshal, U.; Gwee, K.A.; Ng, S.C.; Quigley, E.M. The gut microbiota and irritable bowel syndrome: Friend or foe? Int. J. Inflamm. 2012, 2012, 151085. [Google Scholar] [CrossRef]
- Yadav, A.; Chandra, H.; Maurya, V.K. Probiotics: Recent advances and future prospects. J. Plant Dev. Sci. 2017, 9, 967–975. [Google Scholar]
- Tegegne, B.A.; Kebede, B. Probiotics, their prophylactic and therapeutic applications in human health development: A review of the literature. Heliyon 2022, 8, e09725. [Google Scholar] [CrossRef]
- Kerry, R.G.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef]
- Sikorski, Z.E. Chemical and Functional Properties of Food Components; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Codex Alimentarius Commission. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; Food and Agriculture Organization of the United Nations, World Health Organization, Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy; World Health Organization: Geneva, Switzerland, 2006. [Google Scholar]
- Jäger, R.; Purpura, M.; Farmer, S.; Cash, H.A.; Keller, D. Probiotic Bacillus coagulans GBI-30, 6086 improves protein absorption and utilization. Probiotics Antimicrob. Proteins 2018, 10, 611–615. [Google Scholar] [CrossRef]
- Stephens, R.W.; Arhire, L.; Covasa, M. Gut microbiota: From microorganisms to metabolic organ influencing obesity. Obesity 2018, 26, 801–809. [Google Scholar] [CrossRef]
- Da Ros, A.; Polo, A.; Rizzello, C.G.; Acin-Albiac, M.; Montemurro, M.; Di Cagno, R.; Gobbetti, M. Feeding with sustainably sourdough bread has the potential to promote the healthy microbiota metabolism at the colon level. Microbiol. Spectr. 2021, 9, e00494-21. [Google Scholar] [CrossRef] [PubMed]
- Mackie, A.; Mulet-Cabero, A.I.; Torcello-Gómez, A. Simulating human digestion: Developing our knowledge to create healthier and more sustainable foods. Food Funct. 2020, 11, 9397–9431. [Google Scholar] [CrossRef]
- Fioramonti, J.; Theodorou, V.; Bueno, L. Probiotics: What are they? What are their effects on gut physiology? Best Pract. Res. Clin. Gastroenterol. 2003, 17, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Filannino, P.; Di Cagno, R.; Gobbetti, M. Metabolic and functional paths of lactic acid bacteria in plant foods: Get out of the labyrinth. Curr. Opin. Biotechnol. 2018, 49, 64–72. [Google Scholar] [CrossRef]
- Vernaza, M.G.; Dia, V.P.; De Mejia, E.G.; Chang, Y.K. Antioxidant and antiinflammatory properties of germinated and hydrolysed Brazilian soybean flours. Food Chem. 2012, 134, 2217–2225. [Google Scholar] [CrossRef] [PubMed]
- Sanjukta, S.; Rai, A.K. Production of bioactive peptides during soybean fermentation and their potential health benefits. Trends Food Sci. Technol. 2016, 50, 1–10. [Google Scholar] [CrossRef]
- Ayivi, R.D.; Gyawali, R.; Krastanov, A.; Aljaloud, S.O.; Worku, M.; Tahergorabi, R.; Silva, R.C.D.; Ibrahim, S.A. Lactic acidbacteria: Food safety and human health applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Fernández, M.F.; Boris, S.; Barbes, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 2003, 94, 449–455. [Google Scholar] [CrossRef]
- Zárate, G.; Chaia, A.P.; González, S.; Oliver, G. Viability and β-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 2000, 63, 1214–1221. [Google Scholar] [CrossRef]
- De Angelis, M.; Siragusa, S.; Berloco, M.; Caputo, L.; Settanni, L.; Alfonsi, G.; Amerio, M.; Grandi, A.; Ragni, A.; Gobbetti, M. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 2006, 157, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Nionelli, L.; Coda, R.; Gobbetti, M. Synthesis of the cancer preventive peptide lunasin by lactic acid bacteria during sourdough fermentation. Nutr. Cancer 2012, 64, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, G.L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Shannon, E.; Conlon, M.; Hayes, M. The Prebiotic Effect of Australian Seaweeds on Commensal Bacteria and Short Chain Fatty Acid Production in a Simulated Gut Model. Nutrients 2022, 14, 2163. [Google Scholar] [CrossRef]
- Mangiafico, S.S. Summary and Analysis of Extension Program Evaluation in R; Version 1.18. 1; Rutgers Cooperative Extension: New Brunswick, NJ, USA, 2016. [Google Scholar]
- Alameri, F.; Tarique, M.; Osaili, T.; Obaid, R.; Abdalla, A.; Masad, R.; Al-Sbiei, A.; Fernandez-Cabezudo, M.; Liu, S.Q.; Al-Ramadi, B.; et al. Lactic acid bacteria isolated from fresh vegetable products: Potential probiotic and postbiotic characteristics including immunomodulatory effects. Microorganisms 2022, 10, 389. [Google Scholar] [CrossRef]
- Rodrigues NP, A.; Garcia, E.F.; de Souza, E.L. Selection of lactic acid bacteria with promising probiotic aptitudes from fruit and ability to survive in different food matrices. Braz. J. Microbiol. 2021, 52, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Vitali, B.; Minervini, G.; Rizzello, C.G.; Spisni, E.; Maccaferri, S.; Brigidi, P.; Gobbetti, M.; Di Cagno, R. Novel probiotic candidates for humans isolated from raw fruits and vegetables. Food Microbiol. 2012, 31, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Gallone, B.; Voordeckers, K.; Verstrepen, K.J. Domestication of industrial microbes. Curr. Biol. 2019, 29, R381–R393. [Google Scholar] [CrossRef] [PubMed]
- Polo, A.; Cappello, C.; Carafa, I.; Da Ros, A.; Baccilieri, F.; Di Cagno, R.; Gobbetti, M. A novel functional herbal tea containing probiotic Bacillus coagulans GanedenBC30: An in vitro study using the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). J. Funct. Foods 2022, 88, 104873. [Google Scholar] [CrossRef]
- Ghosh, S.; Chowdhury, R.; Bhattacharya, P. Mixed consortia in bioprocesses: Role of microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 4283–4295. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Calasso, M.; Archetti, G.; Rizzello, C.G. Novel insights on the functional/nutritional features of the sourdough fermentation. Int. J. Food Microbiol. 2019, 302, 103–113. [Google Scholar] [CrossRef]
- Arora, K.; Carafa, I.; Fava, F.; Tuohy, K.M.; Nikoloudaki, O.; Gobbetti, M.; Di Cagno, R. Sourdough performances of the golden cereal Tritordeum: Dynamics of microbial ecology, biochemical and nutritional features. Int. J. Food Microbiol. 2022, 374, 109725. [Google Scholar] [CrossRef]
- Teixeira, J.S.; McNeill, V.; Gänzle, M.G. Levansucrase and sucrose phoshorylase contribute to raffinose, stachyose, and verbascose metabolism by lactobacilli. Food Microbiol. 2012, 31, 278–284. [Google Scholar] [CrossRef]
- Harlé, O.; Falentin, H.; Niay, J.; Valence, F.; Courselaud, C.; Chuat, V.; Maillard, M.B.; Guédon, É.; Deutsch, S.M.; Thierry, A. Diversity of the metabolic profiles of a broad range of lactic acid bacteria in soy juice fermentation. Food Microbiol. 2020, 89, 103410. [Google Scholar] [CrossRef]
- Montemurro, M.; Pontonio, E.; Gobbetti, M.; Rizzello, C.G. Investigation of the nutritional, functional and technological effects of the sourdough fermentation of sprouted flours. Int. J. Food Microbiol. 2019, 302, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; Lavermicocca, P.; De Vincenzi, M.; Giovannini, C.; Faccia, M.; Gobbetti, M. Proteolysis by sourdough lactic acid bacteria: Effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 2002, 68, 623–633. [Google Scholar] [CrossRef]
- Corsetti, A.; Gobbetti, M.; Smacchi, E. Antibacterial activity of sourdough lactic acid bacteria: Isolation of a bacteriocin-like inhibitory substance from Lactobacillus sanfranciscoC57. Food Microbiol. 1996, 13, 447–456. [Google Scholar] [CrossRef]
- Jensen, M.P.; Ardö, Y. Variation in aminopeptidase and aminotransferase activities of six cheese related Lactobacillus helveticus strains. Int. Dairy J. 2010, 20, 149–155. [Google Scholar] [CrossRef]
- Costantini, A.; Da Ros, A.; Nikoloudaki, O.; Montemurro, M.; Di Cagno, R.; Genot, B.; Gobbetti, M.; Rizzello, C.G. How cereal flours, starters, enzymes, and process parameters affect the in vitro digestibility of sourdough bread. Food Res. Int. 2022, 159, 111614. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Liu, H.; Brown, M.A.; Qiao, S. Dietary protein and gut microbiota composition and function. Curr. Protein Pept. Sci. 2019, 20, 145–154. [Google Scholar] [CrossRef]
- McDougall, G.J.; Fyffe, S.; Dobson, P.; Stewart, D. Anthocyanins from red wine–their stability under simulated gastrointestinal digestion. Phytochemistry 2005, 66, 2540–2548. [Google Scholar] [CrossRef]
- Macedo, A.C.; Vieira, M.; Poças, R.; Malcata, F.X. Peptide hydrolase system of lactic acid bacteria isolated from Serra da Estrela cheese. Int. Dairy J. 2000, 10, 769–774. [Google Scholar] [CrossRef]
- Diether, N.E.; Willing, B.P. Microbial fermentation of dietary protein: An important factor in diet–microbe–host interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef]
- Tlais, A.Z.; Da Ros, A.; Filannino, P.; Vincentini, O.; Gobbetti, M.; Di Cagno, R. Biotechnological re-cycling of apple by-products: A reservoir model to produce a dietary supplement fortified with biogenic phenolic compounds. Food Chem. 2021, 336, 127616. [Google Scholar] [CrossRef] [PubMed]
- Tlais AZ, A.; Lemos Junior WJ, F.; Filannino, P.; Campanaro, S.; Gobbetti, M.; Di Cagno, R. How microbiome composition correlates with biochemical changes during sauerkraut fermentation: A focus on neglected bacterial players and functionalities. Microbiol. Spectr. 2022, 10, e00168-22. [Google Scholar] [CrossRef] [PubMed]
- Wojtunik-Kulesza, K.; Oniszczuk, A.; Oniszczuk, T.; Combrzyński, M.; Nowakowska, D.; Matwijczuk, A. Influence of in vitro digestion on composition, bioaccessibility and antioxidant activity of food polyphenols—A non-systematic review. Nutrients 2020, 12, 1401. [Google Scholar] [CrossRef] [PubMed]
- Tlais AZ, A.; Fiorino, G.M.; Polo, A.; Filannino, P.; Di Cagno, R. High-value compounds in fruit, vegetable and cereal byproducts: An overview of potential sustainable reuse and exploitation. Molecules 2020, 25, 2987. [Google Scholar] [CrossRef]
- McFarland, L.V. Efficacy of single-strain probiotics versus multi-strain mixtures: Systematic review of strain and disease specificity. Dig. Dis. Sci. 2021, 66, 694–704. [Google Scholar] [CrossRef]
- Timmerman, H.M.; Koning, C.J.; Mulder, L.; Rombouts, F.M.; Beynen, A.C. Monostrain, multistrain and multispecies probiotics—A comparison of functionality and efficacy. Int. J. Food Microbiol. 2004, 96, 219–233. [Google Scholar] [CrossRef]
- Pringsulaka, O.; Rueangyotchanthana, K.; Suwannasai, N.; Watanapokasin, R.; Amnueysit, P.; Sunthornthummas, S.; Sukkhum, S.; Sarawaneeyaruk, S.; Rangsiruji, A. In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest. Sci. 2015, 174, 66–73. [Google Scholar] [CrossRef]

| Raffinose Hydrolysis | Peptidase Activity | Peptides | Total Free Phenolics | Final Score (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Strains | Residual Raffinose | Leu-p-Na | Pro-p-Na | Bread | Cheese | Chickpea Flour | Bread | Chickpea Flour | Pomegranate | Tomato | Residual Raffinose | Peptidase Activity | Peptides | Total Free Phenolics |
| Lactiplantibacillus plantarum | K2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 0 | 25 |
| P3 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 100 | 0 | 67 | 25 | |
| POM1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 33 | 0 | |
| D9.46 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 100 | 100 | 33 | 50 | |
| K13 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 100 | 0 | 25 | |
| K9 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 100 | 50 | 33 | 25 | |
| D9.40 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 50 | 33 | 0 | |
| Fin10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 50 | 0 | 0 | |
| P1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 33 | 25 | |
| K1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | |
| Fin6 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 33 | 25 | |
| D3.15 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 50 | 0 | 50 | |
| PR3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 50 | 0 | 0 | |
| D9.30 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 50 | 33 | 0 | |
| E3.19 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | |
| 1LS16 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 33 | 75 | |
| E3.13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 75 | |
| IT1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 67 | 50 | |
| ILS9 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 33 | 50 | |
| CIL6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 100 | 0 | 0 | 25 | |
| S1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 33 | 25 | |
| POM20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| POM35 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 50 | 33 | 50 | |
| 11j | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 50 | 33 | 25 | |
| S6w5 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 50 | 33 | 25 | |
| AFI5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 33 | 50 | |
| IT5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 50 | |
| DM | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 100 | 0 | 67 | 25 | |
| C5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 33 | 25 | |
| POM27 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | |
| KI-5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 33 | 25 | |
| PR14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 25 | |
| PR6 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | |
| POM42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| POM43 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 67 | 0 | |
| Lacticaseibacillus paracasei | 31a | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 100 | 67 | 25 |
| Lacticaseibacillus rhamnosus | B6.19 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 |
| Levilactobacillus brevis | MDI9 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 33 | 25 |
| Pediococcus acidilactici | LP39 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 50 | 67 | 0 |
| Pediococcus pentosaceus | 105c | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 100 | 33 | 50 |
| POM10 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 33 | 0 | |
| TLD10-14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 50 | |
| TLD10-5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 33 | 25 | |
| TLD7-12 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cappello, C.; Tlais, A.Z.A.; Acin-Albiac, M.; Lemos Junior, W.J.F.; Pinto, D.; Filannino, P.; Rinaldi, F.; Gobbetti, M.; Di Cagno, R. Identification and Selection of Prospective Probiotics for Enhancing Gastrointestinal Digestion: Application in Pharmaceutical Preparations and Dietary Supplements. Nutrients 2023, 15, 1306. https://doi.org/10.3390/nu15061306
Cappello C, Tlais AZA, Acin-Albiac M, Lemos Junior WJF, Pinto D, Filannino P, Rinaldi F, Gobbetti M, Di Cagno R. Identification and Selection of Prospective Probiotics for Enhancing Gastrointestinal Digestion: Application in Pharmaceutical Preparations and Dietary Supplements. Nutrients. 2023; 15(6):1306. https://doi.org/10.3390/nu15061306
Chicago/Turabian StyleCappello, Claudia, Ali Zein Alabiden Tlais, Marta Acin-Albiac, Wilson José Fernandes Lemos Junior, Daniela Pinto, Pasquale Filannino, Fabio Rinaldi, Marco Gobbetti, and Raffaella Di Cagno. 2023. "Identification and Selection of Prospective Probiotics for Enhancing Gastrointestinal Digestion: Application in Pharmaceutical Preparations and Dietary Supplements" Nutrients 15, no. 6: 1306. https://doi.org/10.3390/nu15061306
APA StyleCappello, C., Tlais, A. Z. A., Acin-Albiac, M., Lemos Junior, W. J. F., Pinto, D., Filannino, P., Rinaldi, F., Gobbetti, M., & Di Cagno, R. (2023). Identification and Selection of Prospective Probiotics for Enhancing Gastrointestinal Digestion: Application in Pharmaceutical Preparations and Dietary Supplements. Nutrients, 15(6), 1306. https://doi.org/10.3390/nu15061306






