Association of Gut Microbiota with Age-Related Macular Degeneration and Glaucoma: A Bidirectional Mendelian Randomization Study
Abstract
:1. Introduction
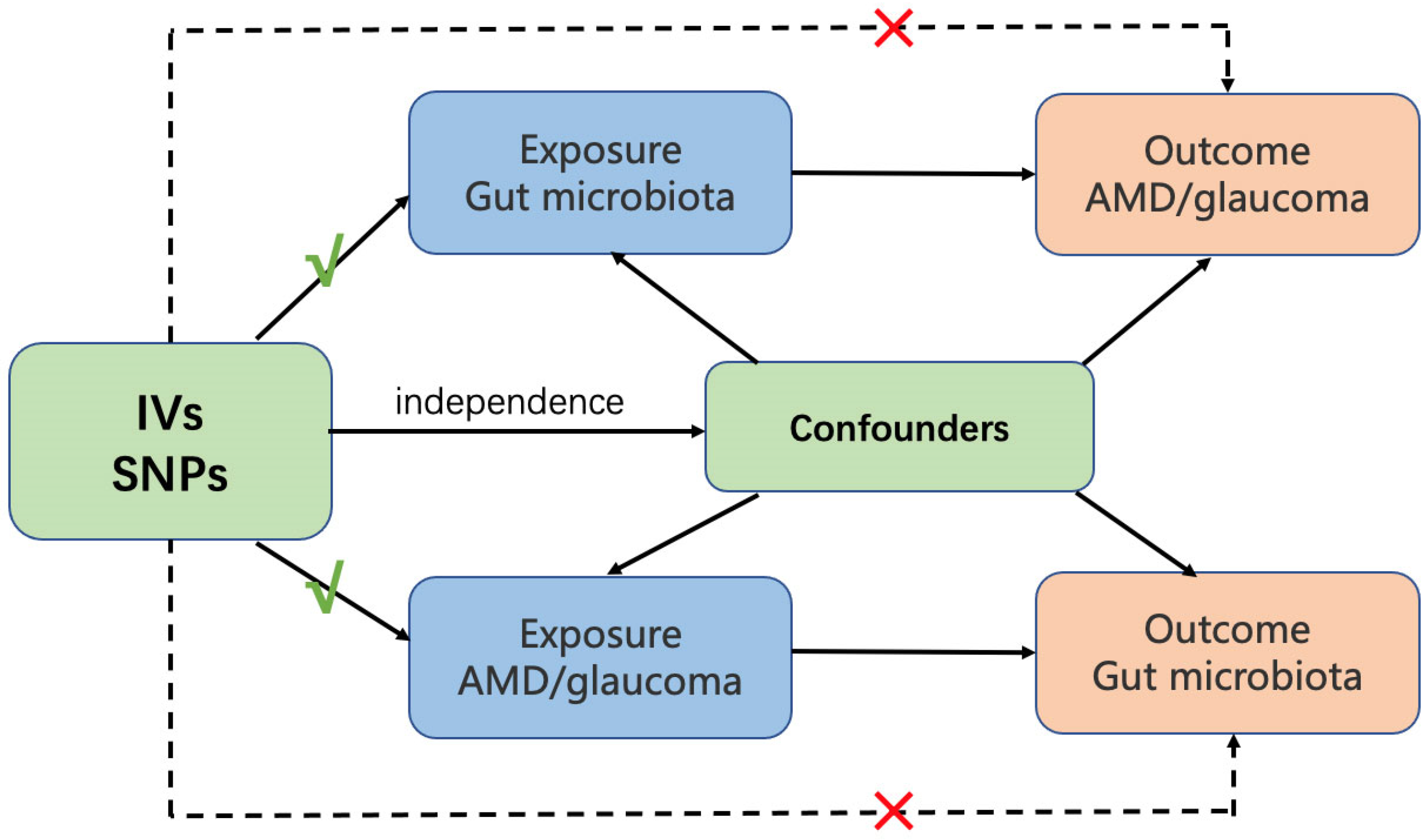
2. Materials and Methods
2.1. Data Sources
2.1.1. Gut Microbiota
2.1.2. AMD
2.1.3. Glaucoma
2.1.4. Instrumental Variables (IVs)
2.2. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.; Xie, Y.; Yuan, Y.; Xiong, R.; Hu, Y.; Ning, K.; Ha, J.; Wang, W.; Han, X.; He, M. The Mediterranean Diet and Age-Related Eye Diseases: A Systematic Review. Nutrients 2023, 15, 2043. [Google Scholar] [PubMed]
- Yao, X.; Yang, H.; Han, H.; Kou, X.; Jiang, Y.; Luo, M.; Zhou, Y.; Wang, J.; Fan, X.; Wang, X.; et al. Genome-wide analysis of genetic pleiotropy and causal genes across three age-related ocular disorders. Hum. Genet. 2023, 142, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.C.; Ma, J.Y.W.; Jobling, A.I.; Brandli, A.; Greferath, U.; Fletcher, E.L.; Vessey, K.A. Exploring the pathogenesis of age-related macular degeneration: A review of the interplay between retinal pigment epithelium dysfunction and the innate immune system. Front. Neurosci. 2022, 16, 1009599. [Google Scholar] [CrossRef] [PubMed]
- Prokosch, V.; Li, P.; Shi, X. Glaucoma as a Neurodegenerative and Inflammatory Disease. Das Glaukom ist eine neurodegenerative und neuroinflammatorische Erkrankung. Klin. Monatsblätter Augenheilkd. 2023, 240, 125–129. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; Natarajan, L.; Knight, R.; Belkaid, Y. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef]
- Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; Largiadèr, C.R.; Wittwer, M.; Wolf, S.; Zinkernagel, M.S. Associations of the intestinal microbiome with the complement system in neovascular age-related macular degeneration. NPJ Genom. Med. 2020, 5, 34. [Google Scholar] [CrossRef]
- Lima-Fontes, M.; Meira, L.; Barata, P.; Falcão, M.; Carneiro, Â. Gut microbiota and age-related macular degeneration: A growing partnership. Surv. Ophthalmol. 2022, 67, 883–891. [Google Scholar] [CrossRef]
- Gong, H.; Zhang, S.; Li, Q.; Zuo, C.; Gao, X.; Zheng, B.; Lin, M. Gut microbiota compositional profile and serum metabolic phenotype in patients with primary open-angle glaucoma. Exp. Eye Res. 2020, 191, 107921. [Google Scholar] [CrossRef]
- Gong, H.; Zeng, R.; Li, Q.; Liu, Y.; Zuo, C.; Ren, J.; Zhao, L.; Lin, M. The profile of gut microbiota and central carbon-related metabolites in primary angle-closure glaucoma patients. Int. Ophthalmol. 2022, 42, 1927–1938. [Google Scholar] [CrossRef]
- Greenland, S. An introduction to instrumental variables for epidemiologists. Int. J. Epidemiol. 2000, 29, 722–729. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Mendelian Randomization: Methods for Causal Inference Using Genetic Variants; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Liu, K.; Zou, J.; Fan, H.; Hu, H.; You, Z. Causal effects of gut microbiota on diabetic retinopathy: A Mendelian randomization study. Front. Immunol. 2022, 13, 930318. [Google Scholar] [CrossRef] [PubMed]
- Nusinovici, S.; Li, H.; Thakur, S.; Baskaran, M.; Tham, Y.-C.; Zhou, L.; Sabanayagam, C.; Aung, T.; Silver, D.; Fan, Q.; et al. High-Density Lipoprotein 3 Cholesterol and Primary Open-Angle Glaucoma: Metabolomics and Mendelian Randomization Analyses. Ophthalmology 2022, 129, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wu, P.; Zou, J.; Fan, H.; Hu, H.; Cheng, Y.; He, F.; Liu, J.; You, Z. Mendelian randomization analysis reveals causal relationships between gut microbiome and optic neuritis. Hum. Genet. 2022, 28, 1139–1148. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Garay, J.A.R.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Winkler, T.W.; Grassmann, F.; Brandl, C.; Kiel, C.; Günther, F.; Strunz, T.; Weidner, L.; Zimmermann, M.E.; Korb, C.A.; Poplawski, A.; et al. Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med. Genom. 2020, 13, 120. [Google Scholar] [CrossRef]
- Craig, J.E.; Han, X.; Qassim, A.; Hassall, M.; Bailey, J.N.C.; Kinzy, T.G.; Khawaja, A.P.; An, J.; Marshall, H.; Gharahkhani, P.; et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat. Genet. 2020, 52, 160–166. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Pocock, S.J.; Simon, R. Sequential treatment assignment with balancing for prognostic factors in the controlled clinical trial. Biometrics 1975, 31, 103–115. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef]
- Scott, N.W.; McPherson, G.C.; Ramsay, C.R.; Campbell, M.K. The method of minimization for allocation to clinical trials. A review. Control. Clin. Trials 2002, 23, 662–674. [Google Scholar] [CrossRef]
- Hemani, G.; Tilling, K.; Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017, 13, e1007081. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Chen, M.; Xu, H. Parainflammation, chronic inflammation, and age-related macular degeneration. J. Leukoc. Biol. 2015, 98, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Horai, R.; Zárate-Bladés, C.R.; Dillenburg-Pilla, P.; Chen, J.; Kielczewski, J.L.; Silver, P.B.; Jittayasothorn, Y.; Chan, C.-C.; Yamane, H.; Honda, K.; et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity 2015, 43, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, X.; Lu, Y. Gut microbiota and derived metabolomic profiling in glaucoma with progressive neurodegeneration. Front. Cell. Infect. Microbiol. 2022, 12, 968992. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Ranjith, K.; Dutta, A.; Pinna, N.K.; Mande, S.S.; Sharma, S.; Garg, P.; Murthy, S.I.; Shivaji, S. Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS ONE 2018, 13, e0199640. [Google Scholar] [CrossRef]
- Moon, J.; Choi, S.H.; Yoon, C.H.; Kim, M.K. Gut dysbiosis is prevailing in Sjögren’s syndrome and is related to dry eye severity. PLoS ONE 2020, 15, e0229029. [Google Scholar] [CrossRef]
- Wang, J.; Qie, J.; Zhu, D.; Zhang, X.; Zhang, Q.; Xu, Y.; Wang, Y.; Mi, K.; Pei, Y.; Liu, Y.; et al. The landscape in the gut microbiome of long-lived families reveals new insights on longevity and aging—Relevant neural and immune function. Gut Microbes 2022, 14, 2107288. [Google Scholar] [CrossRef]
- Jayasudha, R.; Chakravarthy, S.K.; Prashanthi, G.S.; Sharma, S.; Garg, P.; I Murthy, S.; Shivaji, S. Alterations in gut bacterial and fungal microbiomes are associated with bacterial Keratitis, an inflammatory disease of the human eye. J. Biosci. 2018, 43, 835–856. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Sai Prashanthi, G.; Ali, M.H.; Sharma, S.; Tyagi, M.; Shivaji, S. Dysbiosis in the Gut Bacterial Microbiome of Patients with Uveitis, an Inflammatory Disease of the Eye. Indian J. Microbiol. 2018, 58, 457–469. [Google Scholar] [CrossRef]
- Low, L.; Suleiman, K.; Shamdas, M.; Bassilious, K.; Poonit, N.; Rossiter, A.E.; Acharjee, A.; Loman, N.; Murray, P.I.; Wallace, G.R.; et al. Gut Dysbiosis in Ocular Mucous Membrane Pemphigoid. Front. Cell. Infect. Microbiol. 2022, 12, 780354. [Google Scholar] [CrossRef]
- Zinkernagel, M.S.; Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; Leichtle, A.B.; Largiadèr, C.R.; Fiedler, G.M.; Wolf, S. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci. Rep. 2017, 7, 40826. [Google Scholar] [CrossRef] [PubMed]
- Lin, P. Importance of the intestinal microbiota in ocular inflammatory diseases: A review. Clin. Exp. Ophthalmol. 2019, 47, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016, 40, 597–608. [Google Scholar] [CrossRef] [PubMed]
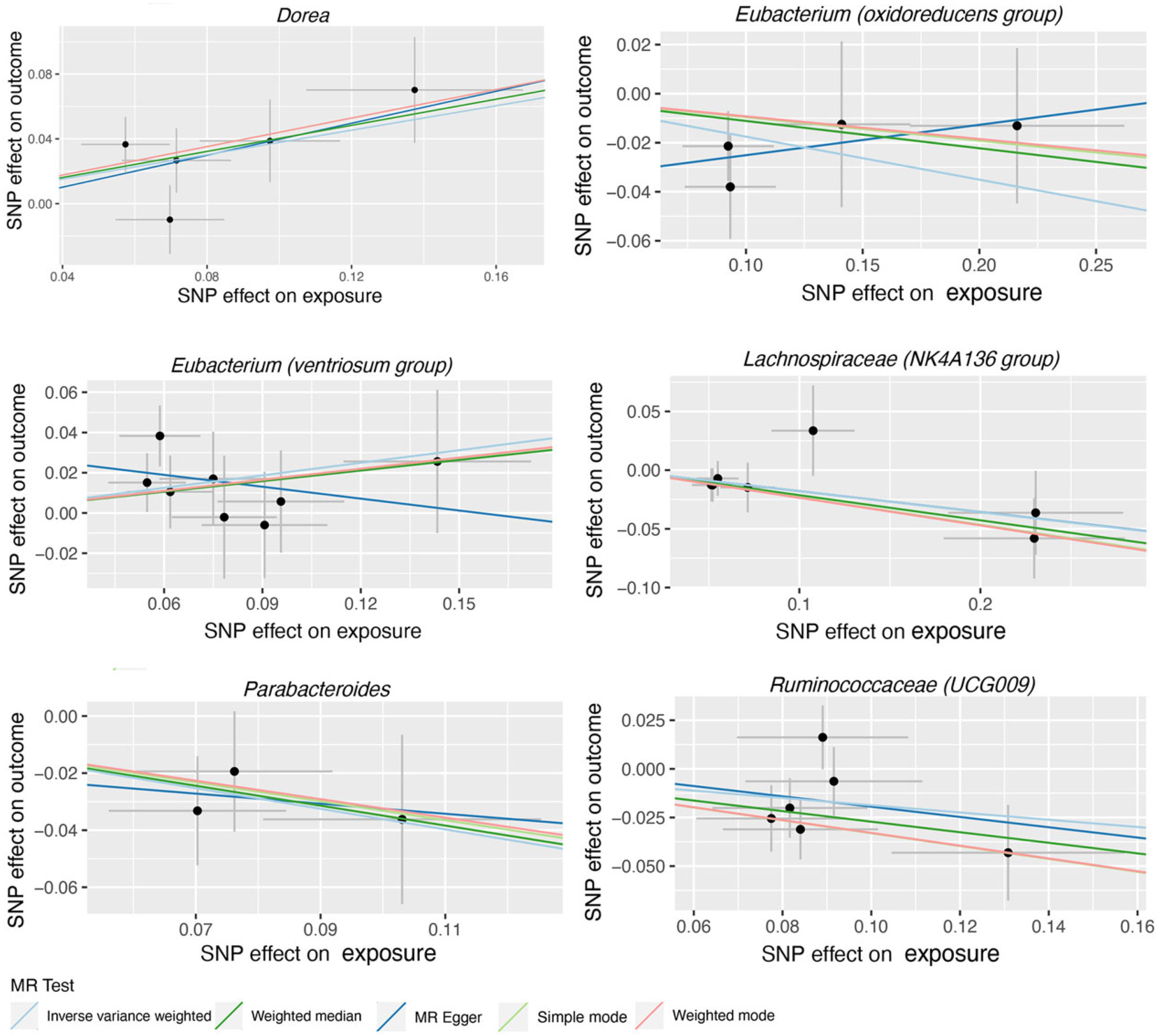
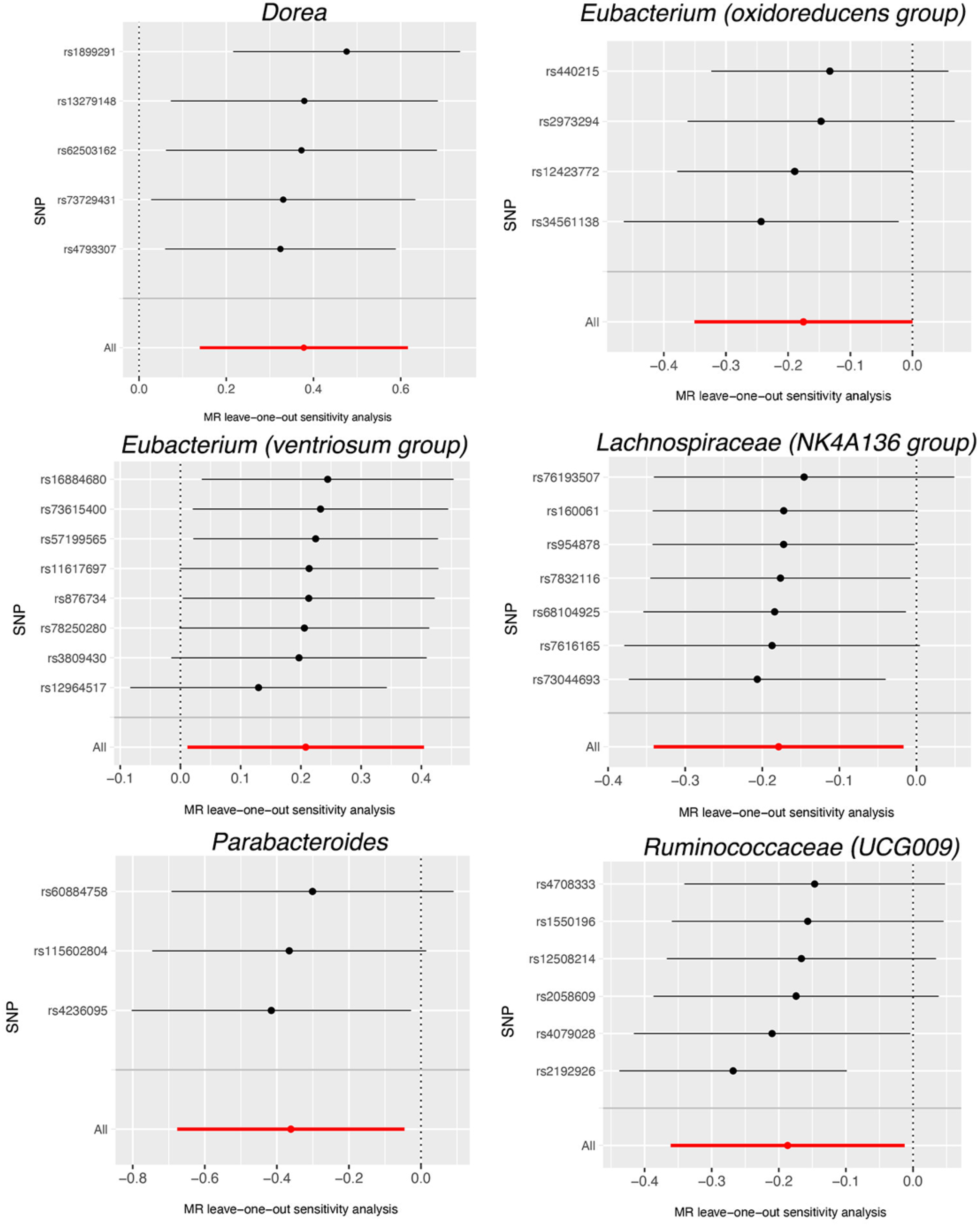
| Bacterial Taxa (Exposures) | Methods | SNPs | OR | 95% CI | p |
|---|---|---|---|---|---|
| Dorea | MR Egger | 5 | 1.64 | 0.64–4.22 | 0.382 |
| Weighted median | 5 | 1.50 | 1.08–2.08 | 0.016 | |
| IVW | 5 | 1.46 | 1.15–1.85 | 0.002 | |
| Simple mode | 5 | 1.55 | 1.01–2.39 | 0.116 | |
| Weighted mode | 5 | 1.55 | 1.04–2.33 | 0.099 | |
| Eubacterium (oxidoreducens group) | MR Egger | 4 | 1.13 | 0.67–1.90 | 0.687 |
| Weighted median | 4 | 0.89 | 0.72–1.11 | 0.318 | |
| IVW | 4 | 0.84 | 0.70–1.00 | 0.049 | |
| Simple mode | 4 | 0.91 | 0.68–1.22 | 0.572 | |
| Weighted mode | 4 | 0.91 | 0.70–1.18 | 0.547 | |
| Eubacterium (ventriosum group) | MR Egger | 8 | 0.82 | 0.41–1.66 | 0.602 |
| Weighted median | 8 | 1.19 | 0.92–1.54 | 0.175 | |
| IVW | 8 | 1.23 | 1.01–1.50 | 0.038 | |
| Simple mode | 8 | 1.20 | 0.82–1.76 | 0.380 | |
| Weighted mode | 8 | 1.20 | 0.84–1.73 | 0.355 | |
| Lachnospiraceae (NK4A136 group) | MR Egger | 7 | 0.84 | 0.63–1.11 | 0.277 |
| Weighted median | 7 | 0.81 | 0.66–0.99 | 0.041 | |
| IVW | 7 | 0.84 | 0.71–0.98 | 0.031 | |
| Simple mode | 7 | 0.79 | 0.61–1.04 | 0.143 | |
| Weighted mode | 7 | 0.79 | 0.63–1.00 | 0.093 | |
| Parabacteroides | MR Egger | 3 | 0.84 | 0.10–6.75 | 0.896 |
| Weighted median | 3 | 0.71 | 0.48–1.04 | 0.080 | |
| IVW | 3 | 0.70 | 0.51–0.96 | 0.025 | |
| Simple mode | 3 | 0.72 | 0.45–1.13 | 0.290 | |
| Weighted mode | 3 | 0.72 | 0.47–1.11 | 0.280 | |
| Ruminococcaceae (UCG009) | MR Egger | 6 | 0.77 | 0.21–2.80 | 0.709 |
| Weighted median | 6 | 0.76 | 0.62–0.94 | 0.011 | |
| IVW | 6 | 0.83 | 0.70–0.99 | 0.036 | |
| Simple mode | 6 | 0.72 | 0.53–0.98 | 0.093 | |
| Weighted mode | 6 | 0.72 | 0.52–0.99 | 0.101 |
| Bacterial Taxa (Exposures) | Methods | SNPs | OR | 95% CI | p |
|---|---|---|---|---|---|
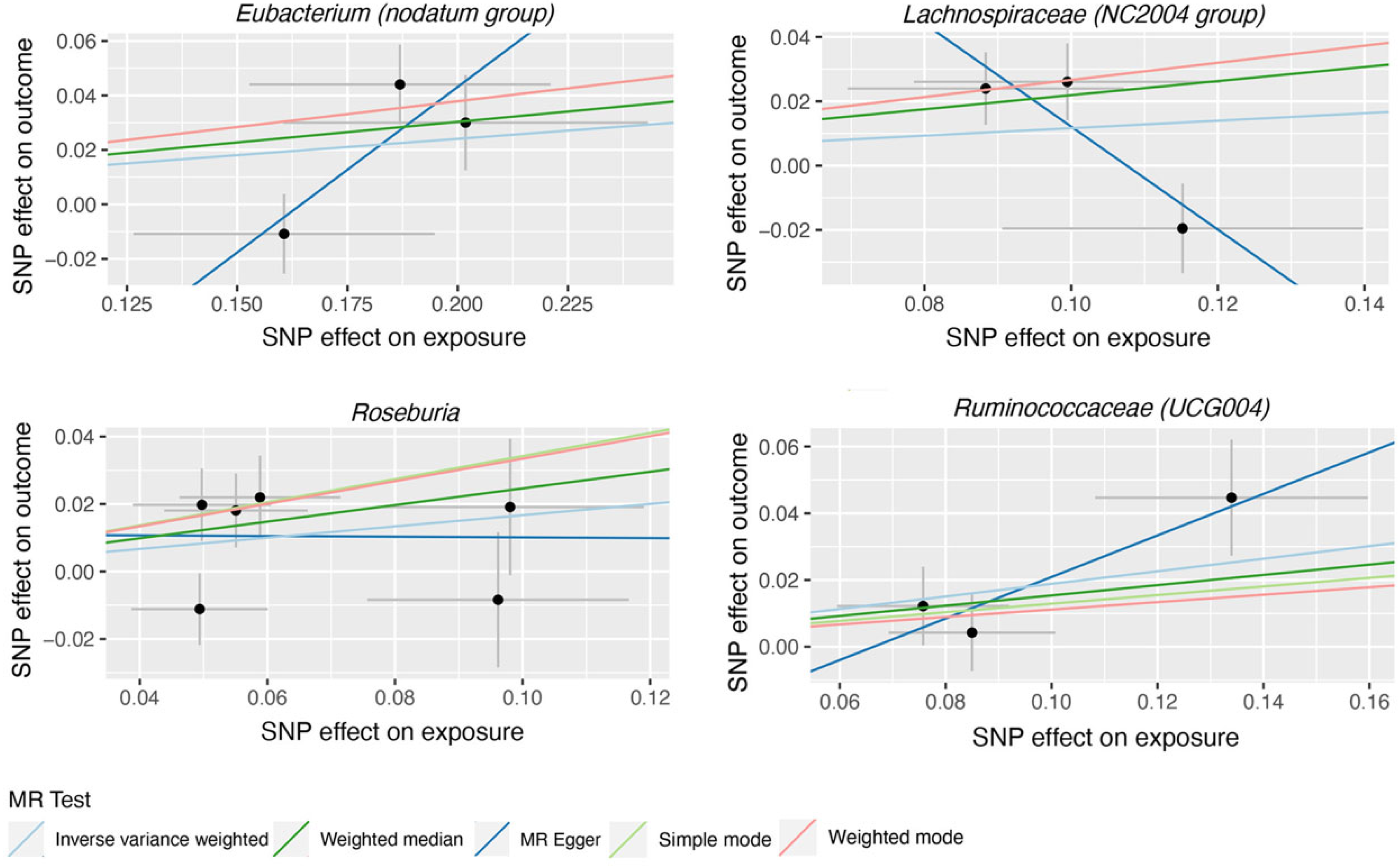
| Eubacterium (nodatum group) | MR Egger | 3 | 3.37 | 0.70–16.2 | 0.371 |
| Weighted median | 3 | 1.16 | 1.01–1.35 | 0.041 | |
| IVW | 3 | 1.13 | 0.95–1.34 | 0.173 | |
| Simple mode | 3 | 1.21 | 1.01–1.45 | 0.176 | |
| Weighted mode | 3 | 1.21 | 1.01–1.45 | 0.179 | |
| Lachnospiraceae (NC2004 group) | MR Egger | 3 | 0.20 | 0.03–1.19 | 0.328 |
| Weighted median | 3 | 1.24 | 1.03–1.51 | 0.026 | |
| IVW | 3 | 1.12 | 0.84–1.50 | 0.427 | |
| Simple mode | 3 | 1.31 | 1.01–1.68 | 0.175 | |
| Weighted mode | 3 | 1.31 | 1.02–1.68 | 0.172 | |
| Roseburia | MR Egger | 6 | 0.99 | 0.41–2.41 | 0.984 |
| Weighted median | 6 | 1.28 | 1.03–1.59 | 0.028 | |
| IVW | 6 | 1.18 | 0.96–1.45 | 0.112 | |
| Simple mode | 6 | 1.41 | 0.98–2.03 | 0.124 | |
| Weighted mode | 6 | 1.40 | 0.95–2.05 | 0.146 | |
| Ruminococcaceae (UCG004) | MR Egger | 3 | 1.86 | 0.93–3.72 | 0.328 |
| Weighted median | 3 | 1.17 | 0.94–1.45 | 0.161 | |
| IVW | 3 | 1.21 | 1.02–1.43 | 0.029 | |
| Simple mode | 3 | 1.14 | 0.88–1.47 | 0.482 |
| Bacterial Taxa (Exposures) | Methods | SNPs | OR | 95% CI | p |
|---|---|---|---|---|---|
| Dorea | MR Egger | 8 | 0.96 | 0.89–1.03 | 0.274 |
| Weighted median | 8 | 0.96 | 0.92–1.01 | 0.152 | |
| IVW | 8 | 0.96 | 0.92–1.01 | 0.098 | |
| Simple mode | 8 | 0.96 | 0.88–1.04 | 0.319 | |
| Weighted mode | 8 | 0.96 | 0.92–1.01 | 0.191 | |
| Eubacterium (oxidoreducens group) | MR Egger | 8 | 1.10 | 0.96–1.26 | 0.209 |
| Weighted median | 8 | 1.02 | 0.93–1.11 | 0.713 | |
| IVW | 8 | 1.02 | 0.95–1.10 | 0.579 | |
| Simple mode | 8 | 1.05 | 0.91–1.23 | 0.518 | |
| Weighted mode | 8 | 1.03 | 0.95–1.13 | 0.471 | |
| Eubacterium (ventriosum group) | MR Egger | 8 | 1.03 | 0.95–1.12 | 0.467 |
| Weighted median | 8 | 1.03 | 0.97–1.09 | 0.330 | |
| IVW | 8 | 1.03 | 0.99–1.08 | 0.163 | |
| Simple mode | 8 | 0.99 | 0.90–1.08 | 0.807 | |
| Weighted mode | 8 | 1.02 | 0.97–1.08 | 0.488 | |
| Lachnospiraceae (NK4A136 group) | MR Egger | 8 | 0.93 | 0.87–1.01 | 0.124 |
| Weighted median | 8 | 0.96 | 0.91–1.01 | 0.114 | |
| IVW | 8 | 0.96 | 0.92–1.01 | 0.086 | |
| Simple mode | 8 | 1.00 | 0.92–1.09 | 0.970 | |
| Weighted mode | 8 | 0.95 | 0.91–1.00 | 0.103 | |
| Parabacteroides | MR Egger | 8 | 0.94 | 0.85–1.04 | 0.302 |
| Weighted median | 8 | 0.98 | 0.94–1.03 | 0.483 | |
| IVW | 8 | 1.00 | 0.94–1.06 | 0.990 | |
| Simple mode | 8 | 0.96 | 0.88–1.04 | 0.307 | |
| Weighted mode | 8 | 0.98 | 0.93–1.03 | 0.419 | |
| Ruminococcaceae (UCG009) | MR Egger | 8 | 1.00 | 0.97–1.16 | 0.964 |
| Weighted median | 8 | 1.03 | 0.95–1.12 | 0.486 | |
| IVW | 8 | 1.04 | 0.96–1.12 | 0.308 | |
| Simple mode | 8 | 0.95 | 0.83–1.08 | 0.430 |
| Bacterial Taxa (Exposures) | Methods | SNPs | OR | 95% CI | p |
|---|---|---|---|---|---|
| Eubacterium (nodatum group) | MR Egger | 81 | 1.06 | 0.87–1.28 | 0.578 |
| Weighted median | 81 | 1.15 | 1.04–1.28 | 0.005 | |
| IVW | 81 | 1.07 | 1.00–1.14 | 0.052 | |
| Simple mode | 81 | 1.27 | 0.97–1.67 | 0.086 | |
| Weighted mode | 81 | 1.24 | 0.99–1.55 | 0.071 | |
| Lachnospiraceae (NC2004 group) | MR Egger | 75 | 0.89 | 0.77–1.03 | 0.113 |
| Weighted median | 75 | 0.95 | 0.88–1.03 | 0.217 | |
| IVW | 75 | 1.01 | 0.95–1.06 | 0.845 | |
| Simple mode | 75 | 0.93 | 0.78–1.11 | 0.433 | |
| Weighted mode | 75 | 0.92 | 0.81–1.06 | 0.263 | |
| Roseburia | MR Egger | 84 | 0.96 | 0.89–1.04 | 0.309 |
| Weighted median | 84 | 1.00 | 0.96–1.05 | 0.906 | |
| IVW | 84 | 1.00 | 0.97–1.03 | 0.996 | |
| Simple mode | 84 | 0.95 | 0.87–1.03 | 0.236 | |
| Weighted mode | 84 | 0.96 | 0.90–1.03 | 0.293 | |
| Ruminococcaceae (UCG004) | MR Egger | 83 | 0.99 | 0.89–1.11 | 0.906 |
| Weighted median | 83 | 1.05 | 0.99–1.12 | 0.113 | |
| IVW | 83 | 1.00 | 0.96–1.04 | 0.886 | |
| Simple mode | 83 | 1.08 | 0.93–1.25 | 0.322 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Lu, P. Association of Gut Microbiota with Age-Related Macular Degeneration and Glaucoma: A Bidirectional Mendelian Randomization Study. Nutrients 2023, 15, 4646. https://doi.org/10.3390/nu15214646
Li C, Lu P. Association of Gut Microbiota with Age-Related Macular Degeneration and Glaucoma: A Bidirectional Mendelian Randomization Study. Nutrients. 2023; 15(21):4646. https://doi.org/10.3390/nu15214646
Chicago/Turabian StyleLi, Chen, and Peirong Lu. 2023. "Association of Gut Microbiota with Age-Related Macular Degeneration and Glaucoma: A Bidirectional Mendelian Randomization Study" Nutrients 15, no. 21: 4646. https://doi.org/10.3390/nu15214646
APA StyleLi, C., & Lu, P. (2023). Association of Gut Microbiota with Age-Related Macular Degeneration and Glaucoma: A Bidirectional Mendelian Randomization Study. Nutrients, 15(21), 4646. https://doi.org/10.3390/nu15214646




