MicroRNA-34a Mediates High-Fat-Induced Hepatic Insulin Resistance by Targeting ENO3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Liver and Serum Samples
2.2. Animal Experiments
2.3. IPGTT
2.4. Serum Lipids, Insulin and Homeostasis Model Assessment (HOMA)-IR
2.5. Hematoxylin–Eosin (HE) and Immunohistochemical (IHC) Staining
2.6. Cell Culture, Treatment and Transfection
2.7. Cellular Glucose Uptake, Glucose Consumption, Glucose Production and Glycogen Content Assays
2.8. Target Prediction and Luciferase Activity Assay
2.9. Western Blot
2.10. Quantitative Real-Time PCR
2.11. Statistical Analysis
3. Results
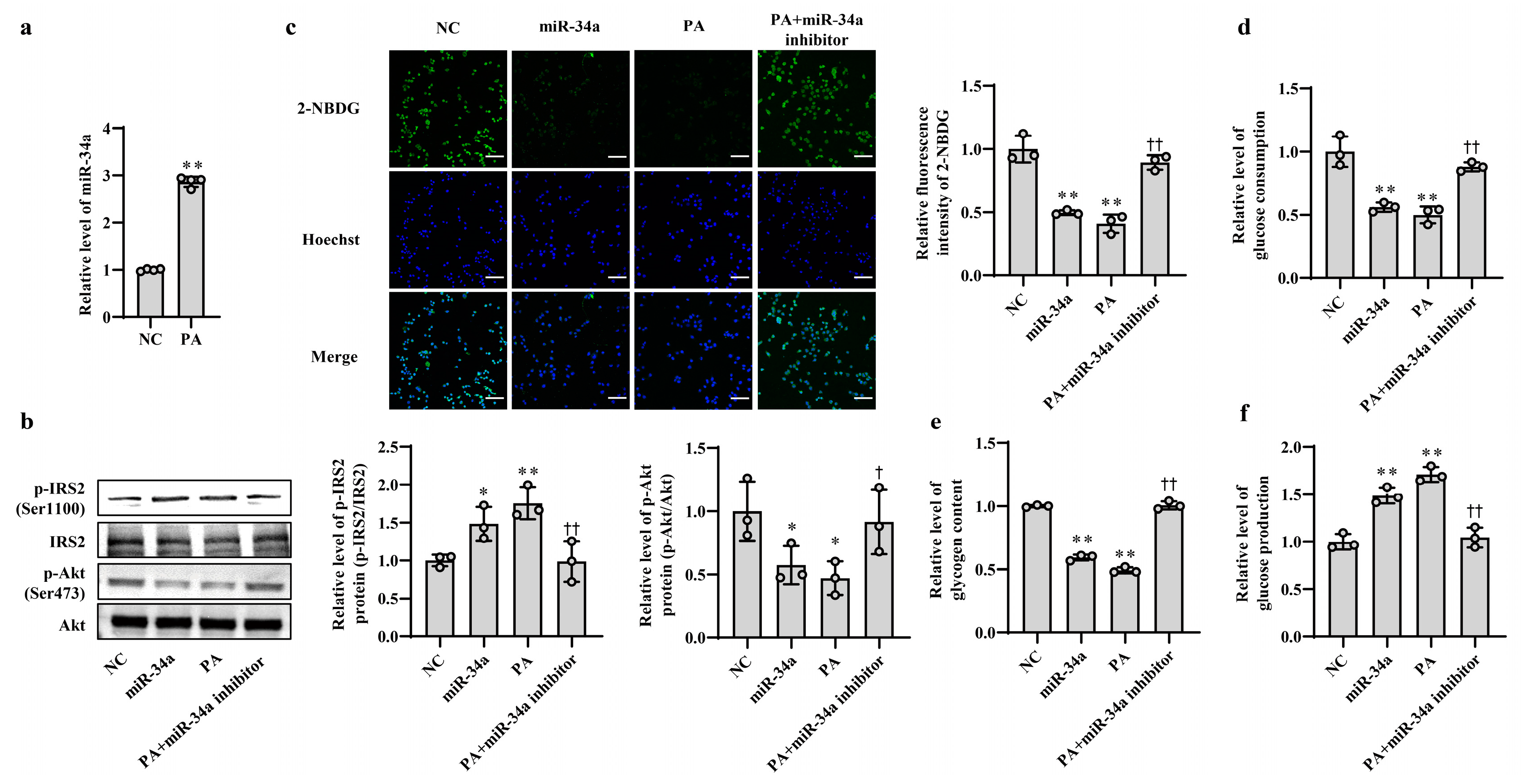
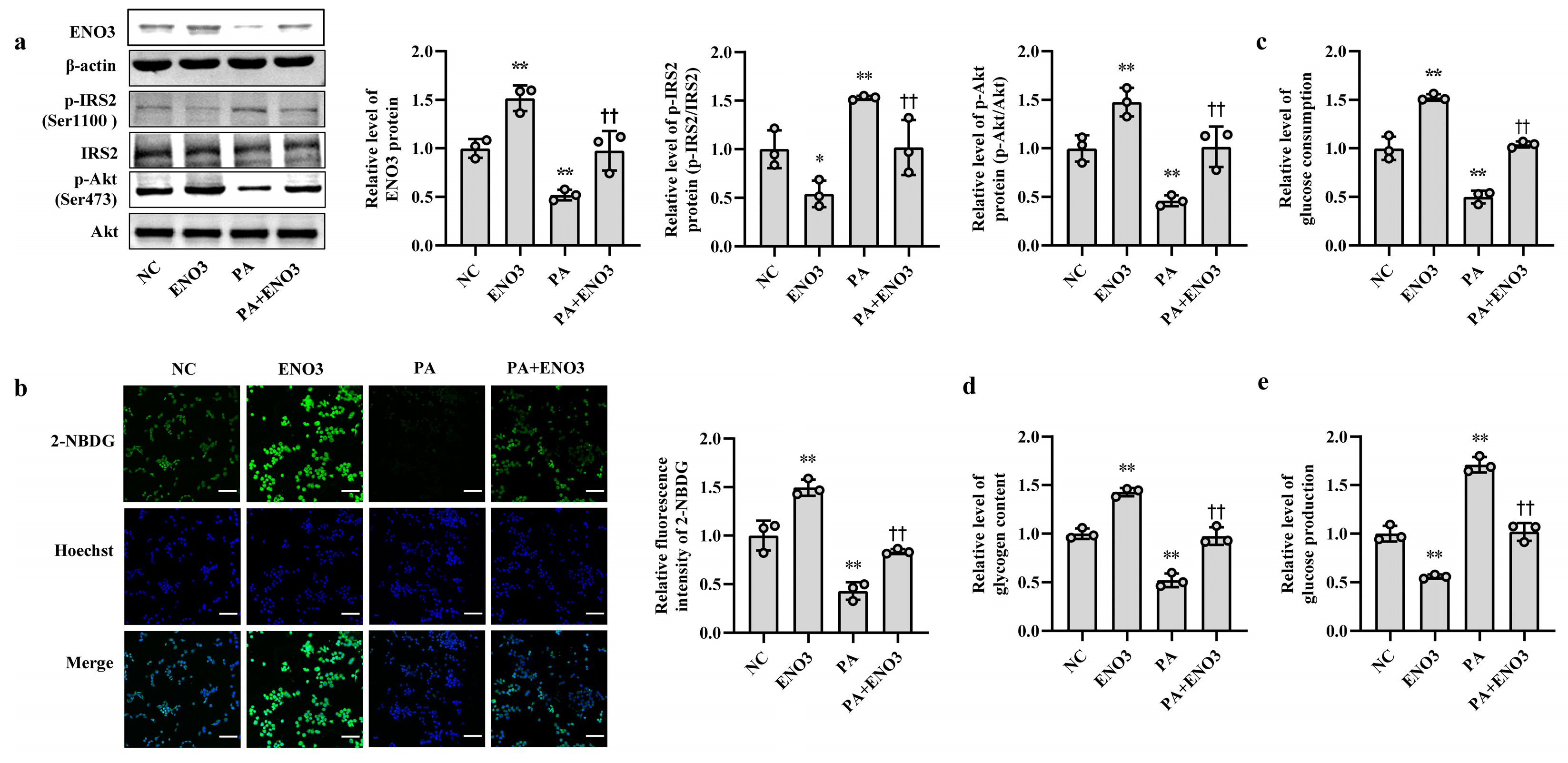
3.1. Increased miR-34a Mediates PA-Induced IR in AML12 Cells
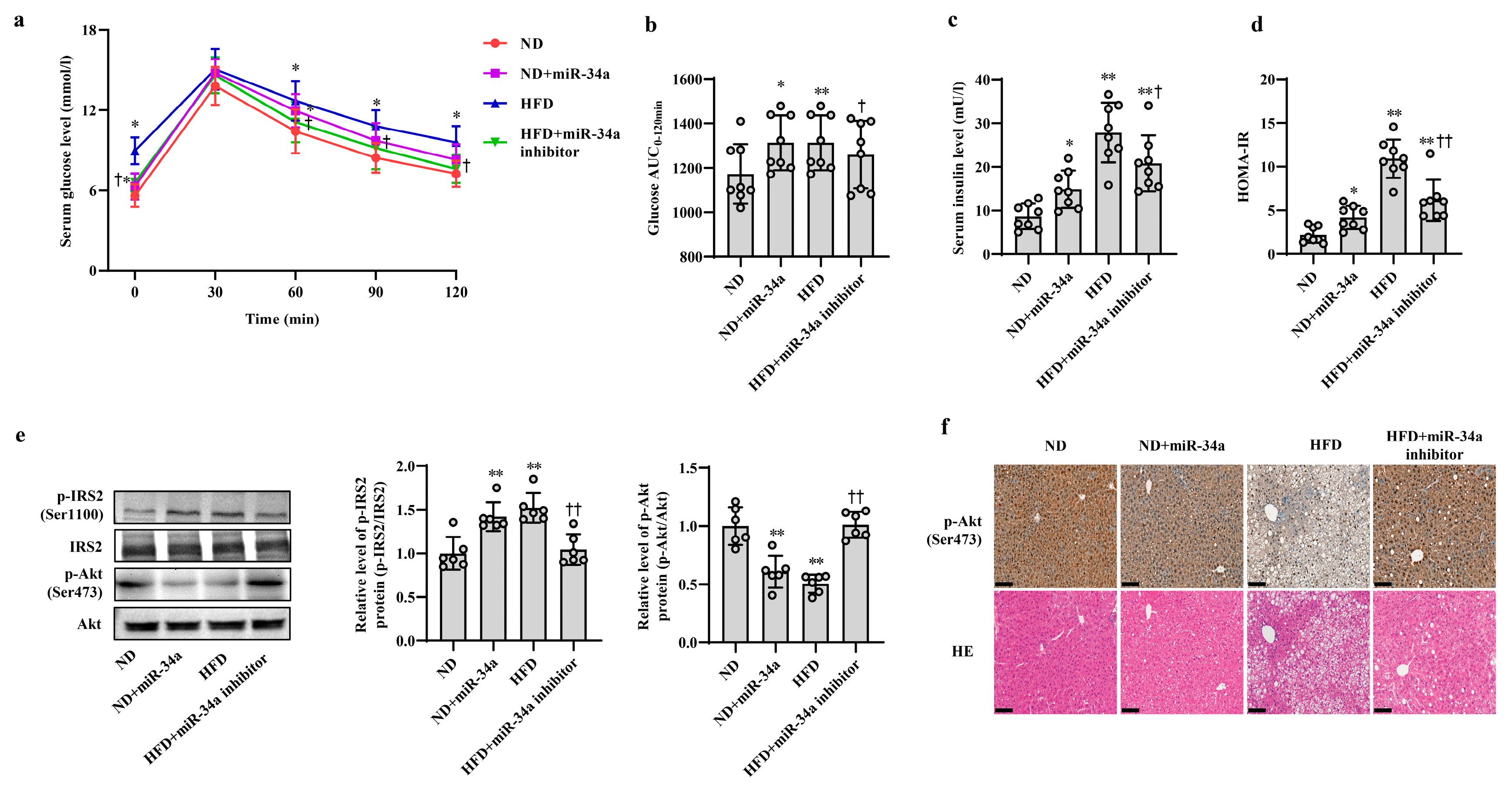
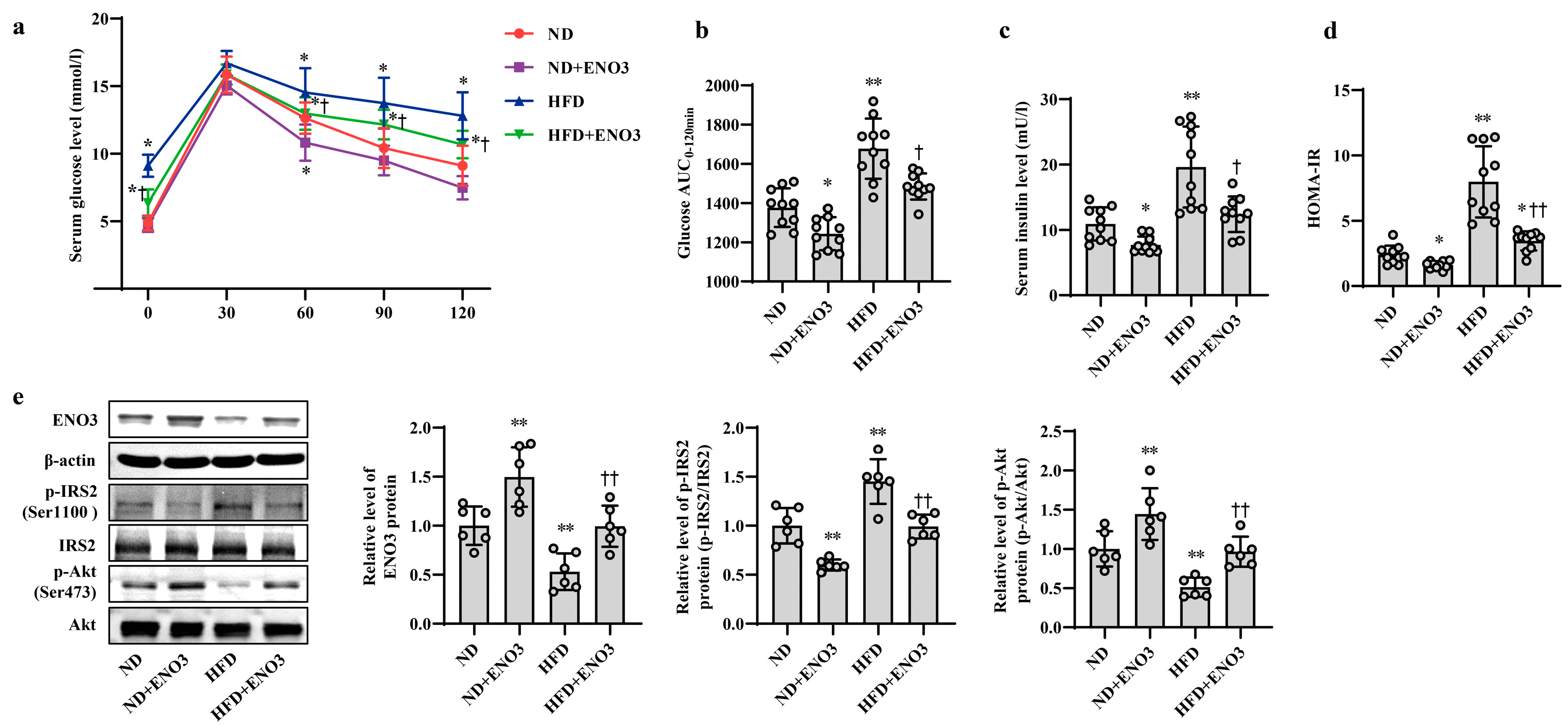
3.2. MiR-34a Contributes to HFD-Induced Hepatic IR in Mice
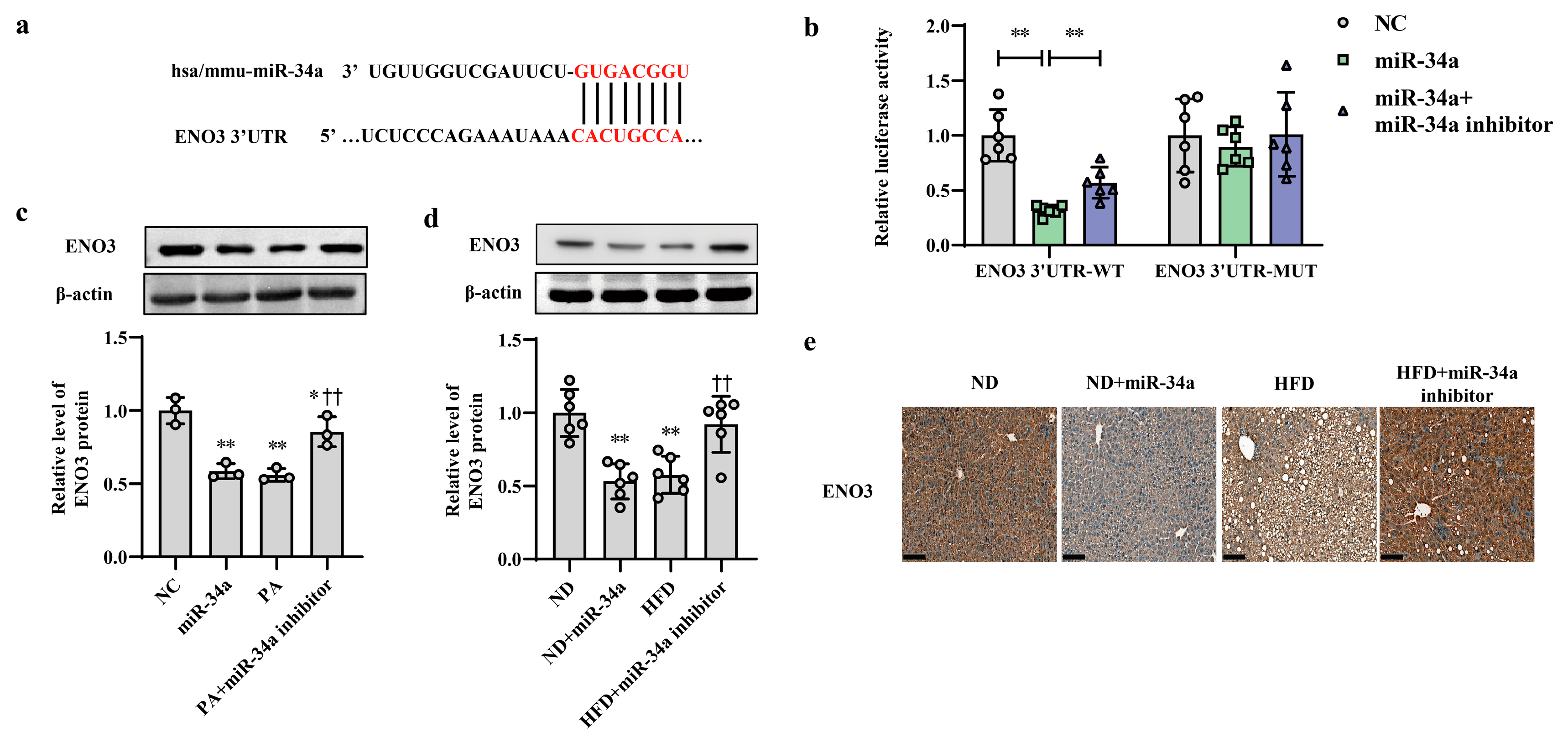
3.3. ENO3 Expression Is Specifically Reduced by miR-34a as a Direct Target Gene
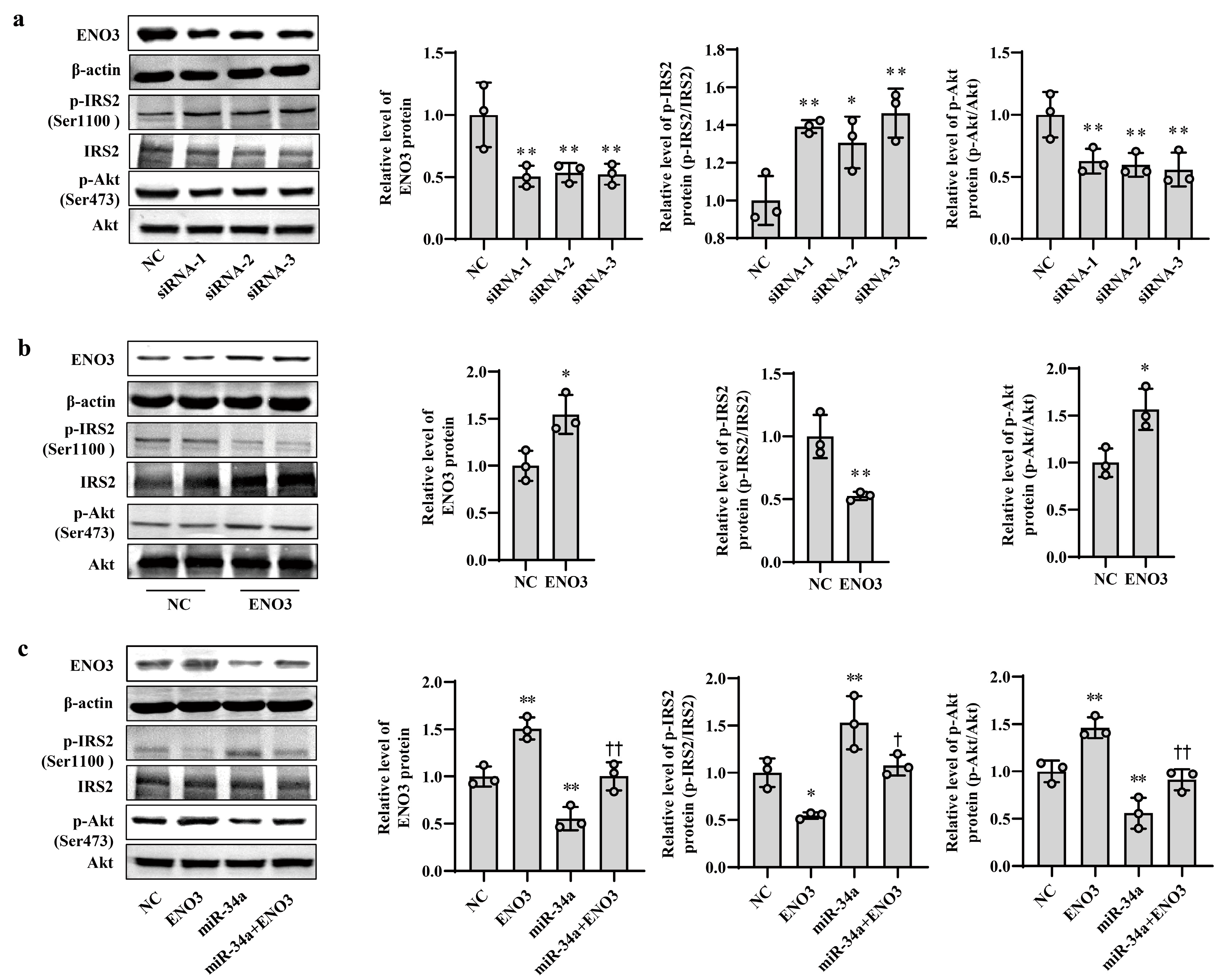
3.4. ENO3 Is Involved in PA- or HFD-Induced Hepatic IR
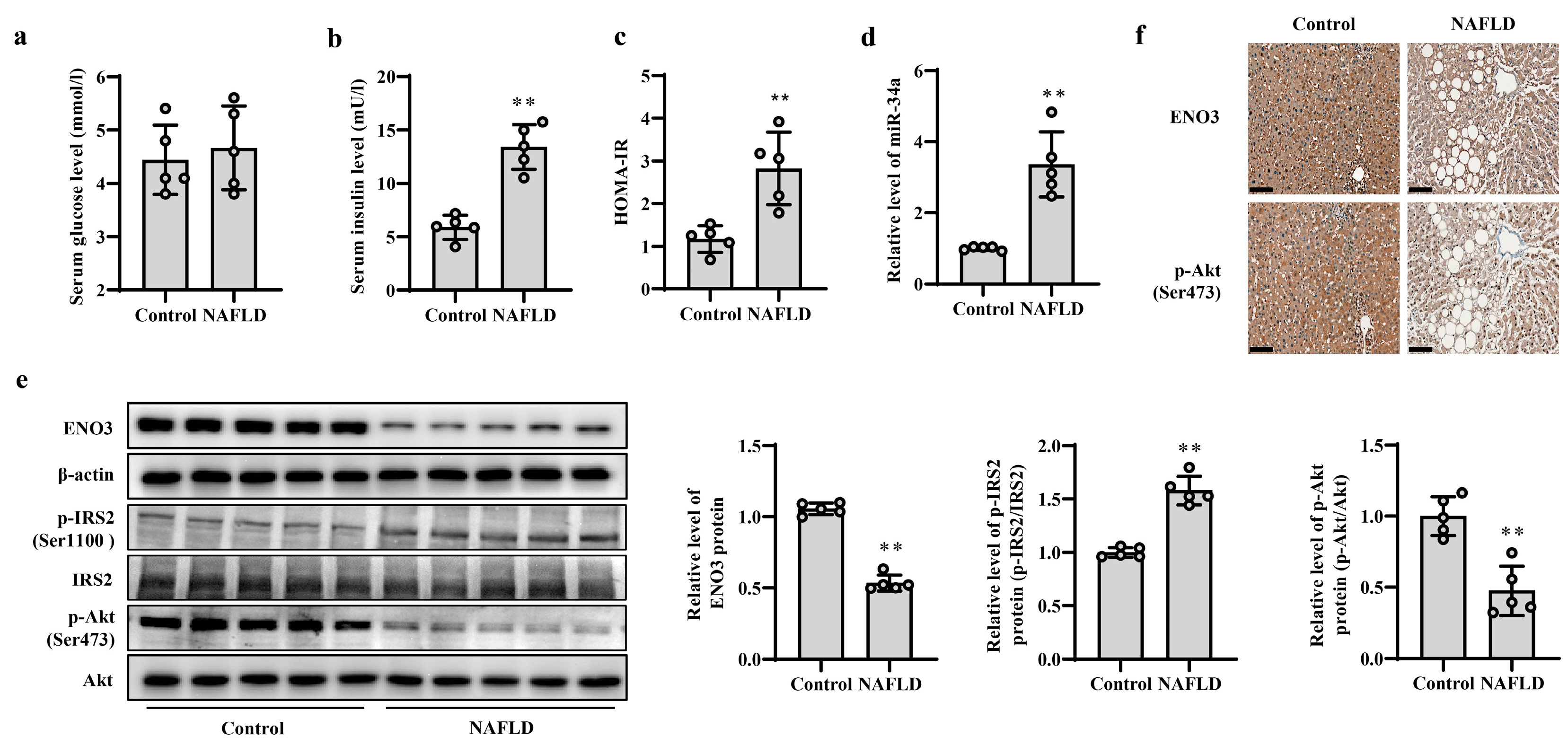
3.5. Validation of Changes in miR-34a/ENO3 Pathway in Subjects with Hepatic IR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samuel, V.T.; Shulman, G.I. Nonalcoholic Fatty Liver Disease as a Nexus of Metabolic and Hepatic Diseases. Cell Metab. 2018, 27, 22–41. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Roles of Diacylglycerols and Ceramides in Hepatic Insulin Resistance. Trends Pharmacol. Sci. 2017, 38, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Birkenfeld, A.L.; Shulman, G.I. Nonalcoholic fatty liver disease, hepatic insulin resistance, and type 2 diabetes. Hepatology 2014, 59, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Chakraborty, C.; Doss, C.G.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA 2014, 5, 697–712. [Google Scholar] [CrossRef]
- Song, Y.; Wu, L.; Li, M.; Xiong, X.; Fang, Z.; Zhou, J.; Yan, G.; Chen, X.; Yang, J.; Li, Y. Down-regulation of MicroRNA-592 in obesity contributes to hyperglycemia and insulin resistance. EBioMedicine 2019, 42, 494–503. [Google Scholar] [CrossRef]
- Du, X.; Li, X.; Chen, L.; Zhang, M.; Lei, L.; Gao, W.; Shi, Z.; Dong, Y.; Wang, Z.; Li, X.; et al. Hepatic miR-125b inhibits insulin signaling pathway by targeting PIK3CD. J. Cell. Physiol. 2018, 233, 6052–6066. [Google Scholar] [CrossRef]
- Xu, Q.; Li, Y.; Shang, Y.F.; Wang, H.L.; Yao, M.X. miRNA-103: Molecular link between insulin resistance and nonalcoholic fatty liver disease. World J. Gastroenterol. 2015, 21, 511–516. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 38, 53. [Google Scholar] [CrossRef]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Roy, S.; Dhas, Y.; Mishra, N. Senescence-associated miR-34a and miR-126 in middle-aged Indians with type 2 diabetes. Clin. Exp. Med. 2020, 20, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Jacobo, R.E.; Uresti-Rivera, E.E.; Portales-Perez, D.P.; Gonzalez-Amaro, R.; Lara-Ramirez, E.E.; Enciso-Moreno, J.A.; Garcia-Hernandez, M.H. Circulating miR-146a, miR-34a and miR-375 in type 2 diabetes patients, pre-diabetic and normal-glycaemic individuals in relation to beta-cell function, insulin resistance and metabolic parameters. Clin. Exp. Pharmacol. Physiol. 2019, 46, 1092–1100. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, Z.; Zhou, Z.; Li, Y.; Wang, Y.; Zhou, Z.; Lu, H.; Sun, C.; Chu, X. High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with high-fat diet-induced hepatic insulin resistance in mice. Genes Nutr. 2019, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, R.; Liu, C.; Ma, R.; Wang, L.; Chen, B.; Li, L.; Niu, J.; Zhao, D.; Mo, F.; et al. Evaluation of Decalcification Techniques for Rat Femurs Using HE and Immunohistochemical Staining. BioMed Res. Int. 2017, 2017, 9050754. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Zhang, W.; Liu, W.; Lv, Z.; Gui, R.; Li, M.; Li, Y.; Sun, X.; Liu, P.; et al. ETNPPL impairs autophagy through regulation of the ARG2-ROS signaling axis, contributing to palmitic acid-induced hepatic insulin resistance. Free Radic. Biol. Med. 2023, 199, 126–140. [Google Scholar] [CrossRef]
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common features of microRNA target prediction tools. Front. Genet. 2014, 5, 23. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol. Biol. 2013, 936, 117–127. [Google Scholar] [CrossRef]
- Lu, N.; Li, Y.; Qin, H.; Zhang, Y.L.; Sun, C.H. Gossypin up-regulates LDL receptor through activation of ERK pathway: A signaling mechanism for the hypocholesterolemic effect. J. Agric. Food Chem. 2008, 56, 11526–11532. [Google Scholar] [CrossRef]
- Do, G.M.; Oh, H.Y.; Kwon, E.Y.; Cho, Y.Y.; Shin, S.K.; Park, H.J.; Jeon, S.M.; Kim, E.; Hur, C.G.; Park, T.S.; et al. Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol. Nutr. Food Res. 2011, 55 (Suppl. S2), S173–S185. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Sales, E.; Jeddi, S.; Ebrahimi-Kalan, A.; Alipour, M.R. Protective Role of trans-Chalcone against the Progression from Simple Steatosis to Non-alcoholic Steatohepatitis: Regulation of miR-122, 21, 34a, and 451. Adv. Pharm. Bull. 2022, 12, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiong, Y.; Sheng, Q.; Zhao, S.; Wattacheril, J.; Flynn, C.R. A micro-RNA expression signature for human NAFLD progression. J. Gastroenterol. 2016, 51, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Chen, J.N.; Sun, F.; Wang, Y.Q.; Pan, Q.; Fan, J.G. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxidative Med. Cell. Longev. 2017, 2017, 3960197. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Qiu, Y.; Yang, Z.; Kim, H.; Qian, Q.; Sun, Q.; Zhang, C.; Yin, L.; Fang, D.; Back, S.H.; et al. IRE1alpha prevents hepatic steatosis by processing and promoting the degradation of select microRNAs. Sci. Signal 2018, 11, eaao4617. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T.M.; Kramer, D.L.; Rondinone, C.M. Serine/threonine phosphorylation of IRS-1 triggers its degradation: Possible regulation by tyrosine phosphorylation. Diabetes 2001, 50, 24–31. [Google Scholar] [CrossRef]
- Kim, B.; Leventhal, P.S.; White, M.F.; Feldman, E.L. Differential regulation of insulin receptor substrate-2 and mitogen-activated protein kinase tyrosine phosphorylation by phosphatidylinositol 3-kinase inhibitors in SH-SY5Y human neuroblastoma cells. Endocrinology 1998, 139, 4881–4889. [Google Scholar] [CrossRef]
- Copps, K.D.; White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 2012, 55, 2565–2582. [Google Scholar] [CrossRef]
- Krause, C.; Geissler, C.; Tackenberg, H.; El Gammal, A.T.; Wolter, S.; Spranger, J.; Mann, O.; Lehnert, H.; Kirchner, H. Multi-layered epigenetic regulation of IRS2 expression in the liver of obese individuals with type 2 diabetes. Diabetologia 2020, 63, 2182–2193. [Google Scholar] [CrossRef]
- Ho, J.A.; Chang, H.C.; Shih, N.Y.; Wu, L.C.; Chang, Y.F.; Chen, C.C.; Chou, C. Diagnostic detection of human lung cancer-associated antigen using a gold nanoparticle-based electrochemical immunosensor. Anal. Chem. 2010, 82, 5944–5950. [Google Scholar] [CrossRef]
- Sun, C.; Xu, B.; Liu, X.; Zhang, Z.; Su, Z. Crystal structure of enolase from Drosophila melanogaster. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Comi, G.P.; Fortunato, F.; Lucchiari, S.; Bordoni, A.; Prelle, A.; Jann, S.; Keller, A.; Ciscato, P.; Galbiati, S.; Chiveri, L.; et al. Beta-enolase deficiency, a new metabolic myopathy of distal glycolysis. Ann. Neurol. 2001, 50, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Munoz, V.R.; Gaspar, R.C.; Esteca, M.V.; Baptista, I.L.; Vieira, R.F.L.; da Silva, A.S.R.; de Moura, L.P.; Cintra, D.E.; Ropelle, E.R.; Pauli, J.R. Physical exercise increases ROCK activity in the skeletal muscle of middle-aged rats. Mech. Ageing Dev. 2020, 186, 111213. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Miotto, P.M.; De Nardo, W.; Montgomery, M.K. The Liver as an Endocrine Organ-Linking NAFLD and Insulin Resistance. Endocr. Rev. 2019, 40, 1367–1393. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Wu, P.; Zhang, Y.; Chen, Z.; Sun, J.; Chen, H. Comprehensive Analysis of NAFLD and the Therapeutic Target Identified. Front. Cell Dev. Biol. 2021, 9, 704704. [Google Scholar] [CrossRef]
- Chalasani, N.; Toden, S.; Sninsky, J.J.; Rava, R.P.; Braun, J.V.; Gawrieh, S.; Zhuang, J.; Nerenberg, M.; Quake, S.R.; Maddala, T. Noninvasive stratification of nonalcoholic fatty liver disease by whole transcriptome cell-free mRNA characterization. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G439–G449. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhao, X.; Zhang, L.; Yang, C.; Zhang, K.; Gu, Z.; Ding, H.; Li, S.; Qin, J.; Chu, X. MicroRNA-34a Mediates High-Fat-Induced Hepatic Insulin Resistance by Targeting ENO3. Nutrients 2023, 15, 4616. https://doi.org/10.3390/nu15214616
Wang Y, Zhao X, Zhang L, Yang C, Zhang K, Gu Z, Ding H, Li S, Qin J, Chu X. MicroRNA-34a Mediates High-Fat-Induced Hepatic Insulin Resistance by Targeting ENO3. Nutrients. 2023; 15(21):4616. https://doi.org/10.3390/nu15214616
Chicago/Turabian StyleWang, Yuanyuan, Xue Zhao, Liuchao Zhang, Chunxiao Yang, Kening Zhang, Zhuo Gu, Haiyan Ding, Shuangshuang Li, Jian Qin, and Xia Chu. 2023. "MicroRNA-34a Mediates High-Fat-Induced Hepatic Insulin Resistance by Targeting ENO3" Nutrients 15, no. 21: 4616. https://doi.org/10.3390/nu15214616
APA StyleWang, Y., Zhao, X., Zhang, L., Yang, C., Zhang, K., Gu, Z., Ding, H., Li, S., Qin, J., & Chu, X. (2023). MicroRNA-34a Mediates High-Fat-Induced Hepatic Insulin Resistance by Targeting ENO3. Nutrients, 15(21), 4616. https://doi.org/10.3390/nu15214616





