Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Nematodes and Growth Conditions
2.3. Lifespan Assay
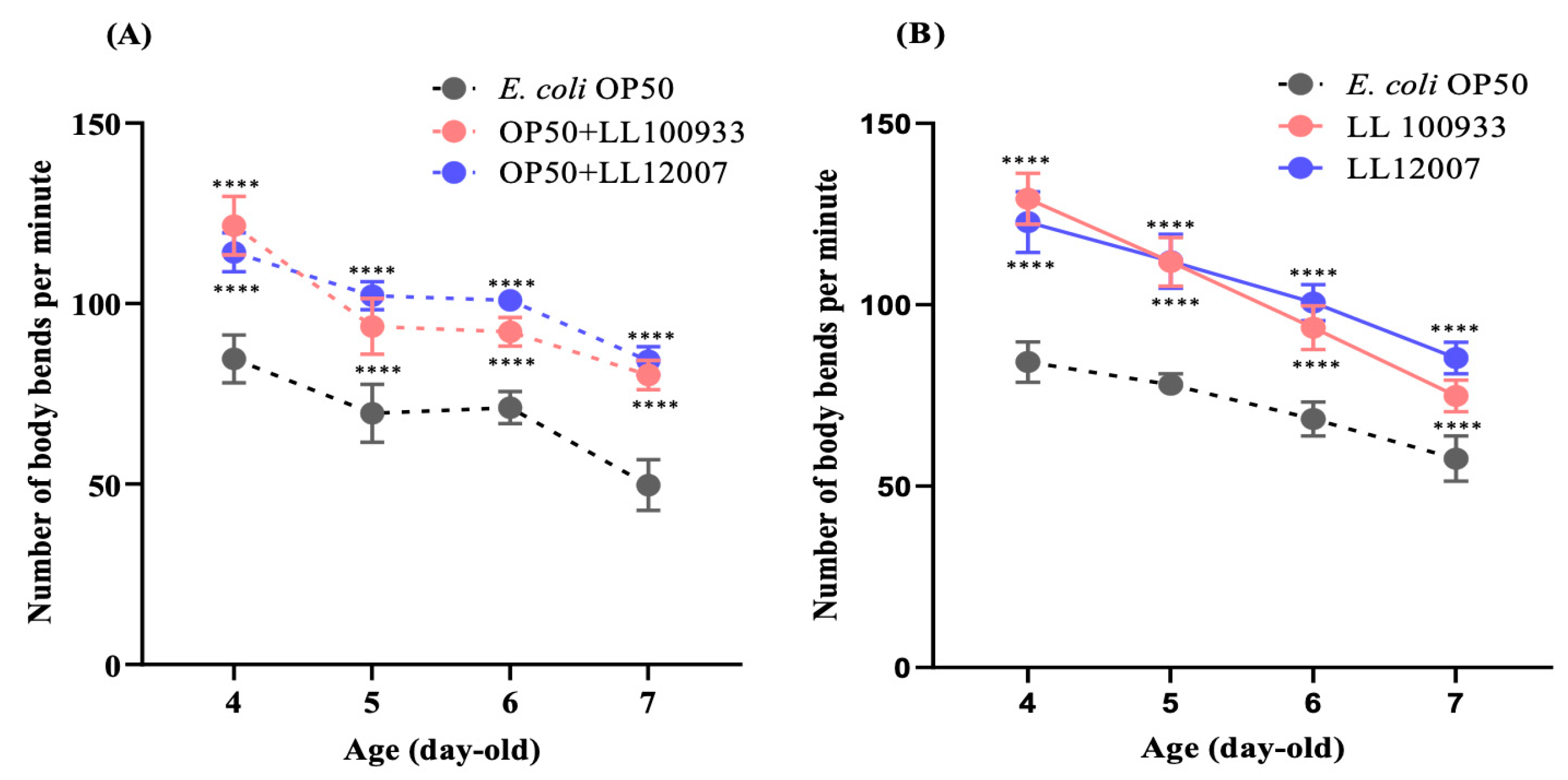
2.4. Locomotion Scoring of Nematodes
2.5. Bacterial Selection Test
2.6. Brood Size
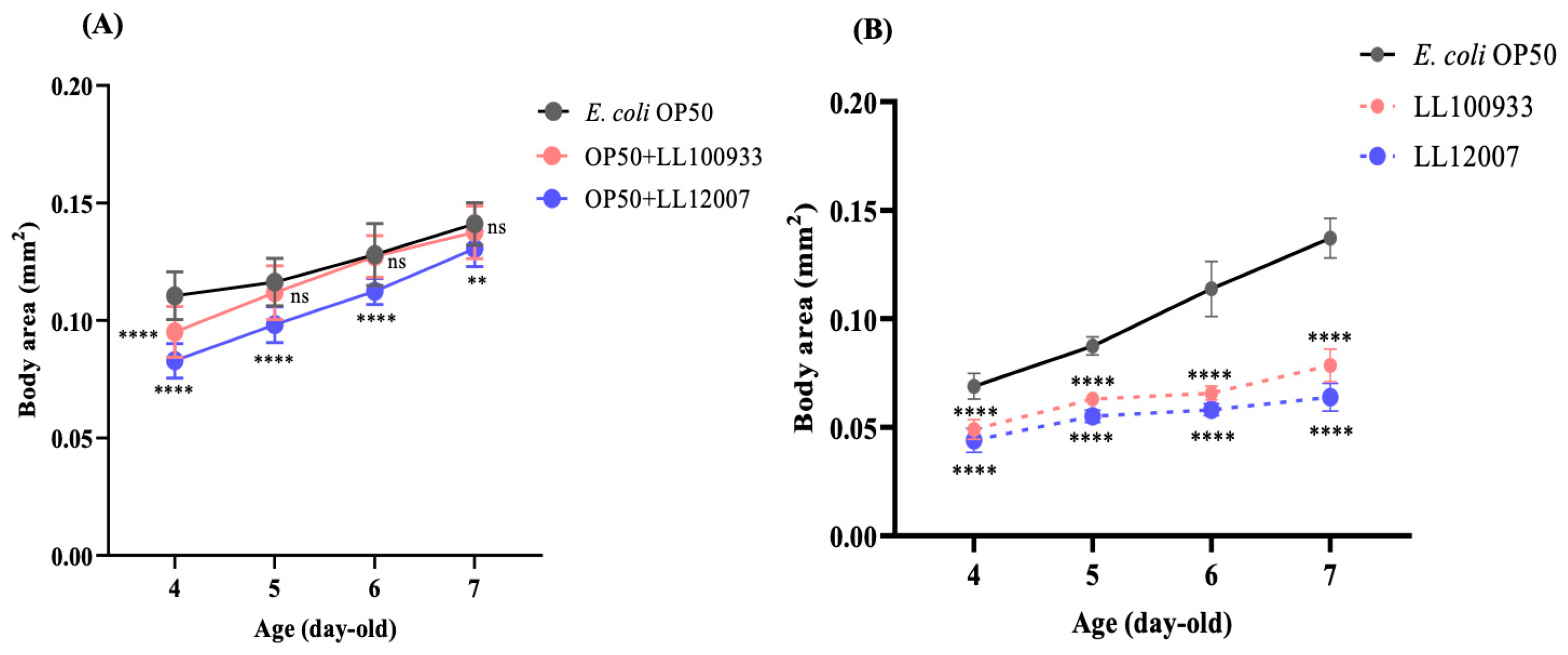
2.7. Body Size
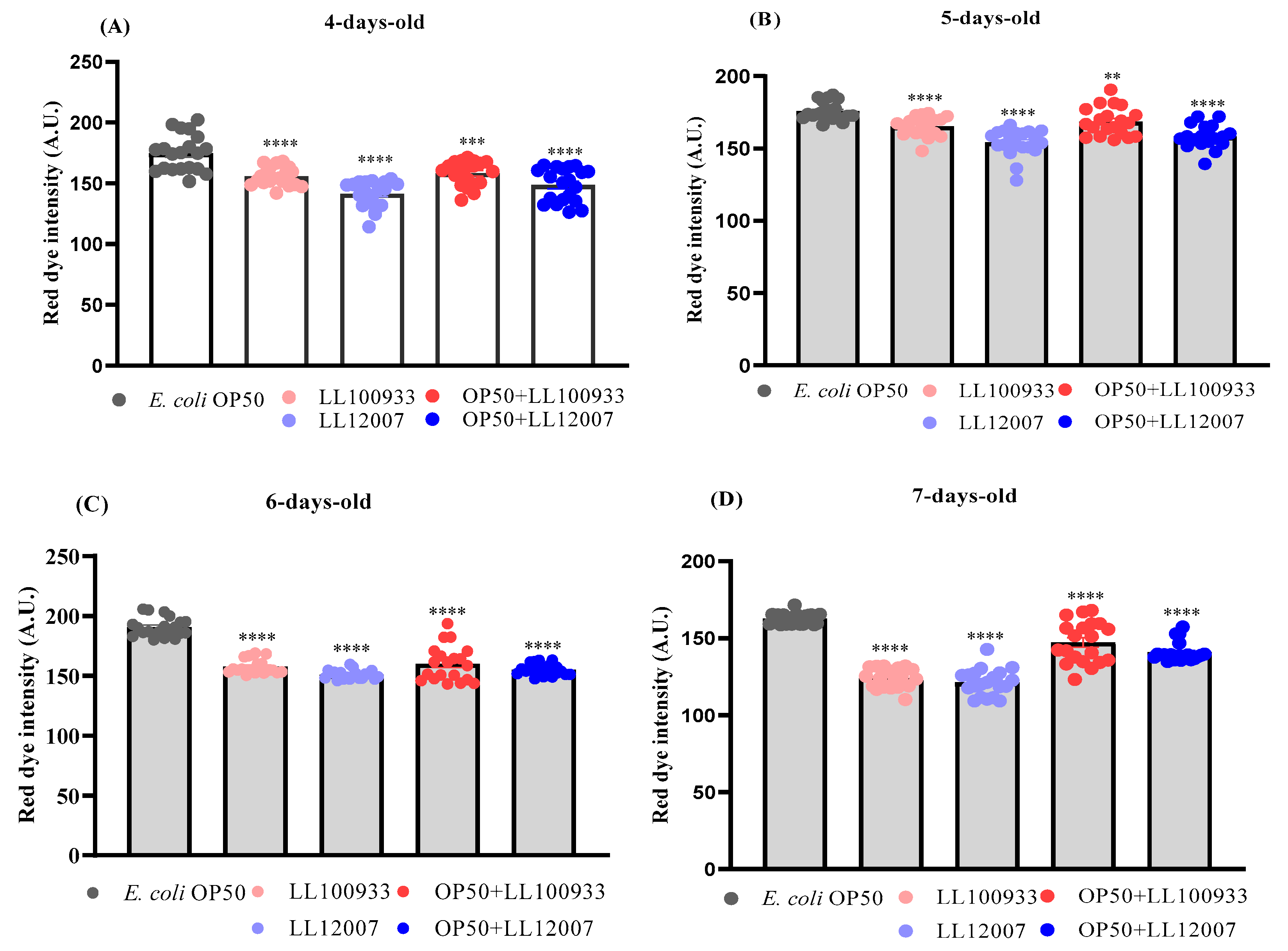
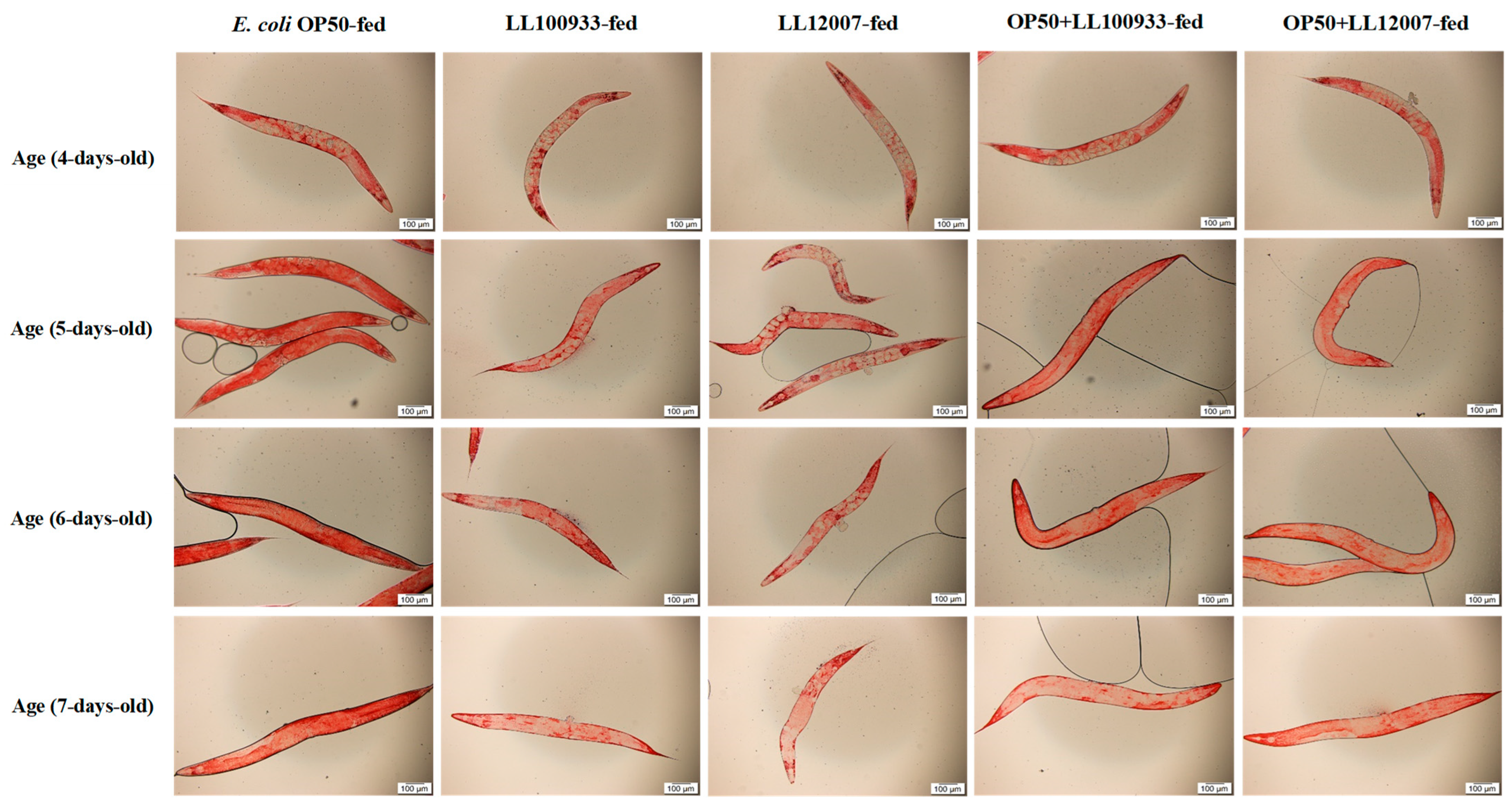
2.8. Lipid Accumulation Staining
2.9. RNA Isolation and Sequencing
2.10. Reverse Transcription and Quantitative Real-Time PCR
2.11. Statistical Analysis
3. Results
3.1. Lifespan and Locomotion of Wild-Type (N2) C. elegans
3.2. Impacts of Lactococcus Feeding in Mutants
3.3. Body and Brood Sizes
3.4. Lipid Accumulation
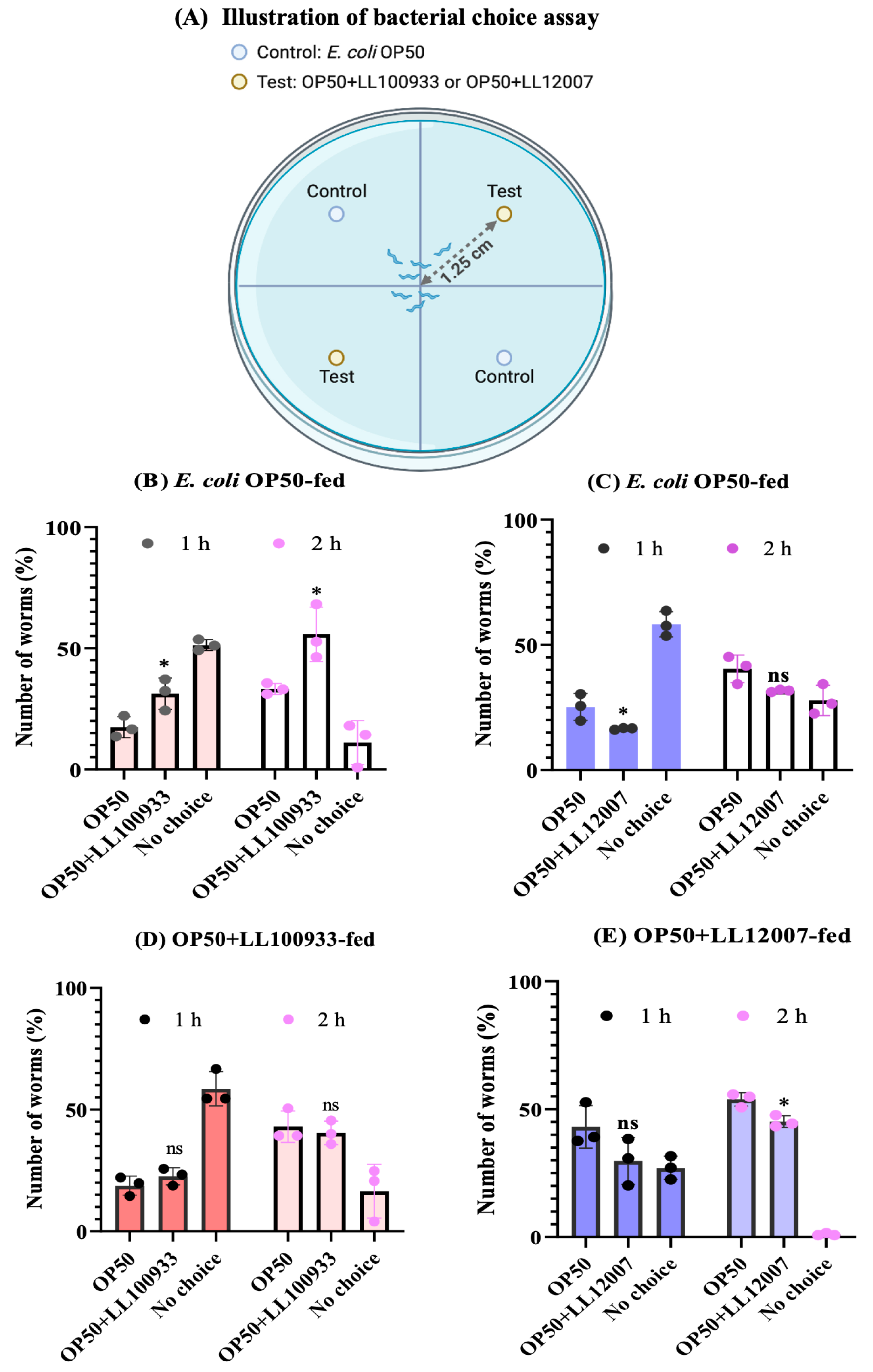
3.5. Bacterial Choice Assay
3.6. Regulation of Genes with Lactococcus Feeding
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, Z.; Hou, K.; Zhao, J.; Wang, H. The Probiotic Properties of Lactic Acid Bacteria and Their Applications in Animal Husbandry. Curr. Microbiol. 2022, 79, 22. [Google Scholar] [CrossRef]
- Sanders, M.E. Probiotics: Definition, Sources, Selection, and Uses. Clin. Infect. Dis. 2008, 46, S58–S61. [Google Scholar] [CrossRef]
- Li, T.T.; Tian, W.L.; Gu, C.T. Elevation of Lactococcus lactis subsp. cremoris to the Species Level as Lactococcus cremoris sp. nov. and Transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 71, 004727. [Google Scholar] [CrossRef]
- Jung, M.Y.; Lee, C.; Seo, M.-J.; Roh, S.W.; Lee, S.H. Characterization of a Potential Probiotic Bacterium Lactococcus Raffinolactis WiKim0068 Isolated from Fermented Vegetable Using Genomic and in Vitro Analyses. BMC Microbiol. 2020, 20, 136. [Google Scholar] [CrossRef]
- Belo, G.A.; Cordeiro, B.F.; Oliveira, E.R.; Braga, M.P.; Da Silva, S.H.; Costa, B.G.; Martins, F.D.S.; Jan, G.; Le Loir, Y.; Gala-García, A.; et al. SlpB Protein Enhances the Probiotic Potential of L. Lactis NCDO 2118 in Colitis Mice Model. Front. Pharmacol. 2021, 12, 755825. [Google Scholar] [CrossRef]
- Bandyopadhyay, B.; Das, S.; Mitra, P.K.; Kundu, A.; Mandal, V.; Adhikary, R.; Mandal, V.; Mandal, N.C. Characterization of Two New Strains of Lactococcus Lactis for Their Probiotic Efficacy over Commercial Synbiotics Consortia. Braz. J. Microbiol. 2022, 53, 903–920. [Google Scholar] [CrossRef] [PubMed]
- Sałański, P.; Kowalczyk, M.; Bardowski, J.K.; Szczepankowska, A.K. Health-Promoting Nature of Lactococcus Lactis IBB109 and Lactococcus Lactis IBB417 Strains Exhibiting Proliferation Inhibition and Stimulation of Interleukin-18 Expression in Colorectal Cancer Cells. Front. Microbiol. 2022, 13, 822912. [Google Scholar] [CrossRef] [PubMed]
- Jawan, R.; Abbasiliasi, S.; Mustafa, S.; Kapri, M.R.; Halim, M.; Ariff, A.B. In Vitro Evaluation of Potential Probiotic Strain Lactococcus Lactis Gh1 and Its Bacteriocin-Like Inhibitory Substances for Potential Use in the Food Industry. Probiotics Antimicro. Prot. 2021, 13, 422–440. [Google Scholar] [CrossRef]
- Wan, X.; Takala, T.M.; Qiao, M.; Saris, P.E.J. Complete Genome Sequence of Nisin-Producing Lactococcus lactis subsp. Lactis N8. Microbiol. Resour. Announc. 2021, 10, e01147-20. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, J.; Zhao, J.; Dorau, R.; Jensen, P.R.; Solem, C. Efficient Production of Nisin A from Low-Value Dairy Side Streams Using a Nonengineered Dairy Lactococcus lactis Strain with Low Lactate Dehydrogenase Activity. J. Agric. Food Chem. 2021, 69, 2826–2835. [Google Scholar] [CrossRef]
- Finch, C.E.; Ruvkun, G. The Genetics of Aging. Annu. Rev. Genom. Hum. Genet. 2001, 2, 435–462. [Google Scholar] [CrossRef] [PubMed]
- Roselli, M.; Schifano, E.; Guantario, B.; Zinno, P.; Uccelletti, D.; Devirgiliis, C. Caenorhabditis elegans and Probiotics Interactions from a Prolongevity Perspective. Int. J. Mol. Sci. 2019, 20, 5020. [Google Scholar] [CrossRef]
- Poupet, C.; Chassard, C.; Nivoliez, A.; Bornes, S. Caenorhabditis elegans, a Host to Investigate the Probiotic Properties of Beneficial Microorganisms. Front. Nutr. 2020, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Dierking, K.; Johnke, J.; Schulenburg, H. Isolation and Characterization of the Natural Microbiota of the Model Nematode Caenorhabditis elegans. JoVE J. Vis. Exp. 2022, 186, 64249. [Google Scholar] [CrossRef]
- Komura, T.; Takemoto, A.; Kosaka, H.; Suzuki, T.; Nishikawa, Y. Prolonged Lifespan, Improved Perception, and Enhanced Host Defense of Caenorhabditis elegans by Lactococcus cremoris subsp. Cremoris. Microbiol. Spectr. 2022, 10, e00454-21. [Google Scholar] [CrossRef]
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Rea, S.; Yashin, A.; Johnson, T. Visualizing Hidden Heterogeneity in Isogenic Populations of C. elegans. Exp. Gerontol. 2006, 41, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Komura, T.; Ikeda, T.; Yasui, C.; Saeki, S.; Nishikawa, Y. Mechanism Underlying Prolongevity Induced by Bifidobacteria in Caenorhabditis elegans. Biogerontology 2013, 14, 73–87. [Google Scholar] [CrossRef]
- Gruber, J.; Ng, L.F.; Fong, S.; Wong, Y.T.; Koh, S.A.; Chen, C.-B.; Shui, G.; Cheong, W.F.; Schaffer, S.; Wenk, M.R.; et al. Mitochondrial Changes in Ageing Caenorhabditis elegans—What Do We Learn from Superoxide Dismutase Knockouts? PLoS ONE 2011, 6, e19444. [Google Scholar] [CrossRef]
- Pompa, L.; Montanari, A.; Tomassini, A.; Bianchi, M.M.; Aureli, W.; Miccheli, A.; Uccelletti, D.; Schifano, E. In Vitro Probiotic Properties and in Vivo Anti-Ageing Effects of Lactoplantibacillus Plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms 2023, 11, 1087. [Google Scholar] [CrossRef]
- Soete, G.; Betist, M.C.; Korswagen, H.C. Regulation of Caenorhabditis elegans Body Size and Male Tail Development by the Novel Gene Lon-8. BMC Dev. Biol. 2007, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, I.; Pani, B.; Gautier, L.; Smolentseva, O.; Eremina, S.; Shamovsky, I.; Katkova-Zhukotskaya, O.; Mironov, A.; Nudler, E. Glycogen Controls Caenorhabditis elegans Lifespan and Resistance to Oxidative Stress. Nat. Commun. 2017, 8, 15868. [Google Scholar] [CrossRef] [PubMed]
- Chow, Y.-L.; Sato, F. Screening of Isoquinoline Alkaloids for Potent Lipid Metabolism Modulation with Caenorhabditis elegans. Biosci. Biotechnol. Biochem. 2013, 77, 2405–2412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.-X.; Tissenbaum, H.A. A Comparative Study of Fat Storage Quantitation in Nematode Caenorhabditis elegans Using Label and Label-Free Methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Nat. Preced. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rubio-Tomás, T.; Tavernarakis, N. Lipid Metabolism and Ageing in Caenorhabditis elegans: A Complex Interplay. Biogerontology 2022, 23, 541–557. [Google Scholar] [CrossRef]
- Abada, E.A.; Sung, H.; Dwivedi, M.; Park, B.-J.; Lee, S.-K.; Ahnn, J. C. elegans Behavior of Preference Choice on Bacterial Food. Mol. Cells 2009, 28, 209–213. [Google Scholar] [CrossRef]
- Malaguarnera, G.; Leggio, F.; Vacante, M.; Motta, M.; Giordano, M.; Biondi, A.; Basile, F.; Mastrojeni, S.; Mistretta, A.; Malaguarnera, M.; et al. Probiotics in the Gastrointestinal Diseases of the Elderly. J. Nutr. Health Aging 2012, 16, 402–410. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. BioMed Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [PubMed]
- Komura, T.; Ikeda, T.; Hoshino, K.; Shibamura, A.; Nishikawa, Y. Caenorhabditis elegans as an Alternative Model to Study Senescence of Host Defense and the Prevention by Immunonutrition. In Recent Advances on Model Hosts; Mylonakis, E., Ausubel, F.M., Gilmore, M., Casadevall, A., Eds.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2012; Volume 710, pp. 19–27. ISBN 978-1-4419-5637-8. [Google Scholar]
- Park, M.R.; Ryu, S.; Maburutse, B.E.; Oh, N.S.; Kim, S.H.; Oh, S.; Jeong, S.-Y.; Jeong, D.-Y.; Oh, S.; Kim, Y. Probiotic Lactobacillus Fermentum Strain JDFM216 Stimulates the Longevity and Immune Response of Caenorhabditis elegans through a Nuclear Hormone Receptor. Sci. Rep. 2018, 8, 7441. [Google Scholar] [CrossRef]
- Oelschlaeger, T.A. Mechanisms of Probiotic Actions—A Review. Int. J. Med. Microbiol. 2010, 300, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Pincus, Z.; Slack, F.J. Developmental Biomarkers of Aging in Caenorhabditis elegans. Dev. Dyn. 2010, 239, 1306–1314. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, G.; Lim, Y.-H. Elucidating the Mechanism of Weissella-Dependent Lifespan Extension in Caenorhabditis elegans. Sci. Rep. 2015, 5, 17128. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Sun, Z.; Sun, T. Lacticaseibacillus Rhamnosus Probio-M9 Extends the Lifespan of Caenorhabditis elegans. Commun. Biol. 2022, 5, 1139. [Google Scholar] [CrossRef] [PubMed]
- Brooks, K.K.; Liang, B.; Watts, J.L. The Influence of Bacterial Diet on Fat Storage in C. elegans. PLoS ONE 2009, 4, e7545. [Google Scholar] [CrossRef]
- Yoon, S.; Cho, H.; Nam, Y.; Park, M.; Lim, A.; Kim, J.-H.; Park, J.; Kim, W. Multifunctional Probiotic and Functional Properties of Lactiplantibacillus plantarum LRCC5314, Isolated from Kimchi. J. Microbiol. Biotechnol. 2022, 32, 72–80. [Google Scholar] [CrossRef]
- Kato, M.; Hamazaki, Y.; Sun, S.; Nishikawa, Y.; Kage-Nakadai, E. Clostridium Butyricum MIYAIRI 588 Increases the Lifespan and Multiple-Stress Resistance of Caenorhabditis elegans. Nutrients 2018, 10, 1921. [Google Scholar] [CrossRef]
- Berdichevsky, A.; Viswanathan, M.; Horvitz, H.R.; Guarente, L. C. elegans SIR-2.1 Interacts with 14-3-3 Proteins to Activate DAF-16 and Extend Life Span. Cell 2006, 125, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. Different Dietary Restriction Regimens Extend Lifespan by Both Independent and Overlapping Genetic Pathways in C. elegans. Aging Cell 2009, 8, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, T.K.; Steinbaugh, M.J.; Hourihan, J.M.; Ewald, C.Y.; Isik, M. SKN-1/Nrf, Stress Responses, and Aging in Caenorhabditis elegans. Free Radic. Biol. Med. 2015, 88, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Choe, K.P.; Leung, C.K.; Miyamoto, M.M. Unique Structure and Regulation of the Nematode Detoxification Gene Regulator, SKN-1: Implications to Understanding and Controlling Drug Resistance. Drug Metab. Rev. 2012, 44, 209–223. [Google Scholar] [CrossRef]
- Spooner, P.M.; Bonner, J.; Maricq, A.V.; Benian, G.M.; Norman, K.R. Large Isoforms of UNC-89 (Obscurin) Are Required for Muscle Cell Architecture and Optimal Calcium Release in Caenorhabditis elegans. PLoS ONE 2012, 7, e40182. [Google Scholar] [CrossRef]
- Park, J.-O.; Pan, J.; Möhrlen, F.; Schupp, M.-O.; Johnsen, R.; Baillie, D.L.; Zapf, R.; Moerman, D.G.; Hutter, H. Characterization of the Astacin Family of Metalloproteases in C. elegans. BMC Dev. Biol. 2010, 10, 14. [Google Scholar] [CrossRef]
- Mahoney, T.R.; Liu, Q.; Itoh, T.; Luo, S.; Hadwiger, G.; Vincent, R.; Wang, Z.-W.; Fukuda, M.; Nonet, M.L. Regulation of Synaptic Transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol. Biol. Cell 2006, 17, 2617–2625. [Google Scholar] [CrossRef]
- Roubin, R.; Naert, K.; Popovici, C.; Vatcher, G.; Coulier, F.; Thierry-Mieg, J.; Pontarotti, P.; Birnbaum, D.; Baillie, D.; Thierry-Mieg, D. Let-756, a C. elegans Fgf Essential for Worm Development. Oncogene 1999, 18, 6741–6747. [Google Scholar] [CrossRef]
- Popovici, C.; Fallet, M.; Marguet, D.; Birnbaum, D.; Roubin, R. Intracellular Trafficking of LET-756, a Fibroblast Growth Factor of C. elegans, Is Controlled by a Balance of Export and Nuclear Signals. Exp. Cell Res. 2006, 312, 1484–1495. [Google Scholar] [CrossRef]
- Jospin, M.; Mariol, M.-C.; Segalat, L.; Allard, B. Patch Clamp Study of the UNC-105 Degenerin and Its Interaction with the LET-2 Collagen in Caenorhabditis elegans Muscle: UNC-105 Degenerin in C. elegans Muscle. J. Physiol. 2004, 557, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Falck, J.R.; Rothe, M.; Schunck, W.-H.; Menzel, R. Role of CYP Eicosanoids in the Regulation of Pharyngeal Pumping and Food Uptake in Caenorhabditis elegans. J. Lipid Res. 2015, 56, 2110–2123. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.S.; Ahmed, S.; Takeuchi, S.; Wada, T.; Kage-Nakadai, E. Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans. Nutrients 2023, 15, 4482. https://doi.org/10.3390/nu15204482
Ali MS, Ahmed S, Takeuchi S, Wada T, Kage-Nakadai E. Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans. Nutrients. 2023; 15(20):4482. https://doi.org/10.3390/nu15204482
Chicago/Turabian StyleAli, Mohammad Shaokat, Shamima Ahmed, Shino Takeuchi, Takayuki Wada, and Eriko Kage-Nakadai. 2023. "Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans" Nutrients 15, no. 20: 4482. https://doi.org/10.3390/nu15204482
APA StyleAli, M. S., Ahmed, S., Takeuchi, S., Wada, T., & Kage-Nakadai, E. (2023). Improvement of Locomotion Caused by Lactococcus lactis subsp. lactis in the Model Organism Caenorhabditis elegans. Nutrients, 15(20), 4482. https://doi.org/10.3390/nu15204482






