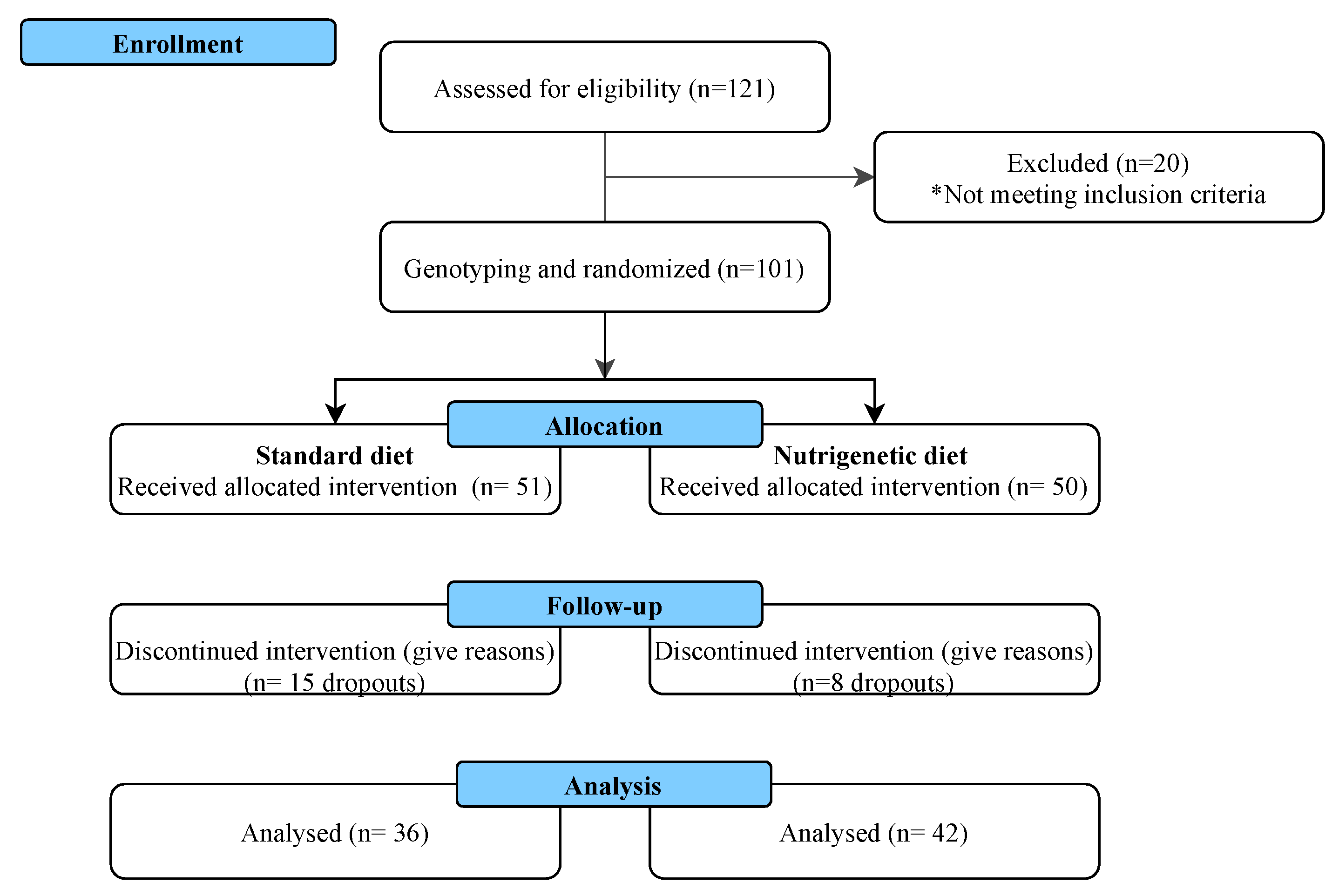
A Nutrigenetic Strategy for Reducing Blood Lipids and Low-Grade Inflammation in Adults with Obesity and Overweight
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping
2.3. Nutrigenetic Portfolios
2.4. Dietary Intake and Assessment
2.5. Anthropometric and Body Composition Measurements
2.6. Biochemical Measurements
2.7. Inflammation Markers
2.8. Statistical Analyses
2.9. Genetic Risk Score (GRS) Calculation
3. Results
3.1. Demographic, Anthropometric, and Body Composition Characteristics of the Study Population
3.2. Biochemical Profile and Inflammation Markers
| Parameter | Total Population (n = 101) | Standard Diet (n = 36) | Nutrigenetic Diet (n = 42) | p Value | ||||||
| Age (years) | 33.5 ± 9.0 | 32.8 ± 8.5 | 34.1 ± 9.5 | NS | ||||||
| Men | 49 (49%) | 26 (51%) | 23 (46%) | NS | ||||||
| Women | 52 (51%) | 25 (49%) | 27 (54%) | NS | ||||||
| Standard diet (n = 36) | Nutrigenetic Diet (n = 42) | |||||||||
| Anthropometrics and Body Composition | ||||||||||
| Parameter | Baseline | 4 Weeks | 8 Weeks | Δ | p Value | Baseline | 4 Weeks | 8 Weeks | Δ | p Value |
| Body weight (kg) | 88.0 ± 2.5 a | 85.65 ± 2.4 b | 84.98 ± 2.4 b | −3.02 ± 0.5 | <0.001 | 84.92 ± 2.3 a | 82.24 ± 2.2 b | 81.04 ± 2.2 c | −4.25 ± 0.5 | <0.001 |
| BMI (kg/m2) | 31.46 ± 0.7 a | 30.64 ± 0.7 b | 30.38 ± 0.7 b | −1.37 ± 0.5 | <0.001 | 29.86 ± 0.5 a | 28.87 ± 0.5 b | 28.44 ± 0.5 c | −1.41 ± 0.1 | <0.001 |
| Body fat (kg) | 33.91 ± 1.5 a | 32.24 ± 1.5 b | 31.35 ± 1.5 b | −2.55 ± 0.3 | <0.001 | 31.30 ± 1.2 a | 29.42 ± 1.2 b | 28.20 ± 1.2 c | −3.09 ± 0.4 | <0.001 |
| Body fat (%) | 38.27 ± 1.1 a | 37.41 ± 1.2 ab | 36.65 ± 1.2 b | −1.62 ± 0.3 | <0.001 | 36.66 ± 1.0 a | 35.49 ± 1.0 b | 34.36 ± 1.1 c | −2.30 ± 0.4 | <0.001 |
| VFAT | 15.22 ± 0.6 a | 14.56 ± 0.6 b | 14.08 ± 0.7 b | −1.13 ± 0.2 | <0.001 | 14.55 ± 0.6 a | 13.67 ± 0.6 b | 13.12 ± 0.6 c | −1.42 ± 0.2 | <0.001 |
| WC (cm) | 96.88 ± 2.2 a | 93.51 ± 1.9 b | 92.95 ± 2.1 b | −3.93 ± 0.7 | <0.001 | 93.31 ± 1.7 a | 90.43 ± 1.4 b | 89.05 ± 1.6 b | −4.25 ± 0.7 | <0.001 |
| HC (cm) | 110.84 ± 1.5 a | 108.88 ± 1.4 b | 107.74 ± 1.4 b | −3.10 ± 0.4 | <0.001 | 109.83 ± 1.7 a | 108.11 ± 1.0 b | 106.94 ± 1.1 b | −2.88 ± 0.4 | <0.001 |
| SMM (kg) | 30.32 ± 1.0 a | 30.00 ± 0.9 a | 30.02 ± 1.0 a | −0.29 ± 0.1 | 0.067 | 30.01 ± 1.0 a | 29.61 ± 1.0 b | 29.51 ± 1.0 b | 0.40 ± 0.1 | 0.057 |
| BMM (kg) | 3.75 ± 0.1 a | 3.71 ± 0.1 a | 3.72 ± 0.1 a | −0.03 ± 0.1 | 0.085 | 3.78 ± 0.1 a | 3.70 ± 0.1 a | 3.71 ± 0.1 a | −0.06 ± 0.1 | 0.074 |
| Standard Diet (n = 36) | Nutrigenetic Diet (n = 42) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Baseline | 4 Weeks | 8 Weeks | Δ | p Value | Baseline | 4 Weeks | 8 Weeks | Δ | p Value |
| TC (mg/dL) | 172.80 ± 5.5 a | 154.97 ± 5.8 b | 158.50 ± 5.3 b | −14.3 ± 3.5 | <0.001 | 177.90 ± 4.4 a | 156.40 ± 4.7 b | 161.31 ± 4.1 b | −16.59 ± 3.0 | <0.001 |
| LDL-c (mg/dL) | 104.58 ± 4.6 a | 93.05 ± 4.6 b | 94.00 ± 4.1 b | −10.58 ± 3.0 | <0.001 | 108.31 ± 3.9 a | 98.16 ± 3.8 b | 99.31 ± 3.59 b | −9.00 ± 3.1 | 0.006 |
| HDL-c (mg/dL) | 35.02 ± 1.6 a | 33.33 ± 1.5 a | 34.94 ± 1.6 a | −0.08 ± 0.8 | 0.922 | 35.0 ± 1.3 a | 35.5 ± 1.0 a | 36.5 ± 1.2 a | +1.5 ± 0.8 | 0.062 |
| TG (mg/dL) * | 166.02 ± 14.1 a | 142.47 ± 14.6 a | 152.25 ± 13.6 a | −13.7 ± 10.7 | 0.209 | 160.69 ± 11.3 a | 129.66 ± 8.9 b | 128.00 ± 8.3 b | −32.69 ± 9.6 | 0.002 * |
| VLDL- (mg/dL) * | 33.19 ± 2.8 a | 28.50 ± 2.9 a | 30.38 ± 2.7 a | −2.8 ± 2.1 | 0.199 | 32.21 ± 2.8 a | 25.90 ± 1.8 b | 25.61 ± 1.6 b | −6.6 ± 1.9 | <0.001 * |
| TG:HDL * | 4.74 ± 0.6 a | 4.27 ± 0.7 a | 4.35 ± 0.6 a | −0.39 ± 0.3 | 0.275 | 4.59 ± 0.4 a | 3.65 ± 0.3 b | 3.50 ± 0.3 b | −1.09 ± 0.4 | 0.002 * |
| Glucose (mg/dL) | 90.08 ± 2.1 a | 85.52 ± 2.1 b | 86.52 ± 2.5 ab | −3.55 ± 1.5 | 0.025 | 89.11 ± 1.6 a | 82.28 ± 1.3 b | 85.38 ± 1.4 ab | −3.73 ± 1.4 | <0.012 |
| Insulin (μUI/mL) | 19.25 ± 1.8 a | 15.00 ± 1.6 ab | 14.69 ± 1.0 b | −4.55 ± 1.7 | 0.014 | 18.33 ± 2.1 a | 14.04 ± 1.2 b | 13.66 ± 1.7 b | −4.66 ± 1.4 | <0.002 |
| HOMA-IR | 4.61 ± 0.4 a | 3.54 ± 0.4 ab | 3.43 ± 0.3 b | −1.18 ± 0.4 | 0.032 | 4.12 ± 0.5 a | 2.89 ± 0.2 b | 2.94 ± 0.4 b | −1.18 ± 0.3 | 0.029 |
| CRP (mg/L) | 11.00 ± 5.0 a | 10.36 ± 4.6 a | 10.08 ± 3.4 a | −0.91 ± 4.9 | 0.276 | 11.21 ± 7.8 a | 10.97 ± 7.06 a | 11.16 ± 8.4 a | −0.04 ± 8.9 | 0.373 |
| IL-6 (pg/mL) (n = 20) | 1.74 ± 0.45 a | 1.30 ± 0.34 a | −0.44 ± 0.20 | 0.86 | 1.69 ± 0.29 a | 0.89 ± 0.18 b | −0.80 ± 0.22 | 0.002 | ||
| TNF-α (pg/mL) (n = 20) | 0.55 ± 0.10 a | 0.34 ± 0.09 a | −0.21 ± 0.12 | 0.109 | 0.84 ± 0.25 a | 0.47 ± 0.26 b | −0.37 ± 0.10 | 0.043 | ||
3.3. Dietary Compliance
| Component | Standard Diet (n = 22) | Nutrigenetic Diet (n = 35) | p Value |
|---|---|---|---|
| Energy | 97.0 ± 2.1 | 98.0 ± 5.8 | 0.89 |
| Protein | 114.4 ± 3.9 | 96.7 ± 6.1 | 0.09 |
| Carbohydrate | 91.2 ± 2.3 | 95.5 ± 6.9 | 0.61 |
| Fat | 100.3 ± 4.0 | 113.9 ± 7.9 | 0.18 |
| Saturated fat | 125.1 ± 9.2 | 181.0 ± 13.8 | 0.005 |
| Monounsaturated fat | 64.1 ± 6.2 | 91.8 ± 7.1 | 0.009 |
| Polyunsaturated fat | 81.5 ± 10.8 | 66.1 ± 6.0 | 0.19 |
3.4. Genetic Risk Score (GRS)
| Nutrigenetic Diet | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | |||||||||
| ΔTGs | ΔVLDL | |||||||||
| Predictors | β Coefficients | Std. Error | p Value a | PC2 | p Value b | β Coefficients | Std. Error | p Value a | PC2 | p Value b |
| LIPC rs1800588 | 34.38 | 14.45 | 0.028 | 0.14 | 0.02 | 6.82 | 2.88 | 0.029 | 0.15 | 0.02 |
| LPL rs13702 | 10.68 | 24.32 | 0.665 | 0.005 | 0.66 | 2.08 | 4.85 | 0.672 | 0.005 | 0.67 |
| FABP2 rs1799883 | −21.62 | 20.44 | 0.294 | 0.03 | 0.29 | −3.90 | 4.00 | 0.341 | 0.02 | 0.34 |
| CETP rs708272 | −47.55 | 24.63 | 0.069 | 0.09 | 0.06 | −9.28 | 4.91 | 0.074 | 0.09 | 0.07 |
| APOE rs7412 and rs429358 | −28.74 | 17.16 | 0.110 | 0.07 | 0.11 | −5.36 | 3.42 | 0.134 | 0.06 | 0.13 |
| SATFAT | 0.98 | 2.23 | 0.663 | 0.005 | 0.66 | 0.18 | 0.44 | 0.680 | 0.004 | 0.68 |
| MUFAs | −1.72 | 2.08 | 0.420 | 0.01 | 0.41 | −0.33 | 0.41 | 0.432 | 0.017 | 0.43 |
| Adj. R2 | 0.49 | 0.03 | 0.29 | 0.04 | ||||||
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chew, N.W.S.; Ng, C.H.; Tan, D.J.H.; Kong, G.; Lin, C.; Chin, Y.H.; Lim, W.H.; Huang, D.Q.; Quek, J.; Fu, C.E.; et al. The Global Burden of Metabolic Disease: Data from 2000 to 2019. Cell Metab. 2023, 35, 414–428.e3. [Google Scholar] [CrossRef]
- Shamah, L.T.; Cuevas, N.L.; Romero, M.M.; Gaona, P.E.B.; Gómez, A.L.M.; Mendoza, A.L.; Méndez, G.H.I.; Rivera, D.J. Encuesta Nacional de Salud y Nutrición 2018–19. Resultados Nacionales, 1st ed.; Reveles, F., Tlapale, A., Gallegos, J., Eds.; Instituto Nacional de Salud Pública: Cuernavaca, México, 2020. [Google Scholar]
- Campos-Nonato, I.; Galván-Valencia, Ó.; Hernández-Barrera, L.; Oviedo-Solís, C.; Barquera, S. Prevalencia de Obesidad y Factores de Riesgo Asociados En Adultos Mexicanos: Resultados de La Ensanut 2022. Salud Publica Mex. 2023, 65, s238–s247. [Google Scholar] [CrossRef]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Després, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease A Scientific Statement From the American Heart Association. Circulation 2021, 143, E984–E1010. [Google Scholar] [CrossRef]
- Kirk, S.F.; Penney, T.L.; McHugh, T.L.; Sharma, A.M. Effective Weight Management Practice: A Review of the Lifestyle Intervention Evidence. Int. J. Obes. 2012, 36, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Samblas, M.; Milagro, F.I.; Martínez, A. DNA Methylation Markers in Obesity, Metabolic Syndrome, and Weight Loss. Epigenetics 2019, 14, 421–444. [Google Scholar] [CrossRef]
- De Toro-Martín, J.; Arsenault, B.J.; Després, J.P.; Vohl, M.C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef]
- Romero-Cortes, T.; Lopez-Perez, P.A.; Toledo, A.K.M.; Perez-Espana, V.H.; Aparicio-Burgos, J.E.; Cuervo-Parra, J.A. Nutrigenomics and Nutrigenetics in Functional Foods. Int. J. Bio-Resour. Stress Manag. 2018, 9, 661–672. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Martinez, J.A. Nutrigenetic Approaches in Obesity and Weight Loss. In Precision Medicine for Investigators, Practitioners and Providers; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 409–415. [Google Scholar] [CrossRef]
- Fall, T.; Mendelson, M.; Speliotes, E.K. Recent Advances in Human Genetics and Epigenetics of Adiposity: Pathway to Precision Medicine? Gastroenterology 2017, 152, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Hannon, B.A.; Khan, N.A.; Teran-Garcia, M. Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism. Nutrients 2018, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Iñiguez, I.; González-Becerra, K.; Ramos-López, O.; Pérez-Beltrán, Y.E.; Mendivil, E. Lipid-Related Genetic Variants for Personalized Dietary Interventions: A Systematic Review. Mol. Nutr. Food Res. 2023, 67, e2200675. [Google Scholar] [CrossRef]
- De Salud, S. Diagnóstico y Tratamiento de Dislipidemias (Hipercolesterolemia) En El Adulto; Secretaría de Salud: Ciudad de México, México, 2016. [Google Scholar]
- 1000 Genomes Project Consortium. 1000 Genome Project. Available online: https://www.internationalgenome.org/ (accessed on 17 September 2020).
- The 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Lutsey, P.L.; Rasmussen-Torvik, L.J.; Pankow, J.S.; Alonso, A.; Smolenski, D.J.; Tang, W.; Coresh, J.; Volcik, K.A.; Ballantyne, C.M.; Boerwinkle, E.; et al. Relation of Lipid Gene Scores to Longitudinal Trends in Lipid Levels and Incidence of Abnormal Lipid Levels among Individuals of European Ancestry the Atherosclerosis Risk in Communities (ARIC) Study. Circ. Cardiovasc. Genet. 2012, 5, 73–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pérez-Beltrán, Y.E.; Rivera-Iñiguez, I.; Gonzalez-Becerra, K.; Pérez-Naitoh, N.; Tovar, J.; Sáyago-Ayerdi, S.G.; Mendivil, E.J. Personalized Dietary Recommendations Based on Lipid-Related Genetic Variants: A Systematic Review. Front. Nutr. 2022, 9, 830283. [Google Scholar] [CrossRef] [PubMed]
- Beckett, E.L.; Jones, P.R.; Veysey, M.; Lucock, M. Nutrigenetics—Personalized Nutrition in the Genetic Age. Explor. Res. Hypothesis Med. 2017, 2, 109–116. [Google Scholar] [CrossRef]
- Langley-Evans, S. Nutrigenetics and the Early Life Origins of Health and Disease: Effects of Protein Restriction. In Principles of Nutrigenetics and Nutrigenomics: Fundamentals for Individualized Nutrition; De Caterina, R., Alfredo Martinez, J., Kohlmeier, M., Eds.; Elsevier: London, UK, 2019; pp. 113–119. [Google Scholar]
- Arnett, D.K.; Blumenthal, R.S.; Albert, M.A.; Buroker, A.B.; Goldberger, Z.D.; Hahn, E.J.; Himmelfarb, C.D.; Khera, A.; Lloyd-Jones, D.; McEvoy, J.W.; et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2019, 140, e596–e646. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.H.; Wu, J.H.Y.; Appel, L.J.; Creager, M.A.; Kris-Etherton, P.M.; Miller, M.; Rimm, E.B.; Rudel, L.L.; Robinson, J.G.; et al. Dietary Fats and Cardiovascular Disease: A Presidential Advisory from the American Heart Association. Circulation 2017, 136, e1–e23. [Google Scholar] [CrossRef]
- NOM-037-SSA2; Norma Oficial Mexicana NOM-037-SSA2-2012 Para La Prevención, Tratamiento y Control de Las Dislipidemias. Diario Oficial de la Federación: Ciudad de México, México, 2012.
- OMS. ENT: Manual STEPS Manual de Vigilancia STEPS de La OMS: El Método STEPwise de La OMS Para La Vigilancia de Los Factores de Riesgo de Las Enfermedades Crónicas. 2018. Available online: https://apps.who.int/iris/handle/10665/43580 (accessed on 20 May 2020).
- Whitworth, J.A.; Chalmers, J. World Health Organisation-International Society of Hypertension (WHO/ISH) Hypertension Guidelines. Clinical and Experimental Hypertension. Clin. Exp. Hypertens. 2004, 26, 747–752. [Google Scholar] [CrossRef]
- Tremblay, A.J.; Morrissette, H.; Gagné, J.-M.; Bergeron, J.; Gagné, C.; Couture, P. Validation of the Friedewald Formula for the Determination of Low-Density Lipoprotein Cholesterol Compared with β-Quantification in a Large Population. Clin. Biochem. 2004, 37, 785–790. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis Model Assessment: Insulin Resistance and β-Cell Function from Fasting Plasma Glucose and Insulin Concentrations in Man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Baez-Duarte, B.G.; Zamora-Gínez, I.; González-Duarte, R.; Torres-Rasgado, E.; Ruiz-Vivanco, G.; Pérez-Fuentes, R.; Guerrero-Romero, F.; Rodriguez-Moran, M.; De La Pena, J.E.; Wacher, N.; et al. Triglyceride/High-Density Lipoprotein Cholesterol (TG/HDL-C) Index as a Reference Criterion of Risk for Metabolic Syndrome (MetS) and Low Insulin Sensitivity in Apparently Healthy Subjects. Gac. Med. Mex. 2017, 153, 152–158. [Google Scholar] [PubMed]
- Hung, C.F.; Breen, G.; Czamara, D.; Corre, T.; Wolf, C.; Kloiber, S.; Bergmann, S.; Craddock, N.; Gill, M.; Holsboer, F.; et al. A Genetic Risk Score Combining 32 SNPs Is Associated with Body Mass Index and Improves Obesity Prediction in People with Major Depressive Disorder. BMC Med. 2015, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Rosberg, V.; Vishram-Nielsen, J.K.; Kristensen, A.M.D.; Pareek, M.; Sehested, T.S.G.; Nilsson, P.M.; Linneberg, A.; Palmieri, L.; Giampaoli, S.; Donfrancesco, C.; et al. Simple Cardiovascular Risk Stratification by Replacing Total Serum Cholesterol with Anthropometric Measures: The MORGAM Prospective Cohort Project. Prev. Med. Rep. 2022, 26, 101700. [Google Scholar] [CrossRef]
- Armenise, C.; Lefebvre, G.; Carayol, J.; Bonnel, S.; Bolton, J.; Di Cara, A.; Gheldof, N.; Descombes, P.; Langin, D.; Saris, W.H.M.; et al. Transcriptome Profiling from Adipose Tissue during a Low-Calorie Diet Reveals Predictors of Weight and Glycemic Outcomes in Obese, Nondiabetic Subjects. Am. J. Clin. Nutr. 2017, 106, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, E.; Garcia-Garcia, M.R.; Gonzalez-Avalos, J.M.; Maldonado-Gonzalez, M.; Ruiz-Madrigal, B.; Vizmanos, B.; Hernandez-Nazara, Z.; Roman, S.; Panduro, A. Effect of Ala54Thr Polymorphism of FABP2 on Anthropometric and Biochemical Variables in Response to a Moderate-Fat Diet. Nutrition 2013, 29, 46–51. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Izaola, O.; De La Fuente, B.; Primo, D.; Romero, E. Role of Fatty Acid-Binding Protein 2 Ala54Thr Genotype on Weight Loss and Cardiovascular Risk Factors after a High-Protein/Low-Carbohydrate versus a Standard Hypocaloric Diet during 9 Months. Ann. Nutr. Metab. 2015, 67, 81–86. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Aller, R.; Izaola, O.; Primo, D.; Urdiales, S.; Romero, E. Effects of a High-Protein/Low-Carbohydrate Diet versus a Standard Hypocaloric Diet on Weight and Cardiovascular Risk Factors: Role of a Genetic Variation in the Rs9939609 FTO Gene Variant. J. Nutrigenet. Nutr. 2015, 8, 128–136. [Google Scholar] [CrossRef]
- Pasiakos, S.M.; Cao, J.J.; Margolis, L.M.; Sauter, E.R.; Whigham, L.D.; McClung, J.P.; Rood, J.C.; Carbone, J.W.; Combs, G.F.; Young, A.J. Effects of High-Protein Diets on Fat-Free Mass and Muscle Protein Synthesis Following Weight Loss: A Randomized Controlled Trial. FASEB J. 2013, 27, 3837–3847. [Google Scholar] [CrossRef]
- De Caterina, R.; Alfredo Martinez, J.; Kohlmeier, M. Principles of Nutrigenetics and Nutrigenomics: Fundamentals for Individualized Nutrition; De Caterina, R., Alfredo Martinez, J., Kohlmeier, M., Eds.; Elsevier: London, UK, 2019. [Google Scholar] [CrossRef]
- Rashid, A.; Jafar, S.; Yaqub, R.K. Role of Peroxisome Proliferator-Activated Receptor (PPAR)-α Gene in Dyslipidemia. Rawal Med. J. 2020, 45, 54–57. [Google Scholar]
- Kohlmeier, M. Nutrigenetics. Applying the Science of Personal Nutrition; Elsevier Inc.: Alpharetta, GA, USA, 2013. [Google Scholar]
- Livingstone, K.M.; Celis-Morales, C.; Navas-Carretero, S.; San-Cristoba, R.; MacReady, A.L.; Fallaize, R.; Forster, H.; Woolhead, C.; O’Donovan, C.B.; Marsaux, C.F.M.; et al. Effect of an Internet-Based, Personalized Nutrition Randomized Trial on Dietary Changes Associated with the Mediterranean Diet: The Food4Me Study. Am. J. Clin. Nutr. 2016, 104, 288–297. [Google Scholar] [CrossRef]
- Gadde, K.M.; Martin, C.K.; Berthoud, H.R.; Heymsfield, S.B. Obesity: Pathophysiology and Management. J. Am. Coll. Cardiol. 2018, 71, 69–84. [Google Scholar] [CrossRef]
- Cunningham, E. How Can I Help My Client Who Is Experiencing a Weight-Loss Plateau? J. Am. Diet. Assoc. 2011, 111, 1966. [Google Scholar] [CrossRef]
- Candi, E.; Campanelli, M.; Sica, G.; Schinzari, F.; Rovella, V.; Di Daniele, N.; Melino, J.; Tesauro, M. Differences in the Vascular and Metabolic Profiles between Metabolically Healthy and Unhealthy Obesity. Endocr. Metab. Sci. 2021, 2, 100077. [Google Scholar] [CrossRef]
- Sameem, M.; Rani, A.; Arshad, M. Association of Rs146292819 Polymorphism in ABCA1 Gene with the Risk of Coronary Artery Disease in Pakistani Population. Biochem. Genet. 2019, 57, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Barter, P.; Gotto, A.M.; LaRosa, J.C.; Maroni, J.; Szarek, M.; Grundy, S.M.; Kastelein, J.J.P.; Bittner, V.; Fruchart, J.-C. HDL Cholesterol, Very Low Levels of LDL Cholesterol, and Cardiovascular Events. N. Engl. J. Med. 2007, 357, 1301–1310. [Google Scholar] [CrossRef]
- Garcia-Rios, A.; Alcala-Diaz, J.F.; Gomez-Delgado, F.; Delgado-Lista, J.; Marin, C.; Leon-Acuña, A.; Camargo, A.; Rodriguez-Cantalejo, F.; Blanco-Rojo, R.; Quintana-Navarro, G.; et al. Beneficial Effect of CETP Gene Polymorphism in Combination with a Mediterranean Diet Influencing Lipid Metabolism in Metabolic Syndrome Patients: CORDIOPREV Study. Clin. Nutr. 2018, 37, 229–234. [Google Scholar] [CrossRef]
- De Luis, D.A.; Izaola, O.; Primo, D.; Aller, R. Role of Rs670 Variant of APOA1 Gene on Metabolic Response after a High Fat vs. a Low Fat Hypocaloric Diets in Obese Human Subjects. J. Diabetes Complicat. 2019, 33, 249–254. [Google Scholar] [CrossRef]
- De Luis Román, D.A.; Izaola Jáuregui, O.; Primo, D.; Aller, R. Influence of Rs670 Variant of APOA1 Gene on Serum HDL Response to an Enriched-Polyunsaturated vs. an Enriched-Monounsaturated Fat Hypocaloric Diet. Nutr. Hosp. 2019, 36, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- Dellis, D.; Tsilingiris, D.; Eleftheriadou, I.; Tentolouris, A.; Sfikakis, P.P.; Dellis, G.; Karanasiou, M.; Meimari, A.; Dimosthenopoulos, C.; Lazarou, S.; et al. Carbohydrate Restriction in the Morning Increases Weight Loss Effect of a Hypocaloric Mediterranean Type Diet: A Randomized, Parallel Group Dietary Intervention in Overweight and Obese Subjects. Nutrition 2020, 71, 110578. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Sorlí, J.V.; Estruch, R.; Coltell, O.; Ortega-Azorín, C.; Portolés, O.; Martínez-González, M.Á.; Bulló, M.; Fitó, M.; Arós, F.; et al. MicroRNA-410 Regulated Lipoprotein Lipase Variant Rs13702 Is Associated with Stroke Incidence and Modulated by Diet in the Randomized Controlled PREDIMED Trial. Am. J. Clin. Nutr. 2014, 100, 719–731. [Google Scholar] [CrossRef]
- Grundy, S.M.; Stone, N.J.; Bailey, A.L.; Beam, C.; Birtcher, K.K.; Blumenthal, R.S.; Braun, L.T.; de Ferranti, S.; Faiella-Tommasino, J.; Forman, D.E.; et al. 2018 Guideline on the Management of Blood Cholesterol. J. Am. Coll. Cardiol. 2018, 73, e285–e350. [Google Scholar] [CrossRef]
- Pavía, L.A.A.; Aguilar, S.C.; Alexanderson, R.E.; Ahumada, A.M.; Alcocer, G.M.; Arenas, J.L.; Arenas, A.L.R.; Borges, V.O.; Benavides, M.A.; Cardona, E.; et al. Consenso de La Sociedad Mexicana de Cardiología En El Diagnóstico y Tratamiento de Las Dislipidemias y Aterosclerosis. Med. Interna Mex. 2020, 36, 390–413. [Google Scholar] [CrossRef]
- Sowmya, S.; Chellam, R.; Arun, A.R.; Madambath, I. Beneficial Effects of Psidium Guajava Leaf Extract on Diabetic Myocardium. Exp. Toxicol. Pathol. 2013, 65, 91–95. [Google Scholar] [CrossRef]
- Rashid, S.; Genest, J. Effect of Obesity on High-Density Lipoprotein Metabolism. Obesity 2007, 15, 2875–2888. [Google Scholar] [CrossRef]
- Li, Y.; He, P.P.; Zhang, D.W.; Zheng, X.L.; Cayabyab, F.S.; Yin, W.D.; Tang, C.K. Lipoprotein Lipase: From Gene to Atherosclerosis. Atherosclerosis 2014, 237, 597–608. [Google Scholar] [CrossRef]
- Richardson, K.; Nettleton, J.A.; Rotllan, N.; Tanaka, T.; Smith, C.E.; Lai, C.Q.; Parnell, L.D.; Lee, Y.C.; Lahti, J.; Lemaitre, R.N.; et al. Gain-of-Function Lipoprotein Lipase Variant Rs13702 Modulates Lipid Traits through Disruption of a MicroRNA-410 Seed Site. Am. J. Hum. Genet. 2013, 92, 5–14. [Google Scholar] [CrossRef]
- NOM-008-SSA3; Norma Oficial Mexicana NOM-008-SSA3-2017, Para El Tratamiento Integral Del Sobrepeso y La Obesidad. Diario Oficial de la Federación: Ciudad de México, México, 2017. [CrossRef]
- Zhu, R.; Fan, Z.; Han, Y.; Li, S.; Li, G.; Wang, L.; Ye, T.; Zhao, W. Acute Effects of Three Cooked Non-Cereal Starchy Foods on Postprandial Glycemic Responses and in Vitro Carbohydrate Digestion in Comparison with Whole Grains: A Randomized Trial. Nutrients 2019, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Tovar, J.; Johansson, M.; Björck, I. A Multifunctional Diet Improves Cardiometabolic-Related Biomarkers Independently of Weight Changes: An 8-Week Randomized Controlled Intervention in Healthy Overweight and Obese Subjects. Eur. J. Nutr. 2016, 55, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Claro-Cala, C.M.; Jiménez-Altayó, F.; Zagmutt, S.; Rodriguez-Rodriguez, R. Molecular Mechanisms Underlying the Effects of Olive Oil Triterpenic Acids in Obesity and Related Diseases. Nutrients 2022, 14, 1606. [Google Scholar] [CrossRef]
- Sung, H.-Y.; Kang, S.-W.; Kim, J.-L.; Li, J.; Lee, E.-S.; Gong, J.-H.; Han, S.J.; Kang, Y.-H. Oleanolic Acid Reduces Markers of Differentiation in 3T3-L1 Adipocytes. Nutr. Res. 2010, 30, 831–839. [Google Scholar] [CrossRef]
- Pérez-Jiménez, A.; Rufino-Palomares, E.E.; Fernández-Gallego, N.; Ortuño-Costela, M.C.; Reyes-Zurita, F.J.; Peragón, J.; García-Salguero, L.; Mokhtari, K.; Medina, P.P.; Lupiáñez, J.A. Target Molecules in 3T3-L1 Adipocytes Differentiation Are Regulated by Maslinic Acid, a Natural Triterpene from Olea Europaea. Phytomedicine 2016, 23, 1301–1311. [Google Scholar] [CrossRef]
- Rodríguez-Cerdeira, C.; Cordeiro-Rodríguez, M.; Carnero-Gregorio, M.; López-Barcenas, A.; Martínez-Herrera, E.; Fabbrocini, G.; Sinani, A.; Arenas-Guzmán, R.; González-Cespón, J.L. Biomarkers of Inflammation in Obesity-Psoriatic Patients. Mediat. Inflamm. 2019, 2019, 7353420. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Romero, E.; Izaola, O.; Primo, D.; Aller, R. Cardiovascular Risk Factors and Insulin Resistance after Two Hypocaloric Diets with Different Fat Distribution in Obese Subjects: Effect of the Rs10767664 Gene Variant in Brain-Derived Neurotrophic Factor. Lifestyle Genom. 2017, 10, 163–171. [Google Scholar] [CrossRef]
- Gomez-Delgado, F.; Alcala-Diaz, J.F.; Garcia-Rios, A.; Delgado-Lista, J.; Ortiz-Morales, A.; Rangel-Zuñiga, O.; Tinahones, F.J.; Gonzalez-Guardia, L.; Malagon, M.M.; Bellido-Muñoz, E.; et al. Polymorphism at the TNF-Alpha Gene Interacts with Mediterranean Diet to Influence Triglyceride Metabolism and Inflammation Status in Metabolic Syndrome Patients: From the CORDIOPREV Clinical Trial. Mol. Nutr. Food Res. 2014, 58, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yu, C.Y.; Deng, W.M. The Role of Pro-Inflammatory Cytokines in Lipid Metabolism of Metabolic Diseases. Int. Rev. Immunol. 2019, 38, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Fan, J.; Su, Q.; Yang, Z. Cytokines and Abnormal Glucose and Lipid Metabolism. Front. Endocrinol. 2019, 10, 703. [Google Scholar] [CrossRef]
- Rodriguez-Ayala, E.; Gallegos-Cabrales, E.C.; Gonzalez-Lopez, L.; Laviada-Molina, H.A.; Salinas-Osornio, R.A.; Nava-Gonzalez, E.J.; Leal-Berumen, I.; Escudero-Lourdes, C.; Escalante-Araiza, F.; Buenfil-Rello, F.A.; et al. Towards Precision Medicine: Defining and Characterizing Adipose Tissue Dysfunction to Identify Early Immunometabolic Risk in Symptom-Free Adults from the GEMM Family Study. Adipocyte 2020, 9, 153–169. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Aller, R.; Izaola, O.; Gonzalez Sagrado, M.; Conde, R. Role of G308 Promoter Variant of Tumor Necrosis Factor Alpha Gene on Weight Loss and Metabolic Parameters after a High Monounsaturated versus a High Polyunsaturated Fat Hypocaloric Diets. Med. Clin. 2013, 141, 189–193. [Google Scholar] [CrossRef]
- Sureda, A.; del Mar Bibiloni, M.; Julibert, A.; Bouzas, C.; Argelich, E.; Llompart, I.; Pons, A.; Tur, J.A. Adherence to the Mediterranean Diet and Inflammatory Markers. Nutrients 2018, 10, 62. [Google Scholar] [CrossRef]
- Campos Mondragón, M.G.; Oliart Ros, R.M.; Angulo Guerrero, J.O. Marcadores Inflamatorios En Pacientes Con Síndrome Metabólico Después de Consumir Ácidos Grasos Omega-3 y Ácido Linoleico Conjugado (CLA). Nutr. Clin. Y Diet. Hosp. 2013, 33, 7–17. [Google Scholar]
- Voisin, S.; Almén, M.S.; Moschonis, G.; Chrousos, G.P.; Manios, Y.; Schiöth, H.B. Dietary Fat Quality Impacts Genome-Wide DNA Methylation Patterns in a Cross-Sectional Study of Greek Preadolescents. Eur. J. Hum. Genet. 2015, 23, 654–662. [Google Scholar] [CrossRef]
- López-Bojorquez, L.N. La Regulación Del Factor de Transcripción NF-ΚB. Un Mediador Molecular En El Proceso Inflamatorio. Rev. Investig. Clin. 2004, 56, 83–92. [Google Scholar]
- National Library of Medicine. I. LPL Lipoprotein Lipase [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/labs/gquery/all/?term=LPL (accessed on 12 February 2023).
- Goodarzi, M.O.; Wong, H.; Quiñones, M.J.; Taylor, K.D.; Guo, X.; Castellani, L.W.; Antoine, H.J.; Yang, H.; Hsueh, W.A.; Rotter, J.I. The 3′ Untranslated Region of the Lipoprotein Lipase Gene: Haplotype Structure and Association with Post-Heparin Plasma Lipase Activity. J. Clin. Endocrinol. Metab. 2005, 90, 4816–4823. [Google Scholar] [CrossRef]
- Hatefi, Z.; Soltani, G.; Khosravi, S.; Kazemi, M.; Salehi, A.; Salehi, R. Micro R-410 Binding Site Single Nucleotide Polymorphism Rs13702 in Lipoprotein Lipase Gene Is Effective to Increase Susceptibility to Type 2 Diabetes in Iranian Population. Adv. Biomed. Res. 2018, 7, 79. [Google Scholar] [CrossRef]
- Daneshpour, M.S.; Faam, B.; Mansournia, M.A.; Hedayati, M.; Halalkhor, S.; Mesbah-Namin, S.A.; Shojaei, S.; Zarkesh, M.; Azizi, F. Haplotype Analysis of Apo AI-CIII-AIV Gene Cluster and Lipids Level: Tehran Lipid and Glucose Study. Endocrine 2012, 41, 103–110. [Google Scholar] [CrossRef]
- De França, E.; Alves, J.G.; Hutz, M.H. APOA1/C3/A4 Gene Cluster Variability and Lipid Levels in Brazilian Children. Braz. J. Med. Biol. Res. 2005, 38, 535–541. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Izaola, O.; Primo, D.; Aller, R. Influencia de La Variante Rs670 Del Gen APOA1 En La Respuesta HDL Sérica a Una Dieta Hipocalórica Enriquecida Con Grasas Poliinsaturadas Frente a Una Enriquecida Con Grasas Monoinsaturadas. Nutr. Hosp. 2019, 36, 1288–1295. [Google Scholar]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; McManus, R.; Hercberg, S.; Lairon, D.; Planells, R.; Roche, H.M. Gene-Nutrient Interactions and Gender May Modulate the Association between ApoA1 and ApoB Gene Polymorphisms and Metabolic Syndrome Risk. Atherosclerosis 2011, 214, 408–414. [Google Scholar] [CrossRef] [PubMed]
- De Luis, D.A.; Izaola, O.; Primo, D.; Aller, R. Role of Rs670 Variant of APOA1 Gene on Lipid Profile, Insulin Resistance and Adipokine Levels in Obese Subjects after Weight Loss with a Dietary Intervention. Diabetes Res. Clin. Pract. 2018, 142, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Guerra-García, M.T.; Moreno-Macías, H.; Ochoa-Guzmán, A.; Ordoñez-Sánchez, M.L.; Rodríguez-Guillen, R.; Vázquez-Cárdenas, P.; Ortíz-Ortega, V.M.; Peimbert-Torres, M.; Aguilar-Salinas, C.A.; Tusié-Luna, M.T. The -514C>T Polymorphism in the LIPC Gene Modifies Type 2 Diabetes Risk through Modulation of HDL-Cholesterol Levels in Mexicans. J. Endocrinol. Invest. 2021, 44, 557–565. [Google Scholar] [CrossRef]
- Posadas-Sánchez, R.; Ocampo-Arcos, W.A.; López-Uribe, Á.R.; Posadas-Romero, C.; Villarreal-Molina, T.; León, E.Á.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.; Cardoso-Saldaña, G.; Medina-Urrutia, A.; et al. Hepatic Lipase (LIPC) C-514T Gene Polymorphism Is Associated with Cardiometabolic Parameters and Cardiovascular Risk Factors but Not with Fatty Liver in Mexican Population. Exp. Mol. Pathol. 2015, 98, 93–98. [Google Scholar] [CrossRef]
- Smith, C.; Van Rompay, M.; Mattei, J.; Garcia, J.; Garcia-Bailo, B.; Lichtenstein, A.; Tucker, K.; Ordovás, J. Dietary Fat Modulation of Hepatic Lipase Variant -514 C/T for Lipids: A Crossover Randomized Dietary Intervention Trial in Caribbean Hispanics. Physiol. Genom. 2017, 49, 592–600. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Cuervo, M.; Goni, L.; Martinez, J.A. Prediction of Blood Lipid Phenotypes Using Obesity-Related Genetic Polymorphisms and Lifestyle Data in Subjects with Excessive Body Weight. Int. J. Genom. 2018, 2018, 4283078. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.T. Quantile-Specific Penetrance of Genes Affecting Lipoproteins, Adiposity and Height. PLoS ONE 2012, 7, e28764. [Google Scholar] [CrossRef]
- Chang, M.H.; Ned, R.M.; Hong, Y.; Yesupriya, A.; Yang, Q.; Liu, T.; Janssens, A.C.J.W.; Dowling, N.F. Racial/Ethnic Variation in the Association of Lipid-Related Genetic Variants with Blood Lipids in the US Adult Population. Circ. Cardiovasc. Genet. 2011, 4, 523–533. [Google Scholar] [CrossRef]
- García-Tascón, M.; Sahelices-Pinto, C.; Mendaña-Cuervo, C.; Magaz-González, A.M. The Impact of the COVID-19 Confinement on the Habits of PA Practice According to Gender (Male/Female): Spanish Case. Int. J. Environ. Res. Public Health 2020, 17, 6961. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, J.Á.; Meza Peña, C.; Grupo de investigación internacional PSYCOVID Nodo México. Ansiedad y Adaptación a La Pandemia En México: A Cross-Sectional Study. Interacciones J. Fam. Clin. Health Psychol. 2022, 8, e242. [Google Scholar] [CrossRef]
- Mitchell, L.J.; Ball, L.E.; Ross, L.J.; Barnes, K.A.; Williams, L.T. Effectiveness of Dietetic Consultations in Primary Health Care: A Systematic Review of Randomized Controlled Trials. J. Acad. Nutr. Diet. 2017, 117, 1941–1962. [Google Scholar] [CrossRef]
- Singh, V. Current Challenges and Future Implications of Exploiting the Omics Data into Nutrigenetics and Nutrigenomics for Personalized Diagnosis and Nutrition-Based Care. Nutrition 2023, 110, 112002. [Google Scholar] [CrossRef]
- Ceriani, F.; Montalvan, M.; Quintero, B.; Suárez, R.; Bautista-Valarezo, E.; Frias-Toral, E. Ethics of the Clinical Practice of Nutrigenetics and Nutrigenomics. Clin. Nutr. Open Sci. 2023, 49, 58–66. [Google Scholar] [CrossRef]
| Standard Diet | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | |||||||||
| ΔTGs | ΔVLDL | |||||||||
| Predictors | β Coefficients | Std. Error | p Value a | PC2 | p Value b | β Coefficients | Std. Error | p Value b | PC2 | p Value b |
| LIPC rs1800588 | −53.92 | 30.48 | 0.127 | 0.06 | 0.12 | −10.02 | 5.85 | 0.138 | 0.05 | 0.13 |
| LPL rs13702 | 159.13 | 45.58 | 0.013 | 0.26 | 0.01 | 31.35 | 8.75 | 0.012 | 0.26 | 0.01 |
| FABP2 rs1799883 | −28.80 | 37.05 | 0.466 | 0.01 | 0.46 | −5.13 | 7.11 | 0.49 | 0.01 | 0.49 |
| CETP rs708272 | 43.32 | 27.69 | 0.169 | 0.05 | 0.16 | 8.69 | 5.31 | 0.153 | 0.05 | 0.15 |
| PPARG rs1801282 | 92.52 | 42.88 | 0.074 | 0.10 | 0.07 | 17.92 | 8.23 | 0.072 | 0.09 | 0.07 |
| APOA1 rs670 | −112.95 | 32.64 | 0.013 | 0.25 | 0.01 | −22.39 | 6.26 | 0.012 | 0.26 | 0.01 |
| SATFAT | 2.61 | 1.88 | 0.216 | 0.04 | 0.21 | 0.51 | 0.36 | 0.20 | 0.04 | 0.20 |
| MUFAs | −2.33 | 1.96 | 0.286 | 0.02 | 0.28 | −0.44 | 0.38 | 0.29 | 0.02 | 0.29 |
| Adj. R2 | 0.69 | 0.03 | 0.71 | 0.02 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Beltrán, Y.E.; González-Becerra, K.; Rivera-Iñiguez, I.; Martínez-López, E.; Ramos-Lopez, O.; Alcaraz-Mejía, M.; Rodríguez-Echevarría, R.; Sáyago-Ayerdi, S.G.; Mendivil, E.J. A Nutrigenetic Strategy for Reducing Blood Lipids and Low-Grade Inflammation in Adults with Obesity and Overweight. Nutrients 2023, 15, 4324. https://doi.org/10.3390/nu15204324
Pérez-Beltrán YE, González-Becerra K, Rivera-Iñiguez I, Martínez-López E, Ramos-Lopez O, Alcaraz-Mejía M, Rodríguez-Echevarría R, Sáyago-Ayerdi SG, Mendivil EJ. A Nutrigenetic Strategy for Reducing Blood Lipids and Low-Grade Inflammation in Adults with Obesity and Overweight. Nutrients. 2023; 15(20):4324. https://doi.org/10.3390/nu15204324
Chicago/Turabian StylePérez-Beltrán, Yolanda E., Karina González-Becerra, Ingrid Rivera-Iñiguez, Erika Martínez-López, Omar Ramos-Lopez, Mildreth Alcaraz-Mejía, Roberto Rodríguez-Echevarría, Sonia G. Sáyago-Ayerdi, and Edgar J. Mendivil. 2023. "A Nutrigenetic Strategy for Reducing Blood Lipids and Low-Grade Inflammation in Adults with Obesity and Overweight" Nutrients 15, no. 20: 4324. https://doi.org/10.3390/nu15204324
APA StylePérez-Beltrán, Y. E., González-Becerra, K., Rivera-Iñiguez, I., Martínez-López, E., Ramos-Lopez, O., Alcaraz-Mejía, M., Rodríguez-Echevarría, R., Sáyago-Ayerdi, S. G., & Mendivil, E. J. (2023). A Nutrigenetic Strategy for Reducing Blood Lipids and Low-Grade Inflammation in Adults with Obesity and Overweight. Nutrients, 15(20), 4324. https://doi.org/10.3390/nu15204324






