The Effects of Prolonged Basic Amino Acid Exposures on Mitochondrial Enzyme Gene Expressions, Metabolic Profiling and Insulin Secretions and Syntheses in Rat INS-1 β-Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Cell Proliferation Assay
2.3. Insulin Release and GSIS Assay
2.4. Glucose Uptake and ATP/ADP Ratio Assay
2.5. Real-Time Quantitative PCR
2.6. Metabolomics Analysis
2.7. Metabolite–Metabolite Interaction Network Analysis
2.8. Western Blotting
2.9. Statistical Analysis
3. Results
3.1. Cell Proliferation, Insulin Release and GSIS
3.2. AA and Glucose Transports
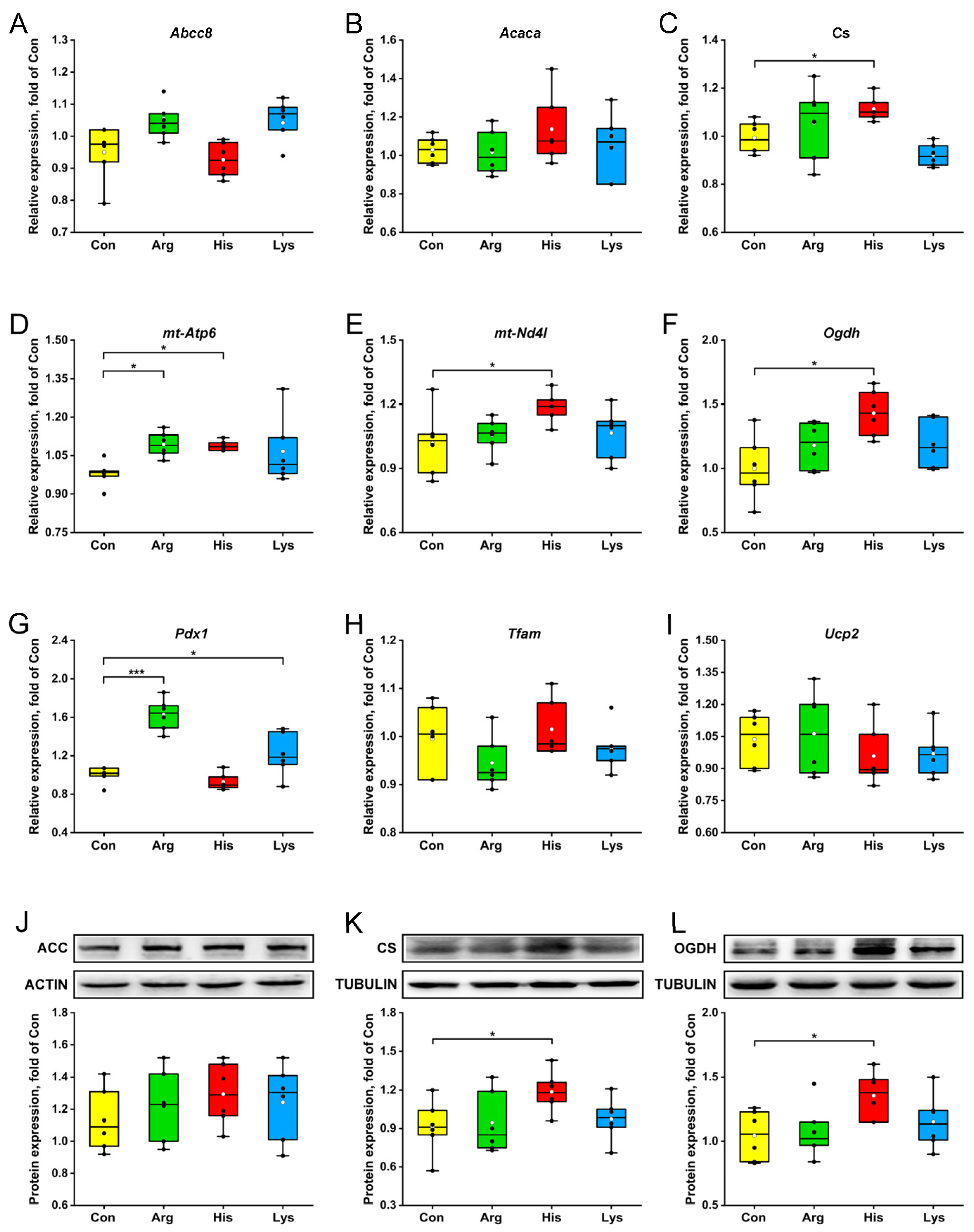
3.3. Mitochondrial Key Enzyme and Transcription Factor Expressions
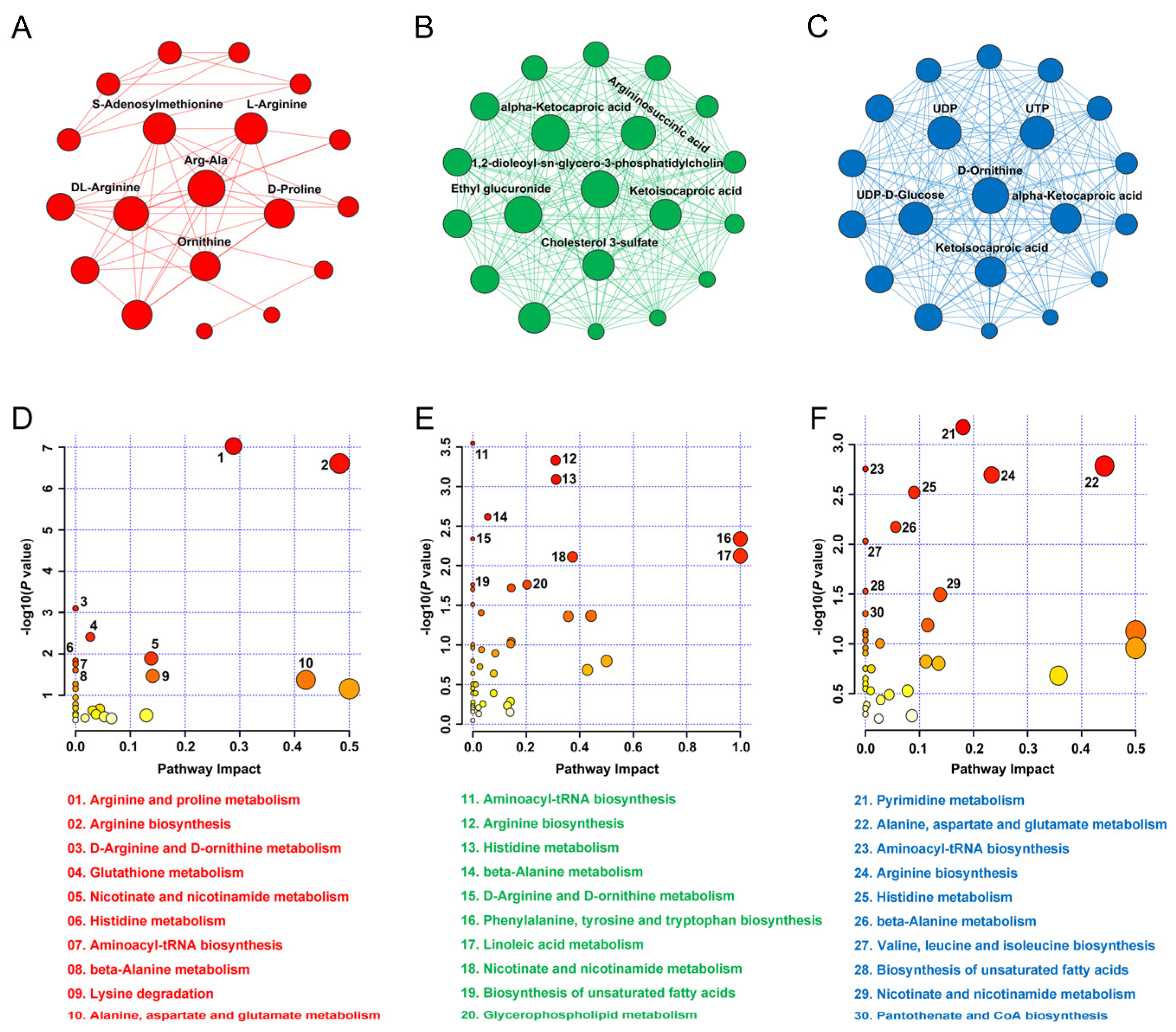
3.4. Metabolomics Analysis
3.5. Hub Metabolite and KEGG Pathway
3.6. Key Genes and Metabolites Related to Insulin Responses
4. Discussion
4.1. Effects of High BAA Treatment on β-Cell Metabolism
4.2. Insulin Response to Excessive Arginine
4.3. Insulin Response to Excessive Histidine
4.4. Insulin Response to Excessive Lysine
4.5. Key Genes and Metabolites Associated with Insulin Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newsholme, P.; Bender, K.; Kiely, A.; Brennan, L. Amino acid metabolism, insulin secretion and diabetes. Biochem. Soc. Trans. 2007, 35, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qiu, L.; Xiao, Q.; Wang, Y.; Meng, X.; Xu, R.; Wang, S.; Na, R. Obesity and diabetes related plasma amino acid alterations. Clin. Biochem. 2013, 46, 1447–1452. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef]
- Calbet, J.A.; MacLean, D.A. Plasma glucagon and insulin responses depend on the rate of appearance of amino acids after ingestion of different protein solutions in humans. J. Nutr. 2002, 132, 2174–2182. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Gaudel, C.; McClenaghan, N.H. Nutrient regulation of insulin secretion and beta-cell functional integrity. Adv. Exp. Med. Biol. 2010, 654, 91–114. [Google Scholar]
- Newsholme, P.; Krause, M. Nutritional regulation of insulin secretion: Implications for diabetes. Clin. Biochem. Rev. 2012, 33, 35–47. [Google Scholar]
- Nolan, C.J.; Prentki, M. The islet beta-cell: Fuel responsive and vulnerable. Trends Endocrinol. Metab. 2008, 19, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Sener, A.; Best, L.C.; Yates, A.P.; Kadiata, M.M.; Olivares, E.; Louchami, K.; Jijakli, H.; Ladrière, L.; Malaisse, W.J. Stimulus-secretion coupling of arginine-induced insulin release: Comparison between the cationic amino acid and its methyl ester. Endocrine 2000, 13, 329–340. [Google Scholar] [CrossRef]
- Sener, A.; Blachier, F.; Rasschaert, J.; Mourtada, A.; Malaisse-Lagae, F.; Malaisse, W.J. Stimulus-secretion coupling of arginine-induced insulin release: Comparison with lysine-induced insulin secretion. Endocrinology 1989, 124, 2558–2567. [Google Scholar] [CrossRef]
- Prentki, M.; Matschinsky, F.M.; Madiraju, S.R. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013, 18, 162–185. [Google Scholar] [CrossRef]
- Ježek, P.; Holendová, B.; Jabůrek, M.; Dlasková, A.; Plecitá-Hlavatá, L. Contribution of mitochondria to insulin secretion by various secretagogues. Antioxid. Redox Signal. 2022, 36, 920–952. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sun, X.; Qiu, L.; Wei, J.; Huang, Q.; Fang, C.; Ye, T.; Kang, M.; Shen, H.; Dong, S. Exposure to bisphenol A induces dysfunction of insulin secretion and apoptosis through the damage of mitochondria in rat insulinoma (INS-1) cells. Cell Death Dis. 2013, 4, e460. [Google Scholar] [CrossRef]
- Spaeth, J.M.; Walker, E.M.; Stein, R. Impact of Pdx1-associated chromatin modifiers on islet β-cells. Diabetes Obes. Metab. 2016, 18, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Hock, M.B.; Kralli, A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009, 71, 177–203. [Google Scholar] [CrossRef]
- McClenaghan, N.H.; Barnett, C.R.; O’Harte, F.P.; Flatt, P.R. Mechanisms of amino acid-induced insulin secretion from the glucose-responsive BRIN-BD11 pancreatic B-cell line. J. Endocrinol. 1996, 151, 349–357. [Google Scholar] [CrossRef]
- McClenaghan, N.H.; Scullion, S.M.; Mion, B.; Hewage, C.; Malthouse, J.P.; Flatt, P.R.; Newsholme, P.; Brennan, L. Prolonged L-alanine exposure induces changes in metabolism, Ca2+ handling and desensitization of insulin secretion in clonal pancreatic beta-cells. Clin. Sci. 2009, 116, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Kiely, A.; McClenaghan, N.H.; Flatt, P.R.; Newsholme, P. Pro-inflammatory cytokines increase glucose, alanine and triacylglycerol utilization but inhibit insulin secretion in a clonal pancreatic beta-cell line. J. Endocrinol. 2007, 195, 113–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, M.; Fu, J.J.; Mao, J.L.; Dong, X.P.; Chen, Y.W. Lipidomics reveals the relationship between lipid oxidation and flavor formation of basic amnio acids participated Low-Sodium cured large yellow croaker. Food Chem. 2023, 429, 136888. [Google Scholar] [CrossRef] [PubMed]
- Gammelsaeter, R.; Coppola, T.; Marcaggi, P.; Storm-Mathisen, J.; Chaudhry, F.A.; Attwell, D.; Regazzi, R.; Gundersen, V. A role for glutamate transporters in the regulation of insulin secretion. PLoS ONE 2011, 6, e22960. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.; Neufeld, Z.; Newsholme, P. Mathematical model of metabolism and electrophysiology of amino acid and glucose stimulated insulin secretion: In vitro validation using a β-cell line. PLoS ONE 2013, 8, e52611. [Google Scholar] [CrossRef]
- Bröer, S.; Gauthier-Coles, G. Amino acid homeostasis in mammalian cells with a focus on amino acid transport. J. Nutr. 2022, 152, 16–28. [Google Scholar] [CrossRef]
- Mullooly, N.; Vernon, W.; Smith, D.M.; Newsholme, P. Elevated levels of branched-chain amino acids have little effect on pancreatic islet cells, but L-arginine impairs function through activation of the endoplasmic reticulum stress response. Exp. Physiol. 2014, 99, 538–551. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, J.T.; Hamid, M.; Guo-Parke, H.; McClenaghan, N.H.; Gomis, R.; Flatt, P.R. Development and functional characterization of insulin-releasing human pancreatic beta cell lines produced by electrofusion. J. Biol. Chem. 2011, 286, 21982–21992. [Google Scholar] [CrossRef] [PubMed]
- Monti, L.D.; Valsecchi, G.; Costa, S.; Sandoli, E.P.; Phan, C.V.; Pontiroli, A.E.; Pozza, G.; Piatti, P.M. Effects of endothelin-1 and nitric oxide on glucokinase activity in isolated rat hepatocytes. Metabolism 2000, 49, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Parkash, J.; Asotra, K. L-histidine sensing by calcium sensing receptor inhibits voltage-dependent calcium channel activity and insulin secretion in β-cells. Life Sci. 2011, 88, 440–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sjöholm, A. Histaminergic regulation of pancreatic beta-cell replication and insulin secretion. Biochem. Biophys. Res. Commun. 1995, 214, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.C., Jr.; Fajans, S.S.; Pek, S.; Thiffault, C.A.; Knopf, R.F.; Conn, J.W. Synergistic effect of essential amino acids and glucose upon insulin secretion in man. Diabetes 1970, 19, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Barisón, M.J.; Damasceno, F.S.; Mantilla, B.S.; Silber, A.M. The active transport of histidine and its role in ATP production in Trypanosoma cruzi. J. Bioenerg. Biomembr. 2016, 48, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, R.; Yan, L.J. Roles of pyruvate, NADH, and mitochondrial complex I in redox balance and imbalance in β cell function and dysfunction. J. Diabetes Res. 2015, 2015, 512618. [Google Scholar] [CrossRef]
- Vangipurapu, J.; Stancáková, A.; Smith, U.; Kuusisto, J.; Laakso, M. Nine amino acids are associated with decreased insulin secretion and elevated glucose levels in a 7.4-year follow-up study of 5,181 finnish men. Diabetes 2019, 68, 1353–1358. [Google Scholar] [CrossRef]
- Nakada, N.; Mikami, T.; Hana, K.; Ichinoe, M.; Yanagisawa, N.; Yoshida, T.; Endou, H.; Okayasu, I. Unique and selective expression of L-amino acid transporter 1 in human tissue as well as being an aspect of oncofetal protein. Histol. Histopathol. 2014, 29, 217–227. [Google Scholar] [PubMed]
- Kobayashi, N.; Okazaki, S.; Sampetrean, O.; Irie, J.; Itoh, H.; Saya, H. CD44 variant inhibits insulin secretion in pancreatic β cells by attenuating LAT1-mediated amino acid uptake. Sci. Rep. 2018, 8, 2785. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, R.L.; Chester, M.W.; Daniels, M.B.; McGarry, J.D.; Stein, D.T. Circulating fatty acids are essential for efficient glucose-stimulated insulin secretion after prolonged fasting in humans. Diabetes 1998, 47, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lin, X.; White, R.R.; Hanigan, M.D.; Hu, Z.; Hou, Q.; Wang, Y.; Wang, Z. Plasma and pancreas islet hormone concentrations in lactating rats are associated with dietary protein amounts. J. Nutr. 2018, 148, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.G.; Francis, N.; Hill, R.; LE Waters, D.; Blanchard, C.L.; Santhakumar, A.B. Coloured rice phenolic extracts increase expression of genes associated with insulin secretion in rat pancreatic insulinoma β-cells. Int. J. Mol. Sci. 2020, 21, 3314. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Shang, W.; Wang, Y.; Sun, X.; Zhou, B.; Xie, Y.; Xu, X.; Liu, T.; Han, F. ALA protects against ERS-mediated apoptosis in a cochlear cell model with low citrate synthase expression. Arch. Biochem. Biophys. 2020, 688, 108402. [Google Scholar] [CrossRef]
- Lipson, K.L.; Fonseca, S.G.; Ishigaki, S.; Nguyen, L.X.; Foss, E.; Bortell, R.; Rossini, A.A.; Urano, F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006, 4, 245–254. [Google Scholar] [CrossRef]
- Nakata, M.; Yada, T.; Nakagawa, S.; Kobayashi, K.; Maruyama, I. Citrulline-argininosuccinate-arginine cycle coupled to Ca2+-signaling in rat pancreatic beta-cells. Biochem. Biophys. Res. Commun. 1997, 235, 619–624. [Google Scholar] [CrossRef]
- Xu, L.; Lin, X.; Li, X.; Hu, Z.; Hou, Q.; Wang, Y.; Wang, Z. Integration of transcriptomics and metabolomics provides metabolic and functional insights into reduced insulin secretion in MIN6 β-cells exposed to deficient and excessive arginine. FASEB J. 2022, 36, e22206. [Google Scholar] [CrossRef]
- Lai, M.C.; Teng, T.H.; Yang, C. The natural PPAR agonist linoleic acid stimulated insulin release in the rat pancreas. J. Vet. Med. Sci. 2013, 75, 1449–1454. [Google Scholar] [CrossRef]
- Nolan, C.J.; Madiraju, M.S.; Delghingaro-Augusto, V.; Peyot, M.L.; Prentki, M. Fatty acid signaling in the beta-cell and insulin secretion. Diabetes 2006, 55, S16–S23. [Google Scholar] [CrossRef] [PubMed]
- Fauziah, R.R.; Chin, R.; Ogita, S.; Yoshino, T.; Yamamoto, Y. Antiadipogenic effects of different molecular forms of conjugated linoleic acid on 3T3-L1 cells: Comparison between free fatty acid and phosphatidylcholine forms. J. Oleo Sci. 2021, 70, 1797–1803. [Google Scholar] [CrossRef] [PubMed]




| Gene | Accession No. | Primers Position | Primers Sequence (5′→3′) |
|---|---|---|---|
| Abcc8 | NM_013039.2 | Forward | CAAGGTCGTAAACCGCAAGC |
| Reverse | AGGGGTCCAGGTGAAGAAGC | ||
| Acaca | NM_022193 | Forward | CGGCTGTGGAAATTGCG |
| Reverse | GAGGCGGATGGGAATCG | ||
| Actb | NM_031144.3 | Forward | TGTCACCAACTGGGACGATA |
| Reverse | GGGGTGTTGAAGGTCTCAAA | ||
| Cs | NM_130755 | Forward | GCCCTCAACAGTGAAAGCA |
| Reverse | TGGCAATCAGGTCCATACAG | ||
| Gapdh | NC_005103.4 | Forward | GACATGCCGCCTGGAGAAAG |
| Reverse | AGCCCAGGATGCCCTTTAGT | ||
| mt-Atp6 | BC059121 | Forward | GCGTCTGGAGGACCTGTTGA |
| Reverse | CCTCAGGACTGGGGTTTGT | ||
| mt-Nd4l | EU104720 | Forward | TCCCAATTACCATTCTAGTTTT |
| Reverse | AGGTTTTGTACGTAGTCTGTTCCGT | ||
| Ogdh | NM_001017461.1 | Forward | CCGTGCCCGCTGACATTAT |
| Reverse | TCTCCCGAAGAGGAAGTGC | ||
| Pdx1 | NM_022852.3 | Forward | CCGCGTTCATCTCCCTTTC |
| Reverse | TGCCCACTGGCTTTTCCA | ||
| Slc2a2 | NM_181090.2 | Forward | GCTGATCTTCATCCTTCCGTCTGC |
| Reverse | CCAATCATCACGACTACGCCACTC | ||
| Slc38a2 | NM_181090.2 | Forward | GCTGATCTTCATCCTTCCGTCTGC |
| Reverse | CCAATCATCACGACTACGCCACTC | ||
| Slc7a7 | NM_031341.1 | Forward | TACCTACGCTGGAAGGAACCTGAC |
| Reverse | GCGATGCCGATGCCGATGAG | ||
| Tfam | NM_031326.1 | Forward | GAAGAGCAAATGGCTGAAGTT |
| Reverse | GTGCCCAATCCCAATGAC | ||
| Ucp2 | NM_019354.2 | Forward | TGTGGTAAAGGTCCGCTTCC |
| Reverse | TTCGGGCAACATTGGGAG |
| Dependent Variable | Independent Variable | RC | p | R2 |
|---|---|---|---|---|
| Insulin secretion, ng/106 cells | Slc7a7 | 791.10 | 0.0023 | 0.67 |
| Pdx1 | −313.65 | <0.0001 | ||
| Tfam | −1030.39 | 0.011 | ||
| Insulin content, nmol/mg protein | Cs | −947.78 | 0.0065 | 0.32 |
| Nd4l | 639.79 | 0.038 | ||
| ISCR | Cs | 0.057 | 0.014 | 0.24 |
| LGSIS, ng/106 cells | Slc7a7 | −82.13 | 0.011 | 0.26 |
| HGSIS, ng/106 cells | Slc38a2 | −155.19 | 0.0009 | 0.53 |
| Ogdh | −67.19 | 0.011 |
| Dependent Variable | Independent Variable (Peak Value) | RC | p | R2 |
|---|---|---|---|---|
| Insulin secretion, ng/106 cells | Argininosuccinic acid | −0.00077 | 0.0074 | 0.29 |
| ISCR | 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine | 0.000043 | 0.0099 | 0.28 |
| LGSIS, ng/106 cells | Oleic acid | −0.000010 | 0.042 | 0.17 |
| HGSIS, ng/106 cells | L-Citrulline | −0.00010 | 0.0001 | 0.89 |
| 1-Methylnicotinamide | 0.00053 | 0.0014 | ||
| 1,2-Benzenedicarboxylic acid | −0.00079 | 0.0059 | ||
| Alpha-D-Glucose | 0.00021 | 0.0044 | ||
| Linoleic acid | 0.000071 | 0.017 | ||
| Dehydroabietic acid | −0.0013 | 0.0019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Cheng, F.; Bu, D.; Li, X. The Effects of Prolonged Basic Amino Acid Exposures on Mitochondrial Enzyme Gene Expressions, Metabolic Profiling and Insulin Secretions and Syntheses in Rat INS-1 β-Cells. Nutrients 2023, 15, 4026. https://doi.org/10.3390/nu15184026
Xu L, Cheng F, Bu D, Li X. The Effects of Prolonged Basic Amino Acid Exposures on Mitochondrial Enzyme Gene Expressions, Metabolic Profiling and Insulin Secretions and Syntheses in Rat INS-1 β-Cells. Nutrients. 2023; 15(18):4026. https://doi.org/10.3390/nu15184026
Chicago/Turabian StyleXu, Lianbin, Fengqi Cheng, Dengpan Bu, and Xiuli Li. 2023. "The Effects of Prolonged Basic Amino Acid Exposures on Mitochondrial Enzyme Gene Expressions, Metabolic Profiling and Insulin Secretions and Syntheses in Rat INS-1 β-Cells" Nutrients 15, no. 18: 4026. https://doi.org/10.3390/nu15184026
APA StyleXu, L., Cheng, F., Bu, D., & Li, X. (2023). The Effects of Prolonged Basic Amino Acid Exposures on Mitochondrial Enzyme Gene Expressions, Metabolic Profiling and Insulin Secretions and Syntheses in Rat INS-1 β-Cells. Nutrients, 15(18), 4026. https://doi.org/10.3390/nu15184026






