Uncovering the Gut–Liver Axis Biomarkers for Predicting Metabolic Burden in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse Models and Data Collection
2.2. Analysis of Liver, Serum, and Urine Metabolites
2.3. RNA Sequencing and Data Processing
2.4. Microbiota Data Analysis
2.5. Bile Acid Quantification
2.6. Human Datasets
2.7. Transcriptomic Feature Selection
2.8. Machine Learning Models
2.9. Pathway and Network Analysis
2.10. Association Analysis
2.11. Statistics
3. Results
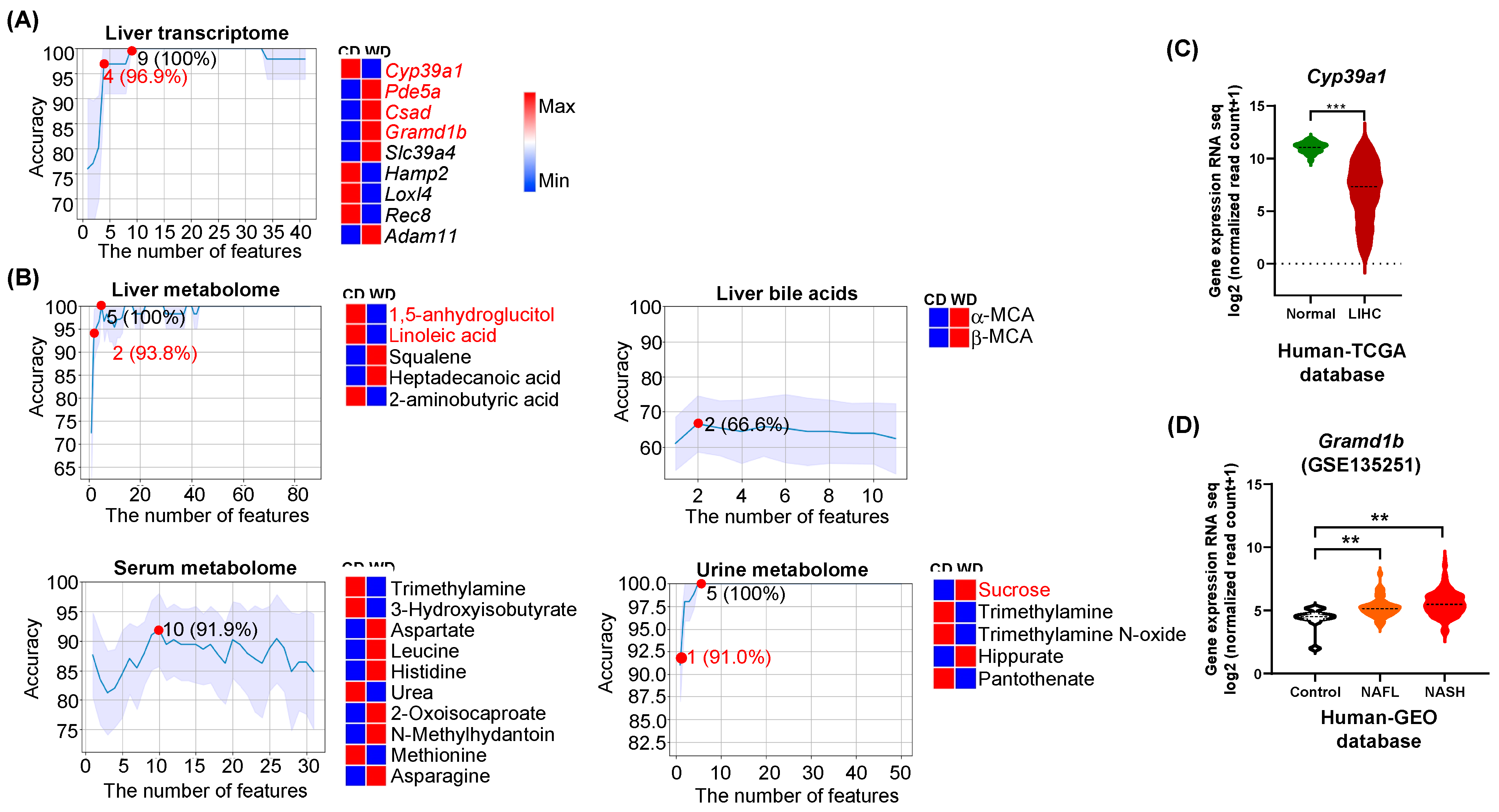
3.1. Predictors of Differential Diet Intakes
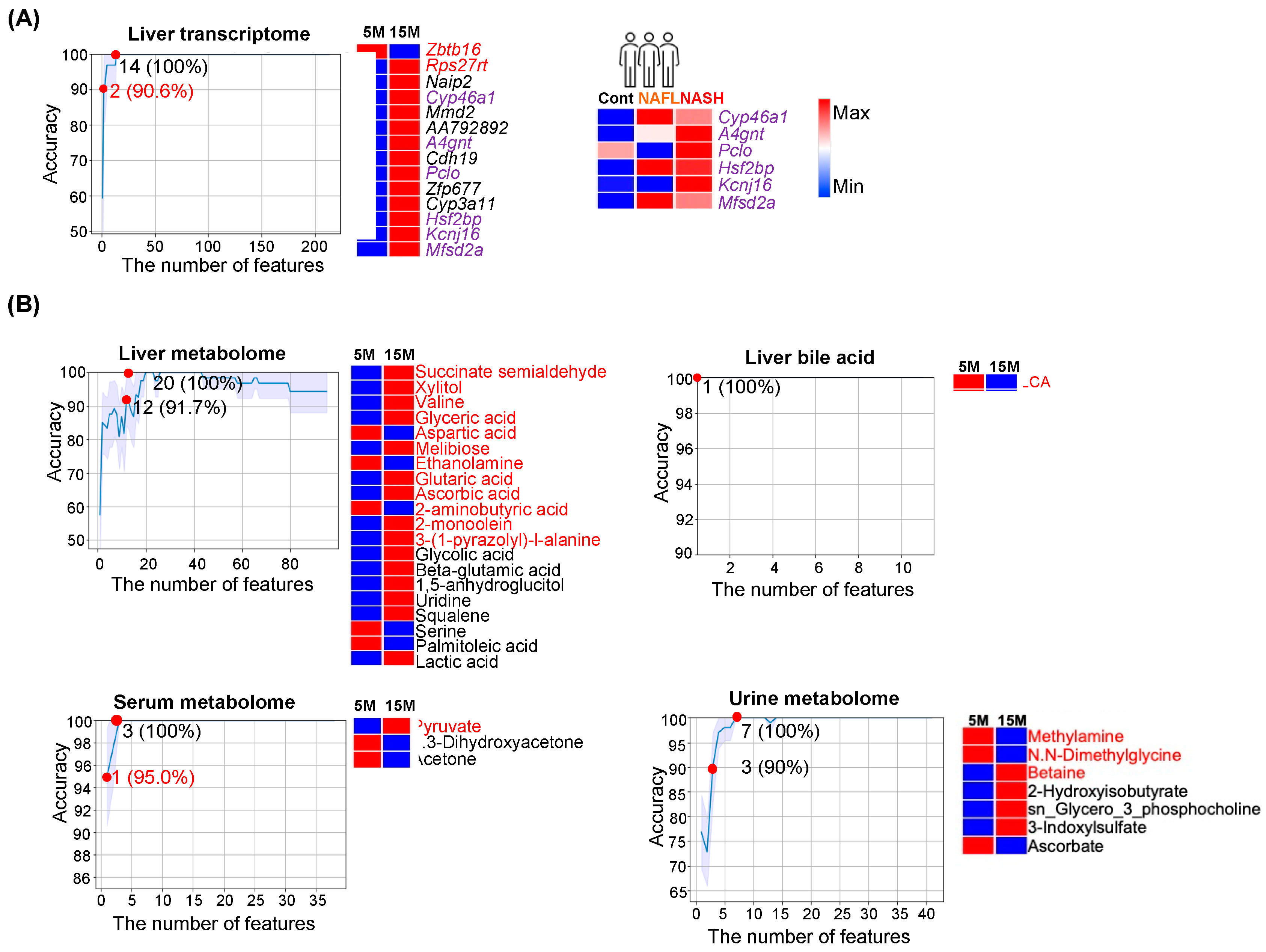
3.2. Age Classification
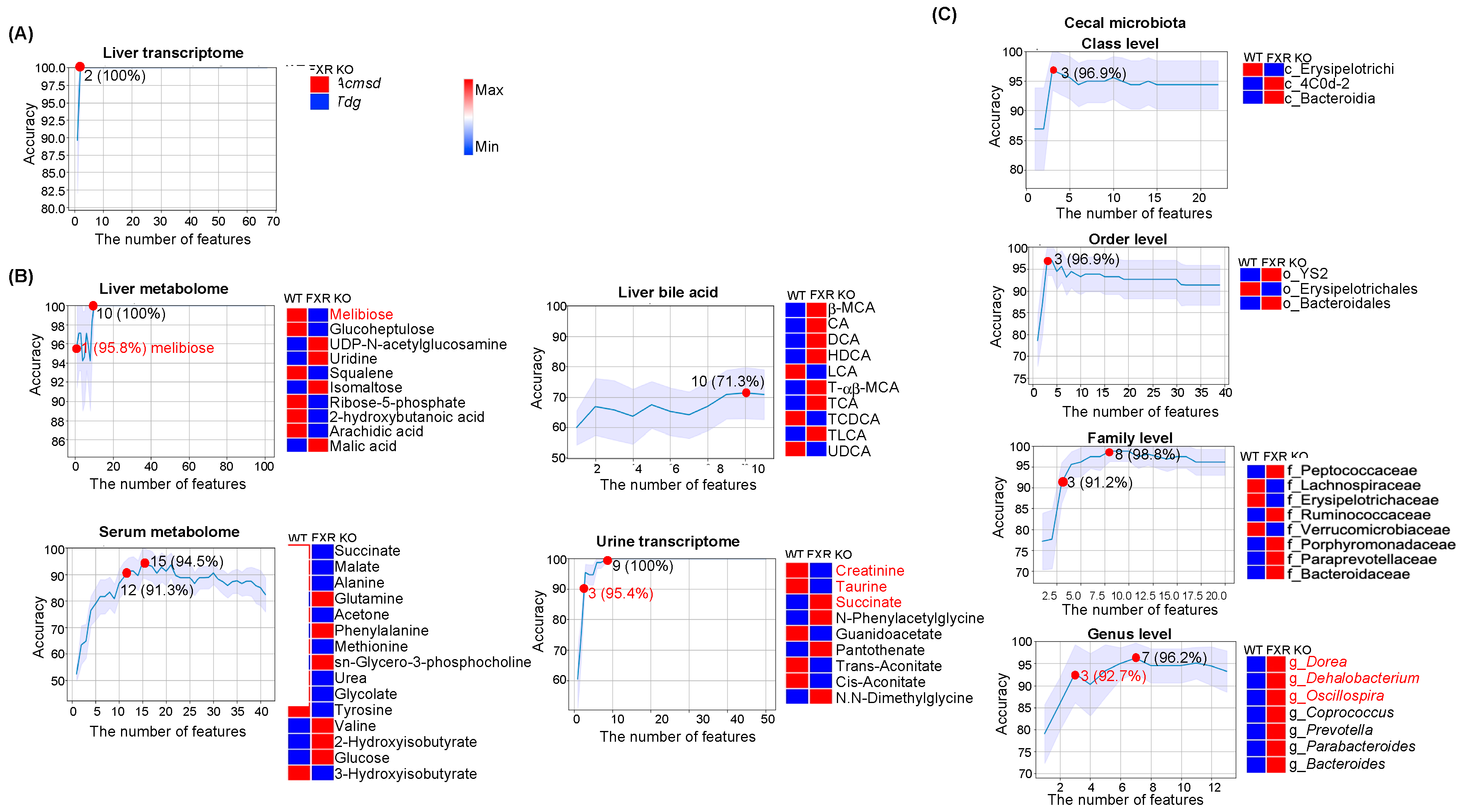
3.3. Predictors for FXR Inactivation
4. Discussion
4.1. Diet Predictors Relate to Metabolic Liver Disease Development
4.2. Features That Classify Ages and Metabolic Liver Diseases
4.3. FXR Inactivation Predictors and Metabolic Liver Diseases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sinal, C.J.; Tohkin, M.; Miyata, M.; Ward, J.M.; Lambert, G.; Gonzalez, F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 2000, 102, 731–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Cai, J.; Gonzalez, F.J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tanaka, N.; Fukami, T.; Xie, C.; Yagai, T.; Kim, D.; Velenosi, T.J.; Yan, T.; Krausz, K.W.; Levi, M.; et al. Role of farnesoid X receptor and bile acids in hepatic tumor development. Hepatol. Commun. 2018, 2, 1567–1582. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Xie, C.; Lv, Y.; Li, J.; Krausz, K.W.; Shi, J.; Brocker, C.N.; Desai, D.; Amin, S.G.; Bisson, W.H.; et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat. Commun. 2015, 6, 10166. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.; Morimura, K.; Shah, Y.; Yang, Q.; Ward, J.M.; Gonzalez, F.J. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 2007, 28, 940–946. [Google Scholar] [CrossRef] [Green Version]
- Sheng, L.; Jena, P.K.; Liu, H.X.; Kalanetra, K.M.; Gonzalez, F.J.; French, S.W.; Krishnan, V.V.; Mills, D.A.; Wan, Y.-J.Y. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci. Rep. 2017, 7, 1748. [Google Scholar] [CrossRef] [Green Version]
- Jena, P.K.; Sheng, L.; Liu, H.X.; Kalanetra, K.M.; Mirsoian, A.; Murphy, W.J.; French, S.W.; Krishnan, V.V.; Mills, D.A.; Wan, Y.-J.Y. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am. J. Pathol. 2017, 187, 1800–1813. [Google Scholar] [CrossRef] [Green Version]
- Sheng, L.; Jena, P.K.; Hu, Y.; Wan, Y.-J.Y. Age-specific microbiota in altering host inflammatory and metabolic signaling as well as metabolome based on the sex. Hepatobiliary Surg. Nutr. 2021, 10, 31–48. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; Di Lucente, J.; Jin, L.W.; Maezawa, I.; Wan, Y.-J.Y. Dysregulated bile acid synthesis and dysbiosis are implicated in Western diet-induced systemic inflammation, microglial activation, and reduced neuroplasticity. FASEB J. 2018, 32, 2866–2877. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Lu, Y.; Li, X.Y. Farnesoid X receptor: A master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol. Sin. 2015, 36, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Wang, X.; Lu, Y.; Wang, E.; Zhang, Z.; Yang, J.; Zhang, H.; Li, X. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. J. Hepatol. 2014, 60, 847–854. [Google Scholar] [CrossRef]
- Sheng, L.; Jena, P.K.; Hu, Y.; Liu, H.X.; Nagar, N.; Kalanetra, K.M.; French, S.W.; French, S.W.; Mills, D.A.; Wan, Y.-J.Y. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J. Pathol. 2017, 243, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Ma, C.; Liu, J.; Li, N.; Gao, M.; Huang, A.; Wang, X.; Huang, W.; Huang, X. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1245–G1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jena, P.K.; Sheng, L.; Nguyen, M.; Di Lucente, J.; Hu, Y.; Li, Y.; Maezawa, I.; Jin, L.W.; Wan, Y.-J.Y. Dysregulated bile acid receptor-mediated signaling and IL-17A induction are implicated in diet-associated hepatic health and cognitive function. Biomark. Res. 2020, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Jena, P.K.; Sheng, L.; Mcneil, K.; Chau, T.Q.; Yu, S.; Kiuru, M.; Fung, M.A.; Hwang, S.T.; Wan, Y.-J.Y. Long-term Western diet intake leads to dysregulated bile acid signaling and dermatitis with Th2 and Th17 pathway features in mice. J. Dermatol. Sci. 2019, 95, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.-J.Y.; Jena, P.K. Precision dietary supplementation based on personal gut microbiota. Nat Rev Gastroenterol Hepatol. 2019, 16, 204–206. [Google Scholar] [CrossRef] [PubMed]
- Jena, P.K.; Setayesh, T.; Sheng, L.; Di Lucente, J.; Jin, L.W.; Wan, Y.-J.Y. Intestinal microbiota remodeling protects mice from Western diet-induced brain inflammation and cognitive decline. Cells 2022, 11, 504. [Google Scholar] [CrossRef]
- Wan, Y.-J.Y.; Jena, P.; Lucente, J.; Jin, L.W. Concomitant improvement of synaptic decline and NASH using fermentable fiber inulin. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Gilbert, M.C.; Setayesh, T.; Wan, Y.-J.Y. The contributions of bacteria metabolites to the development of hepatic encephalopathy. Liver Res. 2022; in press. [Google Scholar] [CrossRef]
- Li, R.; Li, L.; Xu, Y.; Yang, J. Machine learning meets omics: Applications and perspectives. Brief. Bioinform. 2022, 23, bbab460. [Google Scholar] [CrossRef]
- Leung, H.; Long, X.; Ni, Y.; Qian, L.; Nychas, E.; Siliceo, S.L.; Pohl, D.; Hanhineva, K.; Liu, Y.; Xu, A.; et al. Risk assessment with gut microbiome and metabolite markers in NAFLD development. Sci. Transl. Med. 2022, 14, eabk0855. [Google Scholar] [CrossRef]
- Sheng, L.; Jena, P.K.; Liu, H.-X.; Hu, Y.; Nagar, N.; Bronner, D.N.; Settles, M.L.; Baümler, A.J.; Wan, Y.-J.Y. Obesity treatment by epigallocatechin-3-gallate−regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J. 2018, 32, 6371–6384. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Jena, P.K.; Hu, Y.; Sheng, L.; Chen, S.-Y.; Slupsky, C.M.; Davis, R.; Tepper, C.G.; Wan, Y.-J.Y. The essential roles of FXR in diet and age influenced metabolic changes and liver disease development: A multi-omics study. Biomark. Res. 2023, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Chen, S.Y.; Sheng, L.; Jena, P.K.; Kalanetra, K.M.; Mills, D.A.; Wan, Y.Y.; Slupsky, C.M. Long-term effects of western diet consumption in male and female mice. Sci. Rep. 2020, 10, 14686. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinform 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, T.; Liu, H. Recent advances in feature selection and its applications. Knowl. Inf. Syst. 2017, 53, 551–577. [Google Scholar] [CrossRef]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, V.K.; Dahiya, K.; Sharma, A. Problem formulations and solvers in linear SVM: A review. Artif. Intell. Rev. 2019, 52, 803–855. [Google Scholar] [CrossRef]
- Venkatesan, C.; Karthigaikumar, P.; Paul, A.; Satheeskumaran, S.; Kumar, R. ECG signal preprocessing and SVM classifier-based abnormality detection in remote healthcare applications. IEEE Access 2018, 6, 9767–9773. [Google Scholar] [CrossRef]
- Ketu, S.; Mishra, P.K. Scalable kernel-based SVM classification algorithm on imbalance air quality data for proficient healthcare. Complex Intell. Syst. 2021, 7, 2597–2615. [Google Scholar] [CrossRef]
- Razzaghi, T.; Roderick, O.; Safro, I.; Marko, N. Multilevel weighted support vector machine for classification on healthcare data with missing values. PLoS ONE 2016, 11, e0155119. [Google Scholar] [CrossRef] [Green Version]
- Mladenić, D.; Brank, J.; Grobelnik, M.; Milic-Frayling, N. Feature selection using linear classifier weights: Interaction with classification models. In Proceedings of the 27th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA, 25–29 July 2004. [Google Scholar]
- Sindhwani, V.; Bhattacharya, P.; Rakshit, S. Information theoretic feature crediting in multiclass support vector machines. In Proceedings of the 2001 SIAM International Conference on Data Mining, Chicago, IL, USA, 5–7 April 2001; SIAM: Philadelphia, PA, USA, 2001. [Google Scholar]
- Liu, N.; Meng, Z.; Lou, G.; Zhou, W.; Wang, X.; Zhang, Y.; Zhang, L.; Liu, X.; Yen, Y.; Lai, L. Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNF1α regulation of FXR expression. Mol. Endocrinol. 2012, 26, 775–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Yu, T.; Hu, J.; Wu, J.; Feng, S.; Xu, Q.; Zhu, H.; Zhang, X.; Zhang, Y.; Zhou, B.; et al. Downregulation of CYP39A1 serves as a novel biomarker in hepatocellular carcinoma with worse clinical outcome. Oxid. Med. Cell. Longev. 2021, 2021, 5175581. [Google Scholar] [CrossRef]
- Wang, X.; Cai, B.; Yang, X.; Sonubi, O.O.; Zheng, Z.; Ramakrishnan, R.; Shi, H.; Valenti, L.; Pajvani, U.B.; Sandhu, J. Cholesterol stabilizes TAZ in hepatocytes to promote experimental non-alcoholic steatohepatitis. Cell Metab. 2020, 31, 969–986.e7. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Noguchi, S.; Ono, M.; Ochi, T.; Munekage, K.; Okamoto, N.; Suganuma, N.; Saibara, T. High fasting insulin concentrations may be a pivotal predictor for the severity of hepatic fibrosis beyond the glycemic status in non-alcoholic fatty liver disease patients before development of diabetes mellitus. Hepatol. Res. 2017, 47, 983–990. [Google Scholar] [CrossRef]
- Ferrarini, A.; Di Poto, C.; He, S.; Tu, C.; Varghese, R.S.; Kara Balla, A.; Jayatilake, M.; Li, Z.; Ghaffari, K.; Fan, Z.; et al. Metabolomic analysis of liver tissues for characterization of hepatocellular carcinoma. J. Proteome Res. 2019, 18, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Obinata, H.; Hattori, T.; Nakane, S.; Tatei, K.; Izumi, T. Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A. J. Biol. Chem. 2005, 280, 40676–40683. [Google Scholar] [CrossRef] [Green Version]
- Luceri, C.; Caderni, G.; Lodovici, M.; Spagnesi, M.T.; Monserrat, C.; Lancioni, L.; Dolara, P. Urinary excretion of sucrose and fructose as a predictor of sucrose intake in dietary intervention studies. Cancer Epidemiol. Biomark. Prev. 1996, 5, 167–171. [Google Scholar]
- Kim, I.H.; Kisseleva, T.; Brenner, D.A. Aging and liver disease. Curr. Opin. Gastroenterol. 2015, 31, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Qian, J.; Shi, X.; Gao, T.; Liang, T.; Liu, C. Control of hepatic gluconeogenesis by the promyelocytic leukemia zinc finger protein. Mol. Endocrinol. 2014, 28, 1987–1998. [Google Scholar] [CrossRef]
- Bendlová, B.; Vaňková, M.; Hill, M.; Vacínová, G.; Lukášová, P.; VejraŽková, D.; Šedová, L.; Šeda, O.; Včelák, J. ZBTB16 gene variability influences obesity-related parameters and serum lipid levels in Czech adults. Physiol. Res. 2017, 66, S425–S431. [Google Scholar] [CrossRef]
- Goldberg, A.A.; Richard, V.R.; Kyryakov, P.; Bourque, S.D.; Beach, A.; Burstein, M.T.; Glebov, A.; Koupaki, O.; Boukh-Viner, T.; Gregg, C.; et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging 2010, 2, 393–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velazquez-Villegas, L.A.; Perino, A.; Lemos, V.; Zietak, M.; Nomura, M.; Pols, T.W.H.; Schoonjans, K. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat. Commun. 2018, 9, 245. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.X.; Luo, Y.; Wang, D.; Adorini, L.; Pruzanski, M.; Dobrinskikh, E.; Levi, M. A dual agonist of farnesoid X receptor (FXR) and the G protein-coupled receptor TGR5, INT-767, reverses age-related kidney disease in mice. J. Biol. Chem. 2017, 292, 12018–12024. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Atarashi, K.; Plichta, D.R.; Arai, Y.; Sasajima, S.; Kearney, S.M.; Suda, W.; Takeshita, K.; Sasaki, T.; Okamoto, S.; et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature 2021, 599, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Nagao, K.; Inoue, N.; Wang, Y.-M.; Shirouchi, B.; Yanagita, T. Dietary conjugated linoleic acid alleviates nonalcoholic fatty liver disease in Zucker (fa/fa) rats. J. Nutr. 2005, 135, 9–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Jiang, M.; Li, Y.; Su, J.; Li, L.; Qu, X.; Fan, L. 1H-NMR metabolomics analysis of the effect of rubusoside on serum metabolites of golden hamsters on a high-fat diet. Molecules 2020, 25, 1274. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Liu, Y.; Lu, X.; Huang, Y.; Liu, Y.; Cheng, S.; Duan, Y. A cross-sectional study of breath acetone based on diabetic metabolic disorders. J. Breath Res. 2015, 9, 016005. [Google Scholar] [CrossRef]
- Katsyuba, E.; Mottis, A.; Zietak, M.; De Franco, F.; van der Velpen, V.; Gariani, K.; Ryu, D.; Cialabrini, L.; Matilainen, O.; Liscio, P.; et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature 2018, 563, 354–359. [Google Scholar] [CrossRef]
- Allison, S.J. Targeting NAD+ synthesis to boost mitochondrial function and protect the kidney. Nat. Rev. Nephrol. 2019, 15, 1. [Google Scholar] [CrossRef]
- Hassan, H.M.; Isovic, M.; Kolendowski, B.; Bauer-Maison, N.; Onabote, O.; Cecchini, M.; Haig, A.; Maleki Vareki, S.; Underhill, T.M.; Torchia, J. Loss of thymine dna glycosylase causes dysregulation of bile acid homeostasis and hepatocellular carcinoma. Cell Rep. 2020, 31, 107475. [Google Scholar] [CrossRef]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bonder, M.J.; Cenit, M.C.; Tigchelaar, E.F.; Maatman, A.; Dekens, J.A.; Brandsma, E.; Marczynska, J.; Imhann, F.; Weersma, R.K.; et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 2015, 117, 817–824. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Z.; Deng, Q.; Huang, Q.; Wang, X.; Huang, F. Beneficial effects of flaxseed polysaccharides on metabolic syndrome via gut microbiota in high-fat diet fed mice. Food Res. Int. 2020, 131, 108994. [Google Scholar] [CrossRef]
- Carter, C.E.; Gansevoort, R.T.; Scheven, L.; Heerspink, H.J.L.; Shlipak, M.G.; de Jong, P.E.; Ix, J.H. Influence of urine creatinine on the relationship between the albumin-to-creatinine ratio and cardiovascular events. Clin. J. Am. Soc. Nephrol. 2012, 7, 595–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Q.; Guo, J.; Zhang, Y.; Chen, W. The beneficial effects of taurine in alleviating fatty liver disease. J. Funct. Foods 2021, 77, 104351. [Google Scholar] [CrossRef]
- Harber, K.J.; de Goede, K.E.; Verberk, S.G.; Meinster, E.; de Vries, H.E.; van Weeghel, M.; de Winther, M.P.; Van den Bossche, J. Succinate is an inflammation-induced immunoregulatory metabolite in macrophages. Metabolites 2020, 10, 372. [Google Scholar] [PubMed]
- Mills, E.; O’Neill, L.A. Succinate: A metabolic signal in inflammation. Trends Cell Biol. 2014, 24, 313–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, F.; Zhang, J.; Liu, N.; Gu, Y.; Zhang, Y.; Huang, P.; Zhang, N.; Lin, S.; Pan, R.; Meng, Z.; et al. Blocking hepatocarcinogenesis by a cytochrome P450 family member with female-preferential expression. Gut 2022, 71, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.; Li, J.; Liu, L.; Wu, Q.; Fan, L.; Ma, N.; Yu, C.; Lu, H.; Zhang, X.; Chen, J.; et al. CSAD Ameliorates Lipid Accumulation in High-Fat Diet-Fed Mice. Int. J. Mol. Sci. 2022, 23, 15931. [Google Scholar] [CrossRef]
- Weaver, B.P.; Zhang, Y.; Hiscox, S.; Guo, G.L.; Apte, U.; Taylor, K.M.; Sheline, C.T.; Wang, L.; Andrews, G.K. Zip4 (Slc39a4) Expression is Activated in Hepatocellular Carcinomas and Functions to Repress Apoptosis, Enhance Cell Cycle and Increase Migration. PLoS ONE 2010, 5, e13158. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, B.; Kuo, Y.-M.; Zemansky, J.; Gitschier, J. A Novel Member of a Zinc Transporter Family Is Defective in Acrodermatitis Enteropathica. Am. J. Hum. Genet. 2002, 71, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Küry, S.; Dréno, B.; Bézieau, S.; Giraudet, S.; Kharfi, M.; Kamoun, R.; Moisan, J.-P. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet. 2002, 31, 239–240. [Google Scholar] [CrossRef]
- Nakano, A.; Nakano, H.; Toyomaki, Y.; Hanada, K.; Nomura, K. Novel SLC39A4 Mutations in Acrodermatitis Enteropathica. J. Investig. Dermatol. 2003, 120, 963–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, H.-Y.; Wang, N.; Zhang, C.; Chan, Y.-T.; Yuen, M.-F.; Feng, Y. Lysyl Oxidase-Like 4 Fosters an Immunosuppressive Microenvironment During Hepatocarcinogenesis. Hepatology 2021, 73, 2326–2341. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Bai, Y.; Wang, J.; Xie, X.-L.; Li, A.-D.; Ding, Q.; Cui, Z.-J.; Yin, J.; Jiang, X.-Y.; Jiang, H.-Q. REC8 promotes tumor migration, invasion and angiogenesis by targeting the PKA pathway in hepatocellular carcinoma. Clin. Exp. Med. 2021, 21, 479–492. [Google Scholar] [CrossRef]
- Palomer, E.; Martín-Segura, A.; Baliyan, S.; Ahmed, T.; Balschun, D.; Venero, C.; Martin, M.G.; Dotti, C.G. Aging Triggers a Repressive Chromatin State at Bdnf Promoters in Hippocampal Neurons. Cell Rep. 2016, 16, 2889–2900. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.Y.; Chioza, B.A.; Rajab, A.; Schmitz-Abe, K.; Al-Khayat, A.; Al-Turki, S.; Baple, E.L.; Patton, M.A.; Al-Memar, A.Y.; Hurles, M.E.; et al. Loss of PCLO function underlies pontocerebellar hypoplasia type III. Neurology 2015, 84, 1745–1750. [Google Scholar] [CrossRef] [Green Version]
- Kong, F.; Kong, D.; Yang, X.; Yuan, D.; Zhang, N.; Hua, X.; You, H.; Zheng, K.; Tang, R. Integrative analysis of highly mutated genes in hepatitis B virus-related hepatic carcinoma. Cancer Med. 2020, 9, 2462–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, T.; Noji, K.; Shinozaki, S.; Suzuki, S.; Umegaki, K.; Shimokado, K. Diet-induced non-alcoholic fatty liver disease affects expression of major cytochrome P450 genes in a mouse model. J. Pharm. Pharmacol. 2016, 68, 1567–1576. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Z.-Y.; Gan, X.; Dai, H.; Cai, D.; Liu, R.-H.; Zhou, J.-M.; Zhang, H.-L.; Li, Z.-H.; Luo, Q.-Q.; et al. A novel four-gene signature for predicting the prognosis of hepatocellular carcinoma. Scand. J. Gastroenterol. 2022, 57, 1227–1237. [Google Scholar] [CrossRef]
- Schlingmann, K.P.; Renigunta, A.; Hoorn, E.J.; Forst, A.-L.; Renigunta, V.; Atanasov, V.; Mahendran, S.; Barakat, T.S.; Gillion, V.; Godefroid, N.; et al. Defects in KCNJ16 Cause a Novel Tubulopathy with Hypokalemia, Salt Wasting, Disturbed Acid-Base Homeostasis, and Sensorineural Deafness. J. Am. Soc. Nephrol. 2021, 32, 1498–1512. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Kan, J.; Su, A.; Liu, Q.-D.; Wang, K.; Cai, X.; Dong, J. The prognostic value of major facilitator superfamily domain-containing protein 2A in patients with hepatocellular carcinoma. Aging 2019, 11, 8474–8483. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, S.; Ohta, M. Serum 1,5-anhydro-d-glucitol levels in liver cirrhosis. Acta Diabetol. 1998, 35, 65–66. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Murai, J.; Saito, H.; Mukai, M.; Toya, D.; Tanaka, N.; Kanehara, H.; Bando, Y.; Kasayama, S. 1,5-Anhydroglucitol levels are low irrespective of plasma glucose levels in patients with chronic liver disease. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2010, 48, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhao, Y.; Xie, Z.; Xiao, L.; Hu, Q.; Li, Q.; Tang, S.; Wang, J.; Li, L. 1,5-Anhydroglucitol Predicts Mortality in Patients with HBV-Related Acute-on-chronic Liver Failure. J. Clin. Transl. Hepatol. 2022, 10, 651–659. [Google Scholar] [CrossRef]
- Lou-Bonafonte, J.M.; Martínez-Beamonte, R.; Sanclemente, T.; Surra, J.C.; Herrera-Marcos, L.V.; Sanchez-Marco, J.; Arnal, C.; Osada, J. Current Insights into the Biological Action of Squalene. Mol. Nutr. Food Res. 2018, 62, e1800136. [Google Scholar] [CrossRef]
- Jenkins, B.; West, J.A.; Koulman, A. A Review of Odd-Chain Fatty Acid Metabolism and the Role of Pentadecanoic Acid (C15:0) and Heptadecanoic Acid (C17:0) in Health and Disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef] [Green Version]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Bjune, M.S.; Lindquist, C.; Stafsnes, M.H.; Bjørndal, B.; Bruheim, P.; Aloysius, T.A.; Nygård, O.; Skorve, J.; Madsen, L.; Dankel, S.N.; et al. Plasma 3-hydroxyisobutyrate (3-HIB) and methylmalonic acid (MMA) are markers of hepatic mitochondrial fatty acid oxidation in male Wistar rats. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 158887. [Google Scholar] [CrossRef]
- Rao, Y.; Kuang, Z.; Li, C.; Guo, S.; Xu, Y.; Zhao, D.; Hu, Y.; Song, B.; Jiang, Z.; Ge, Z.; et al. Gut Akkermansia muciniphila ameliorates metabolic dysfunction-associated fatty liver disease by regulating the metabolism of L-aspartate via gut-liver axis. Gut Microbes 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Dos Santos, A.L.S.; Anastácio, L.R. The impact of L-branched-chain amino acids and L-leucine on malnutrition, sarcopenia, and other outcomes in patients with chronic liver disease. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 181–194. [Google Scholar] [CrossRef] [PubMed]
- DiNicolantonio, J.J.; Mccarty, M.F.; Okeefe, J.H. Role of dietary histidine in the prevention of obesity and metabolic syndrome. Open Heart 2018, 5, e000676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagamani, S.C.; Ali, S.; Izem, R.; Schady, D.; Masand, P.; Shneider, B.L.; Leung, D.H.; Burrage, L.C. Biomarkers for liver disease in urea cycle disorders. Mol. Genet. Metab. 2021, 133, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Trupp, M.; Jonsson, P.; Ohrfelt, A.; Zetterberg, H.; Obudulu, O.; Malm, L.; Wuolikainen, A.; Linder, J.; Moritz, T.; Blennow, K.; et al. Metabolite and peptide levels in plasma and CSF differentiating healthy controls from patients with newly diagnosed Parkinson’s disease. J. Parkinsons. Dis. 2014, 4, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Y.; Tang, W. Methionine cycle in nonalcoholic fatty liver disease and its potential applications. Biochem. Pharmacol. 2022, 200, 115033. [Google Scholar] [CrossRef]
- Wang, S.; Ding, Y.; Dong, R.; Wang, H.; Yin, L.; Meng, S. Canagliflozin Improves Liver Function in Rats by Upregulating Asparagine Synthetase. Pharmacology 2021, 106, 606–615. [Google Scholar] [CrossRef]
- Chung, M.; Ma, J.; Patel, K.; Berger, S.; Lau, J.; Lichtenstein, A.H. Fructose, high-fructose corn syrup, sucrose, and nonalcoholic fatty liver disease or indexes of liver health: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2014, 100, 833–849. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Ke, Y.; Zhan, R.; Liu, C.; Zhao, M.; Zeng, A.; Shi, X.; Ji, L.; Cheng, S.; Pan, B.; et al. Trimethylamine-N -oxide promotes brain aging and cognitive impairment in mice. Aging Cell 2018, 17, e12768. [Google Scholar] [CrossRef]
- Tan, X.; Liu, Y.; Long, J.; Chen, S.; Liao, G.; Wu, S.; Li, C.; Wang, L.; Ling, W.; Zhu, H. Trimethylamine N-Oxide Aggravates Liver Steatosis through Modulation of Bile Acid Metabolism and Inhibition of Farnesoid X Receptor Signaling in Nonalcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2019, 63, e1900257. [Google Scholar] [CrossRef]
- Lee, K.S.; Cho, Y.; Kim, H.; Hwang, H.; Cho, J.W.; Lee, Y.-H.; Lee, S.-G. Association of Metabolomic Change and Treatment Response in Patients with Non-Alcoholic Fatty Liver Disease. Biomedicines 2022, 10, 1216. [Google Scholar] [CrossRef]
- Jian, H.; Miao, S.; Liu, Y.; Wang, X.; Xu, Q.; Zhou, W.; Li, H.; Dong, X.; Zou, X. Dietary Valine Ameliorated Gut Health and Accelerated the Development of Nonalcoholic Fatty Liver Disease of Laying Hens. Oxidative Med. Cell. Longev. 2021, 2021, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, O.P.; Lehti, M.; Kyröläinen, H.; Kainulainen, H. Heme Oxygenase-1 and Blood Bilirubin Are Gradually Activated by Oral D-Glyceric Acid. Antioxidants 2022, 11, 2319. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhang, L.; Shi, H.; Xue, C.; Yanagita, T.; Zhang, T.; Wang, Y. The Protective Effect of Dietary EPA-Enriched Ethanolamine Plasmalogens against Hyperlipidemia in Aged Mice. Eur. J. Lipid Sci. Technol. 2020, 122, 2000179. [Google Scholar] [CrossRef]
- Lee, H.; Ahn, J.; Shin, S.S.; Yoon, M. Ascorbic acid inhibits visceral obesity and nonalcoholic fatty liver disease by activating peroxisome proliferator-activated receptor α in high-fat-diet-fed C57BL/6J mice. Int. J. Obes. 2018, 43, 1620–1630. [Google Scholar] [CrossRef]
- Zheng, W.V.; Li, Y.; Cheng, X.; Xu, Y.; Zhou, T.; Li, D.; Xiong, Y.; Wang, S.; Chen, Z. Uridine alleviates carbon tetrachloride-induced liver fibrosis by regulating the activity of liver-related cells. J. Cell. Mol. Med. 2021, 26, 840–854. [Google Scholar] [CrossRef]
- Sim, W.-C.; Lee, W.; Sim, H.; Lee, K.-Y.; Jung, S.-H.; Choi, Y.-J.; Kim, H.Y.; Kang, K.W.; Lee, J.-Y.; Choi, Y.J.; et al. Downregulation of PHGDH expression and hepatic serine level contribute to the development of fatty liver disease. Metabolism 2019, 102, 154000. [Google Scholar] [CrossRef] [Green Version]
- Souza, C.O.; Teixeira, A.A.; Biondo, L.A.; Silveira, L.S.; Breda, C.N.D.S.; Braga, T.T.; Camara, N.O.; Belchior, T.; Festuccia, W.T.; Diniz, T.A.; et al. Palmitoleic acid reduces high fat diet-induced liver inflammation by promoting PPAR-γ-independent M2a polarization of myeloid cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158776. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.O.; Teixeira, A.A.S.; Biondo, L.A.; Lima Junior, E.A.; Batatinha, H.A.P.; Rosa Neto, J.C. Palmitoleic Acid Improves Metabolic Functions in Fatty Liver by PPARα-Dependent AMPK Activation. J. Cell. Physiol. 2017, 232, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yang, B.; Yan, Y.; Tong, W.; Zhou, R.; Zhang, J.; Mi, J.; Li, D. Palmitoleic acid protects against hypertension by inhibiting NF-κB-mediated inflammation. Mol. Nutr. Food Res. 2021, 65, e2001025. [Google Scholar] [CrossRef]
- Shao, J.; Ge, T.; Tang, C.; Wang, G.; Pang, L.; Chen, Z. Synergistic anti-inflammatory effect of gut microbiota and lithocholic acid on liver fibrosis. Inflamm. Res. 2022, 71, 1389–1401. [Google Scholar] [CrossRef]
- Veskovic, M.; Mladenovic, D.; Milenkovic, M.; Tosic, J.; Borozan, S.; Gopcevic, K.; Labudovic-Borovic, M.; Dragutinovic, V.; Vucevic, D.; Jorgacevic, B.; et al. Betaine modulates oxidative stress, inflammation, apoptosis, autophagy, and Akt/mTOR signaling in methionine-choline deficiency-induced fatty liver disease. Eur. J. Pharmacol. 2019, 848, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Monacelli, F.; Acquarone, E.; Giannotti, C.; Borghi, R.; Nencioni, A. Vitamin C, Aging and Alzheimer’s Disease. Nutrients 2017, 9, 670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawaguchi, T.; Nakano, D.; Oriishi, T.; Torimura, T. Effects of isomaltulose on insulin resistance and metabolites in patients with non-alcoholic fatty liver disease: A metabolomic analysis. Mol. Med. Rep. 2018, 18, 2033–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciou, S.C.; Chou, Y.T.; Liu, Y.L.; Nieh, Y.C.; Lu, J.W.; Huang, S.F.; Chou, Y.T.; Cheng, L.H.; Lo, J.F.; Chen, M.J.; et al. Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signaling. Int. J. Cancer 2015, 137, 104–115. [Google Scholar] [CrossRef]
- Moisés Laparra, J.; Díez-Municio, M.; Javier Moreno, F.; Herrero, M. Kojibiose ameliorates arachidic acid-induced metabolic alterations in hyperglycaemic rats. Br. J. Nutr. 2015, 114, 1395–1402. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.-N.; Zhao, C.-Z.; Wang, C.-Z.; Rao, J.-B.; Qiu, Y.-W.; Gao, Y.-P.; Wang, X.-Y.; Zhang, Y.-M.; Wu, G.; Chen, J.; et al. Dataset for liver metabolomic profile of highland barley Monascus purpureus went extract-treated golden hamsters with nonalcoholic fatty liver disease. Data Brief 2022, 40, 107773. [Google Scholar] [CrossRef]
- Mills, E.L.; Harmon, C.; Jedrychowski, M.P.; Xiao, H.; Garrity, R.; Tran, N.V.; Bradshaw, G.A.; Fu, A.; Szpyt, J.; Reddy, A.; et al. UCP1 governs liver extracellular succinate and inflammatory pathogenesis. Nat. Metab. 2021, 3, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-H.; Kim, H. The Roles of Glutamine in the Intestine and Its Implication in Intestinal Diseases. Int. J. Mol. Sci. 2017, 18, 1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakaria, M.; Azam, S.; Haque, M.E.; Jo, S.-H.; Uddin, M.S.; Kim, I.-S.; Choi, D.-K. Taurine and its analogs in neurological disorders: Focus on therapeutic potential and molecular mechanisms. Redox Biol. 2019, 24, 101223. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.K.; Chung, H.W.; Lee, H.-S.; Lim, J.; Park, J.H.; Lim, S.C.; Kim, J.M.; Hong, S.-S.; Kwon, S.W. Investigation of metabolite alteration in dimethylnitrosamine-induced liver fibrosis by GC–MS. Bioanalysis 2013, 5, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhou, Y.; Huang, F.-J.; Tang, H.-D.; Xu, X.-H.; Liu, J.-J.; Wang, Y.; Deng, Y.-L.; Ren, R.-J.; Xu, W.; et al. Plasma Metabolite Profiles of Alzheimer’s Disease and Mild Cognitive Impairment. J. Proteome Res. 2014, 13, 2649–2658. [Google Scholar] [CrossRef] [PubMed]
| Risk Prediction | Western Diet Intake | Aging | Bile Acid Receptor Inactivation | |||
|---|---|---|---|---|---|---|
| Features | Accuracy | Features | Accuracy | Features | Accuracy | |
| Hepatic Transcripts | 9 (4) | 100% (96.9%) | 14 (2) | 100% (90.6%) | 2 | 100% |
| Metabolites | ||||||
| Bile acids (liver) | 2 | 66.6% | 1 | 100% | 10 | 71.3% |
| Liver | 5 (2) | 100% (93.8%) | 20 (12) | 100% (91.7%) | 10 (1) | 100% (95.8%) |
| Serum | 10 | 91.9% | 3 (1) | 100% (95.0%) | 15 (12) | 94.5% (91.3%) |
| Urine | 5 (1) | 100% (91.0%) | 7 (3) | 100% (90.0%) | 9 (3) | 100% (95.4%) |
| Microbiota | ||||||
| Phylum level | 8 | 61.9% | 4 | 70.0% | 6 | 90.2% |
| Class level | 9 | 62.6% | 9 | 82.8% | 3 | 96.9% |
| Order level | 26 | 62.5% | 13 | 82.8% | 3 | 96.9% |
| Family level | 10 | 76.8% | 7 | 80.4% | 8 (3) | 98.8% (91.2%) |
| Genus level | 6 | 68.8% | 7 | 82.0% | 7 (3) | 96.2% (92.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Liu, R.; Rezaei, S.; Liu, X.; Wan, Y.-J.Y. Uncovering the Gut–Liver Axis Biomarkers for Predicting Metabolic Burden in Mice. Nutrients 2023, 15, 3406. https://doi.org/10.3390/nu15153406
Yang G, Liu R, Rezaei S, Liu X, Wan Y-JY. Uncovering the Gut–Liver Axis Biomarkers for Predicting Metabolic Burden in Mice. Nutrients. 2023; 15(15):3406. https://doi.org/10.3390/nu15153406
Chicago/Turabian StyleYang, Guiyan, Rex Liu, Shahbaz Rezaei, Xin Liu, and Yu-Jui Yvonne Wan. 2023. "Uncovering the Gut–Liver Axis Biomarkers for Predicting Metabolic Burden in Mice" Nutrients 15, no. 15: 3406. https://doi.org/10.3390/nu15153406
APA StyleYang, G., Liu, R., Rezaei, S., Liu, X., & Wan, Y.-J. Y. (2023). Uncovering the Gut–Liver Axis Biomarkers for Predicting Metabolic Burden in Mice. Nutrients, 15(15), 3406. https://doi.org/10.3390/nu15153406







