Infant Formula Supplemented with Five Human Milk Oligosaccharides Shifts the Fecal Microbiome of Formula-Fed Infants Closer to That of Breastfed Infants
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort Characteristics
2.2. Sample Collection
2.3. Fecal DNA Extraction and Sequencing
2.4. Microbiome Taxonomic Annotation and Profiling
2.5. Microbiome Functional Annotation and Profiling
2.6. Microbiome Ecological Measures and Statistical Analysis
2.7. Machine Learning Predictions of Clinical Variables
3. Results
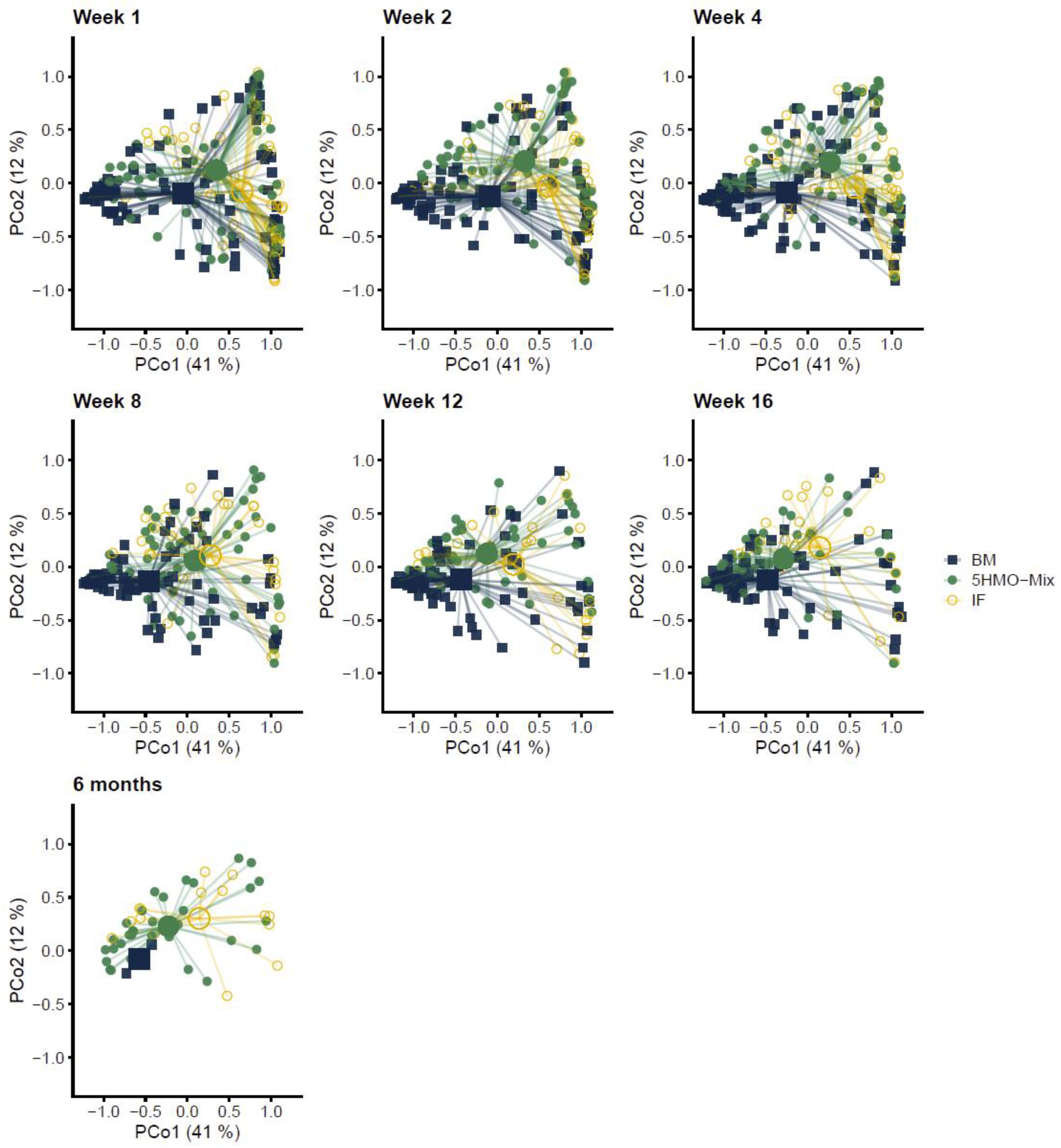
3.1. The Fecal Microbial Composition Differs among Feeding Groups
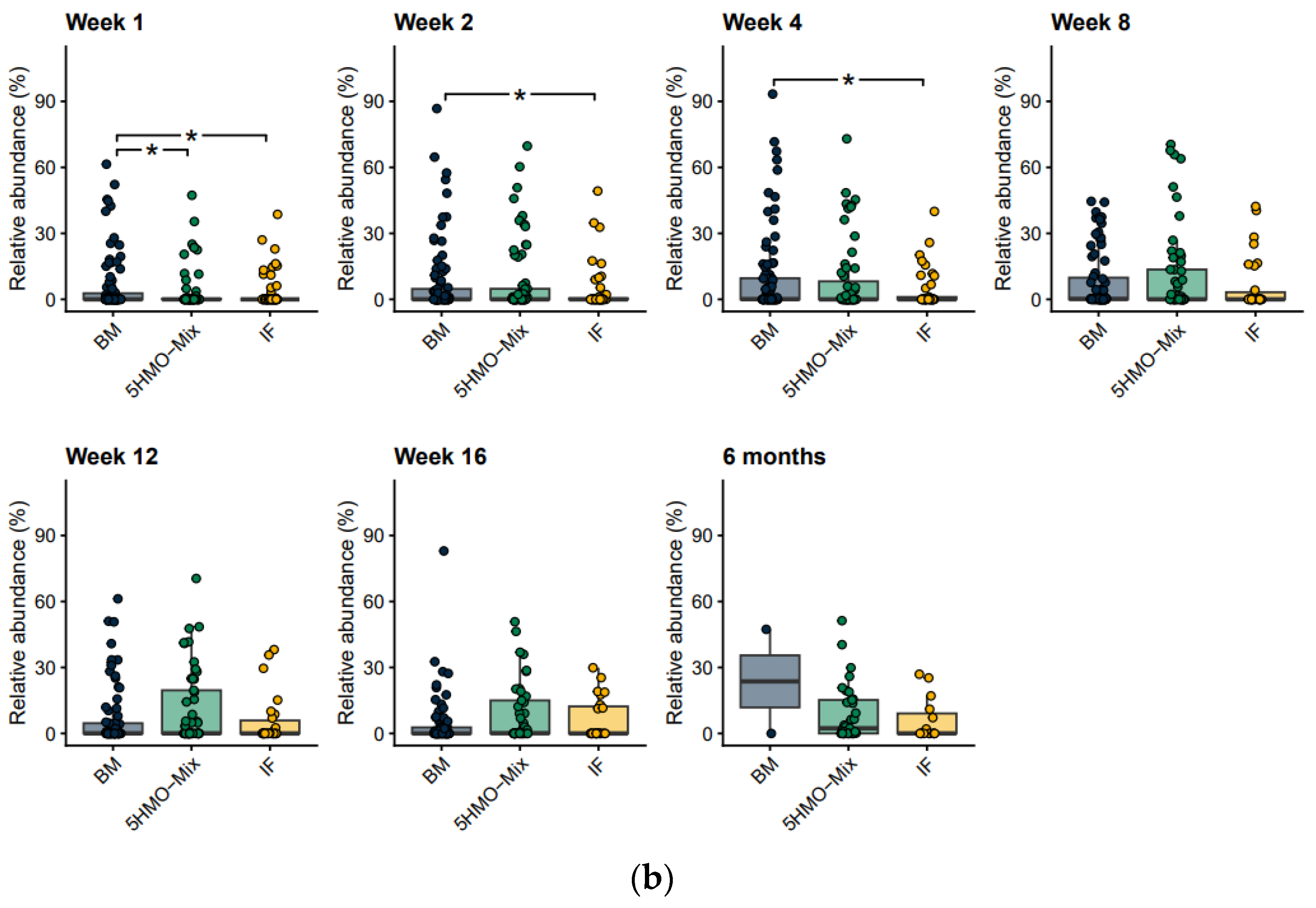
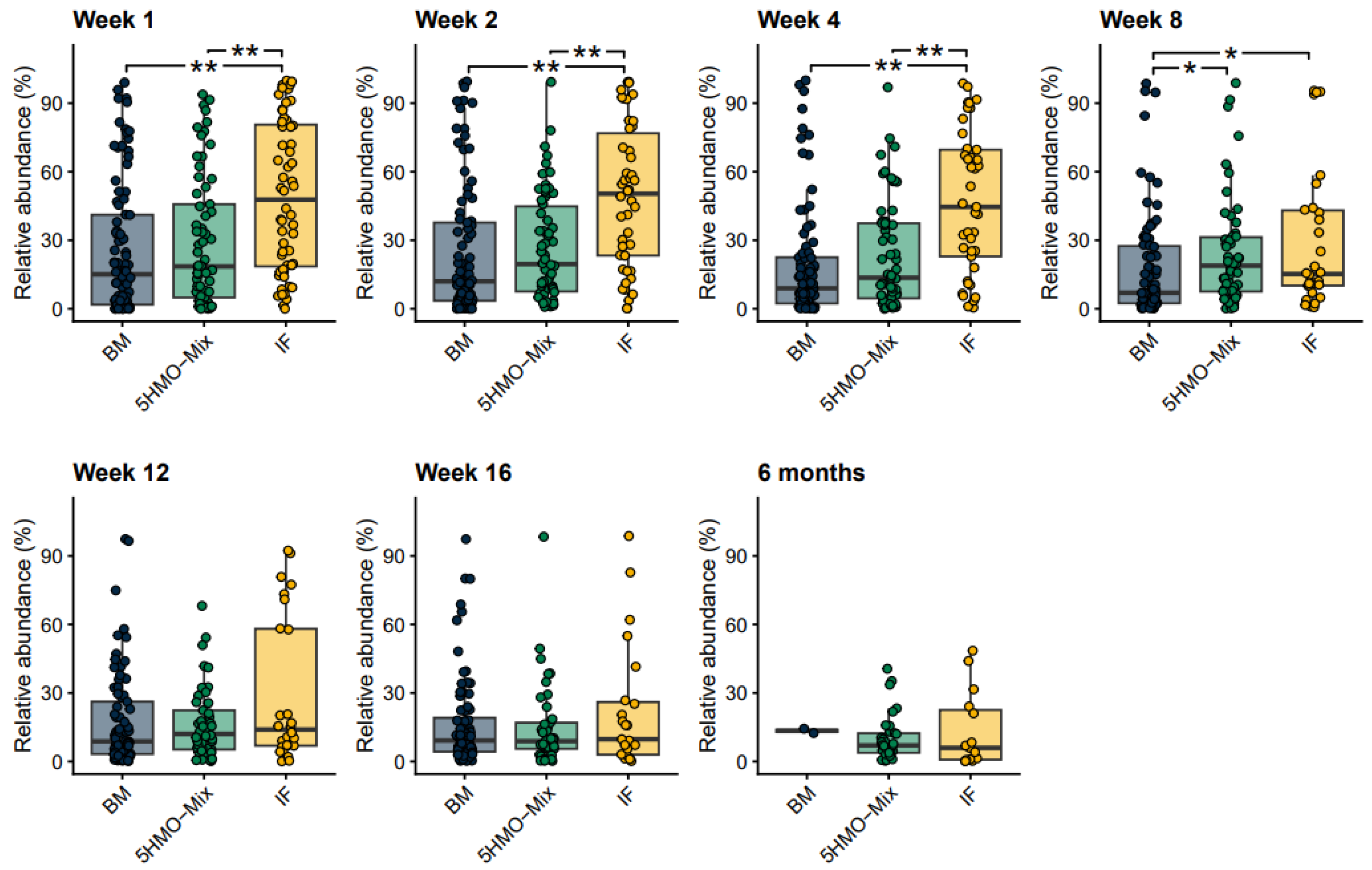
3.2. The Relative Abundance of Bifidobacteria Increases in the 5HMO-Mix Feeding Group
3.3. The 5HMO-Mix Supports Bifidobacterium Species That Are Able to Produce Aromatic Lactic Acids
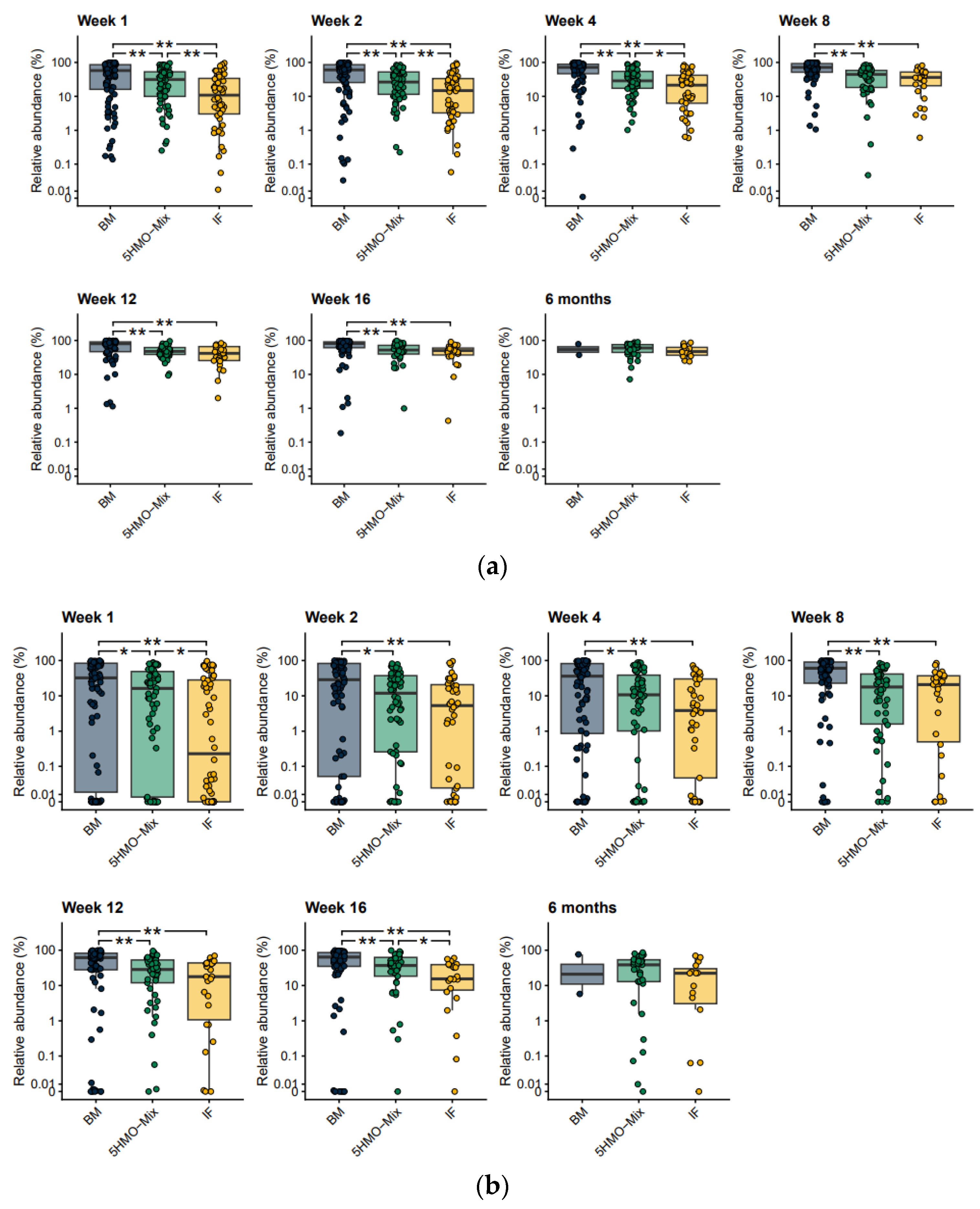
3.4. The Relative Abundance of Opportunistic Pathogens Is Reduced by 5HMO-Mix
3.5. 5HMO-Mix Directs the Functional Potential of the Microbiome towards That of Breastfed Infants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization, Breastfeeding Recommendations. Available online: https://www.who.int/health-topics/breastfeeding (accessed on 1 June 2023).
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health. J. Funct. Foods 2020, 72, 104074. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Sonnenburg, J.L. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012, 18 (Suppl. 4), 12–15. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Rytz, A.; Keddani, S.; Bergonzelli, G.; Garcia-Rodenas, C.L. Blends of human milk oligosaccharides confer intestinal epithelial barrier protection in vitro. Nutrients 2020, 12, 3047. [Google Scholar] [CrossRef]
- Berger, B.; Porta, N.; Foata, F.; Grathwohl, D.; Delley, M.; Moine, D.; Charpagne, A.; Siegwald, L.; Descombes, P.; Alliet, P.; et al. Linking human milk oligosaccharides, infant fecal community types, and later risk to require antibiotics. mBio 2020, 11, e03196-19. [Google Scholar] [CrossRef]
- Asadpoor, M.; Peeters, C.; Henricks, P.A.J.; Varasteh, S.; Pieters, R.J.; Folkerts, G.; Braber, S. Anti-pathogenic functions of non-digestible oligosaccharides in vitro. Nutrients 2020, 12, 1789. [Google Scholar] [CrossRef]
- Newburg, D.S.; Ruiz-Palacios, G.M.; Morrow, A.L. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005, 25, 37–58. [Google Scholar] [CrossRef]
- Ramani, S.; Stewart, C.J.; Laucirica, D.R.; Ajami, N.J.; Robertson, B.; Autran, C.A.; Shinge, D.; Rani, S.; Anandan, S.; Hu, L.; et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018, 9, 5010. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Cervantes, L.E.; Ramos, P.; Chavez-Munguia, B.; Newburg, D.S. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 2003, 278, 14112–14120. [Google Scholar] [CrossRef]
- Chen, X. Human milk oligosaccharides (HMOS): Structure, function, and enzyme-catalyzed synthesis. Adv. Carb. Chem. Biochem. 2015, 72, 113–190. [Google Scholar] [CrossRef]
- Siziba, L.P.; Mank, M.; Stahl, B.; Gonsalves, J.; Blijenberg, B.; Rothenbacher, D.; Genuneit, J. Human milk oligosaccharide profiles over 12 months of lactation: The Ulm SPATZ Health Study. Nutrients 2021, 13, 1973. [Google Scholar] [CrossRef] [PubMed]
- Soyyilmaz, B.; Mikš, M.H.; Röhrig, C.H.; Matwiejuk, M.; Meszaros-Matwiejuk, A.; Vigsnæs, L.K. The mean of milk: A review of human milk oligosaccharide concentrations throughout lactation. Nutrients 2021, 13, 2737. [Google Scholar] [CrossRef] [PubMed]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.T.M.; Man, W.H.; Chu, M.L.J.N.; Arp, K.; Watson, R.L.; Sanders, E.A.M.; Fuentes, S.; et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 4997. [Google Scholar] [CrossRef]
- Stewart, C.J.C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.V.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021, 15, 2574–2590. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.; Kitaoka, M.; Katayama, T. Varied pathways of infant gut-associated bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with Bifidobacteria-rich microbiota formation. Nutrients 2020, 12, 71. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. From lab bench to formulated ingredient: Characterization, production, and commercialization of human milk oligosaccharides. J. Funct. Foods 2020, 72, 104052. [Google Scholar] [CrossRef]
- Marriage, B.J.; Buck, R.H.; Goehring, K.C.; Oliver, J.S.; Williams, J.A. Infants fed a lower calorie formula with 2′ FL show growth and 2′ FL uptake like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 2015, 61, 649–658. [Google Scholar] [CrossRef]
- Puccio, G.; Alliet, P.; Cajozzo, C.; Janssens, E.; Corsello, G.; Sprenger, N.; Wernimont, S.; Egli, D.; Gosoniu, L.; Steenhout, P. Effects of Infant formula with human milk oligosaccharides on growth and morbidity: A randomized multicenter trial. J. Pediatr. Gastroenterol. Nutr. 2017, 64, 624–631. [Google Scholar] [CrossRef]
- Storm, H.M.; Shepard, J.; Czerkies, L.M.; Kineman, B.; Cohen, S.S.; Reichert, H.; Carvalho, R. 2′-Fucosyllactose is well tolerated in a 100% whey, partially hydrolyzed infant formula with Bifidobacterium lactis: A randomized controlled trial. Glob. Pediatr. Health 2019, 6, 2333794X19833995. [Google Scholar] [CrossRef] [PubMed]
- Wallingford, J.C.; Neve Myers, P.; Barber, C.M. Effects of addition of 2-fucosyllactose to infant formula on growth and specific pathways of utilization by Bifidobacterium in healthy term infants. Front. Nutr. 2022, 9, 961526. [Google Scholar] [CrossRef] [PubMed]
- Bosheva, M.; Tokodi, I.; Krasnow, A.; Pedersen, H.K.; Lukjancenko, O.; Eklund, A.C.; Grathwohl, D.; Sprenger, N.; Berger, B.; Cercamondi, C.I. Infant formula with a specific blend of five human milk oligosaccharides drives the gut microbiota development and improves gut maturation markers: A randomized controlled trial. Front. Nutr. 2022, 9, 920362. [Google Scholar] [CrossRef] [PubMed]
- Lasekan, J.; Choe, Y.; Dvoretskiy, S.; Devitt, A.; Zhang, S.; Mackey, A.; Wulf, K.; Buck, R.; Steele, C.; Johnson, M.; et al. Growth and gastrointestinal tolerance in healthy term infants fed milk-based infant formula supplemented with five human milk oligosaccharides (HMOs): A randomized multicenter trial. Nutrients 2022, 14, 2625. [Google Scholar] [CrossRef]
- Parschat, K.; Melsaether, C.; Jäpelt, K.R.; Jennewein, S. Clinical evaluation of 16-week supplementation with 5HMO-Mix in healthy-term human infants to determine tolerability, safety, and effect on growth. Nutrients 2021, 13, 2871. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.K.; Martin, F.P.; Donnicola, D.; Julita, M.; Berger, B.; Sprenger, N. Human milk oligosaccharide-stimulated Bifidobacterium species contribute to prevent later respiratory tract infections. Microorganisms 2021, 9, 1939. [Google Scholar] [CrossRef]
- Martin, F.P.; Tytgat, H.L.P.; Krogh Pedersen, H.; Moine, D.; Eklund, A.C.; Berger, B.; Sprenger, N. Host–microbial co-metabolites modulated by human milk oligosaccharides relate to reduced risk of respiratory tract infections. Front. Nutr. 2022, 9, 1742. [Google Scholar] [CrossRef]
- Conze, D.B.; Kruger, C.L.; Symonds, J.M.; Lodder, R.; Schönknecht, Y.B.; Ho, M.; Derya, S.M.; Parkot, J.; Parschat, K. Weighted analysis of 2′-fucosyllactose, 3-fucosyllactose, lacto-N-tetraose, 3′-sialyllactose, and 6′-sialyllactose concentrations in human milk. Food Chem. Toxicol. 2022, 163, 112877. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Garcia Yunta, R.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016, 1, 16088. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast Genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Dedon, L.R.; Özcan, E.; Rani, A.; Sela, D.A. Bifidobacterium infantis metabolizes 2’fucosyllactose-derived and free fucose through a common catabolic pathway resulting in 1,2-propanediol secretion. Front. Nutr. 2020, 7, 583397. [Google Scholar] [CrossRef] [PubMed]
- Plumbridge, J.; Vimr, E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 1999, 181, 47–54. [Google Scholar] [CrossRef]
- Egan, M.; Motherway, M.O.C.; Ventura, M.; van Sinderen, D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2014, 80, 4414–4426. [Google Scholar] [CrossRef]
- Lang, M.; Binder, M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coors, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B. Mlr3: A modern object-oriented machine learning framework in R. J. Open Source Softw. 2019, 4, 1903. [Google Scholar] [CrossRef]
- Wirbel, J.; Zych, K.; Essex, M.; Karcher, N.; Kartal, E.; Salazar, G.; Bork, P.; Sunagawa, S.; Zeller, G. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021, 22, 93. [Google Scholar] [CrossRef]
- Ikuta, K.S.; Swetschinski, L.R.; Robles Aguilar, G.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Davis Weaver, N.; Wool, E.E.; Han, C.; Gershberg Hayoon, A.; et al. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Xu, J.; Leip, D.D.; Chen, C.H.; Westover, B.P.; Weatherford, J.; Buhler, J.D.; Gordon, J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005, 307, 1955–1959. [Google Scholar] [CrossRef] [PubMed]
- Bolte, E.E.; Moorshead, D.; Aagaard, K.M. Maternal and early life exposures and their potential to influence development of the microbiome. Genome Med. 2022, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Borewicz, K.; Gu, F.; Saccenti, E.; Hechler, C.; Beijers, R.; de Weerth, C.; van Leeuwen, S.S.; Schols, H.A.; Smidt, H. The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020, 10, 4270. [Google Scholar] [CrossRef]
- Hermes, G.D.A.; Eckermann, H.A.; de Vos, W.M.; de Weerth, C. Does entry to center-based childcare affect gut microbial colonization in young infants? Sci. Rep. 2020, 10, 10235. [Google Scholar] [CrossRef]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef]
- Ehrlich, A.M.; Pacheco, A.R.; Henrick, B.M.; Taft, D.; Xu, G.; Huda, M.N.; Mishchuk, D.; Goodson, M.L.; Slupsky, C.; Barile, D.; et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020, 20, 357. [Google Scholar] [CrossRef]
- He, X.; Parenti, M.; Grip, T.; Domellöf, M.; Lönnerdal, B.; Hernell, O.; Timby, N.; Slupsky, C.M. Metabolic phenotype of breast-fed infants, and infants fed standard formula or bovine MFGM supplemented formula: A randomized controlled trial. Sci. Rep. 2019, 9, 339. [Google Scholar] [CrossRef]
- Ames, S.R.; Lotoski, L.C.; Azad, M.B. Comparing early life nutritional sources and human milk feeding practices: Personalized and dynamic nutrition supports infant gut microbiome development and immune system maturation. Gut Microbes 2023, 15, 2190305. [Google Scholar] [CrossRef]
- Gold, M.S.; Quinn, P.J.; Campbell, D.E.; Peake, J.; Smart, J.; Robinson, M.; O’Sullivan, M.; Vogt, J.K.; Pedersen, H.K.; Liu, X.; et al. Effects of an amino acid-based formula supplemented with two human milk oligosaccharides on growth, tolerability, safety, and gut microbiome in infants with cow’s milk protein allergy. Nutrients 2022, 14, 2297. [Google Scholar] [CrossRef]
- Ojima, M.N.; Jiang, L.; Arzamasov, A.A.; Yoshida, K.; Odamaki, T.; Xiao, J.; Nakajima, A.; Kitaoka, M.; Hirose, J.; Urashima, T.; et al. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. ISME J. 2022, 16, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Saturio, S.; Nogacka, A.M.; Alvarado-Jasso, G.M.; Salazar, N.; de los Reyes-Gavilán, C.G.; Gueimonde, M.; Arboleya, S. Role of Bifidobacteria on infant health. Microorganisms 2021, 9, 2415. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Kirjavainen, P.; Eerola, E.; Kero, P.; Salminen, S.; Isolauri, E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 2001, 107, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Sarwar, P.F.; Potter, C.; Comstock, S.S.; Klepac-Ceraj, V. Role of human milk oligosaccharide metabolizing bacteria in the development of atopic dermatitis/eczema. Front. Pediatr. 2023, 11, 1090048. [Google Scholar] [CrossRef]
- He, F.; Ouwehand, A.C.; Isolauri, E.; Hashimoto, H.; Benno, Y.; Salminen, S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol. Med. Microbiol. 2001, 30, 43–47. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Stokholm, J.; Blaser, M.J.; Thorsen, J.; Rasmussen, M.A.; Waage, J.; Vinding, R.K.; Schoos, A.M.M.; Kunøe, A.; Fink, N.R.; Chawes, B.L.; et al. Maturation of the gut microbiome and risk of asthma in childhood. Nature Commun. 2018, 9, 141. [Google Scholar] [CrossRef]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- Forbes, J.D.; Azad, M.B.; Vehling, L.; Tun, H.M.; Konya, T.B.; Guttman, D.S.; Field, C.J.; Lefebvre, D.; Sears, M.R.; Becker, A.B.; et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018, 172, e181161. [Google Scholar] [CrossRef]
- Pärnänen, K.M.M.; Hultman, J.; Markkanen, M.; Satokari, R.; Rautava, S.; Lamendella, R.; Wright, J.; McLimans, C.J.; Kelleher, S.L.; Virta, M.P. Early-life formula feeding is associated with infant gut microbiota alterations and an increased antibiotic resistance load. Am. J. Clin. Nutr. 2022, 115, 407–421. [Google Scholar] [CrossRef]
- Sillner, N.; Walker, A.; Lucio, M.; Maier, T.V.; Bazanella, M.; Rychlik, M.; Haller, D.; Schmitt-Kopplin, P. Longitudinal profiles of dietary and microbial metabolites in formula- and breastfed infants. Front. Mol. Biosci. 2021, 8, 490. [Google Scholar] [CrossRef] [PubMed]
- Victora, C.G.; Bahl, R.; Barros, A.J.D.; França, G.V.A.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Brink, L.R.; Mercer, K.E.; Piccolo, B.D.; Chintapalli, S.V.; Elolimy, A.; Bowlin, A.K.; Matazel, K.S.; Pack, L.; Adams, S.H.; Shankar, K.; et al. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am. J. Clin. Nutr. 2020, 111, 1190–1202. [Google Scholar] [CrossRef]
- Raimondi, S.; Calvini, R.; Candeliere, F.; Leonardi, A.; Ulrici, A.; Rossi, M.; Amaretti, A. Multivariate analysis in microbiome description: Correlation of human gut protein degraders, metabolites, and predicted metabolic functions. Front. Microbiol. 2021, 12, 2639. [Google Scholar] [CrossRef]
- Roager, H.M.; Stanton, C.; Hall, L.J. Microbial metabolites as modulators of the infant gut microbiome and host-microbial interactions in early life. Gut Microbes 2023, 15, 2192151. [Google Scholar] [CrossRef]
- Yao, C.K.; Muir, J.G.; Gibson, P.R. Review article: Insights into colonic protein fermentation, its modulation and potential health implications. Aliment. Pharmacol. Ther. 2016, 43, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef]
- Turroni, F.; Bottacini, F.; Foroni, E.; Mulder, I.; Kim, J.H.; Zomer, A.; Sánchez, B.; Bidossi, A.; Ferrarini, A.; Giubellini, V.; et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl Acad. Sci. USA 2010, 107, 19514–19519. [Google Scholar] [CrossRef]
- Holst, A.Q.; Jois, H.; Laursen, M.F.; Sommer, M.O.A.; Licht, T.R.; Bahl, M.I. Human milk oligosaccharides induce acute yet reversible compositional changes in the gut microbiota of conventional mice linked to a reduction of butyrate levels. MicroLife 2022, 3, uqac006. [Google Scholar] [CrossRef]
- Ryan, J.J.; Monteagudo-Mera, A.; Contractor, N.; Gibson, G.R. Impact of 2’-fucosyllactose on gut microbiota composition in adults with chronic gastrointestinal conditions: Batch culture fermentation model and pilot clinical trial findings. Nutrients 2021, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Barboza, M.; Sonnenburg, E.D.; Pudlo, N.; Martens, E.C.; Desai, P.; Lebrilla, C.B.; Weimer, B.C.; Mills, D.A.; German, J.B.; et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 2011, 10, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Shuoker, B.; Pichler, M.J.; Jin, C.; Sakanaka, H.; Wu, H.; Gascueña, A.M.; Liu, J.; Nielsen, T.S.; Holgersson, J.; Karlsson, E.N.; et al. Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria. Nat. Commun. 2023, 14, 1833. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.S.; Ticer, T.D.; Engevik, M.A. Characterizing the mucin-degrading capacity of the human gut microbiota. Sci. Rep. 2022, 12, 8456. [Google Scholar] [CrossRef] [PubMed]



| Sample Point | Breastmilk (BM) Group | 5HMO-mix Supplement Group | Infant Formula (IF) Control Group |
|---|---|---|---|
| Week 1 | 90 | 65 | 56 |
| Week 2 | 89 | 66 | 46 |
| Week 4 | 85 | 59 | 41 |
| Week 8 | 79 | 54 | 30 |
| Week 12 | 81 | 49 | 26 |
| Week 16 | 80 | 41 | 23 |
| Month 6 | 2 | 33 | 15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holst, A.Q.; Myers, P.; Rodríguez-García, P.; Hermes, G.D.A.; Melsaether, C.; Baker, A.; Jensen, S.R.; Parschat, K. Infant Formula Supplemented with Five Human Milk Oligosaccharides Shifts the Fecal Microbiome of Formula-Fed Infants Closer to That of Breastfed Infants. Nutrients 2023, 15, 3087. https://doi.org/10.3390/nu15143087
Holst AQ, Myers P, Rodríguez-García P, Hermes GDA, Melsaether C, Baker A, Jensen SR, Parschat K. Infant Formula Supplemented with Five Human Milk Oligosaccharides Shifts the Fecal Microbiome of Formula-Fed Infants Closer to That of Breastfed Infants. Nutrients. 2023; 15(14):3087. https://doi.org/10.3390/nu15143087
Chicago/Turabian StyleHolst, Andrea Q., Pernille Myers, Paula Rodríguez-García, Gerben D. A. Hermes, Cathrine Melsaether, Adam Baker, Stina R. Jensen, and Katja Parschat. 2023. "Infant Formula Supplemented with Five Human Milk Oligosaccharides Shifts the Fecal Microbiome of Formula-Fed Infants Closer to That of Breastfed Infants" Nutrients 15, no. 14: 3087. https://doi.org/10.3390/nu15143087
APA StyleHolst, A. Q., Myers, P., Rodríguez-García, P., Hermes, G. D. A., Melsaether, C., Baker, A., Jensen, S. R., & Parschat, K. (2023). Infant Formula Supplemented with Five Human Milk Oligosaccharides Shifts the Fecal Microbiome of Formula-Fed Infants Closer to That of Breastfed Infants. Nutrients, 15(14), 3087. https://doi.org/10.3390/nu15143087






