Gene–Diet Interactions on Metabolic Disease-Related Outcomes in Southeast Asian Populations: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
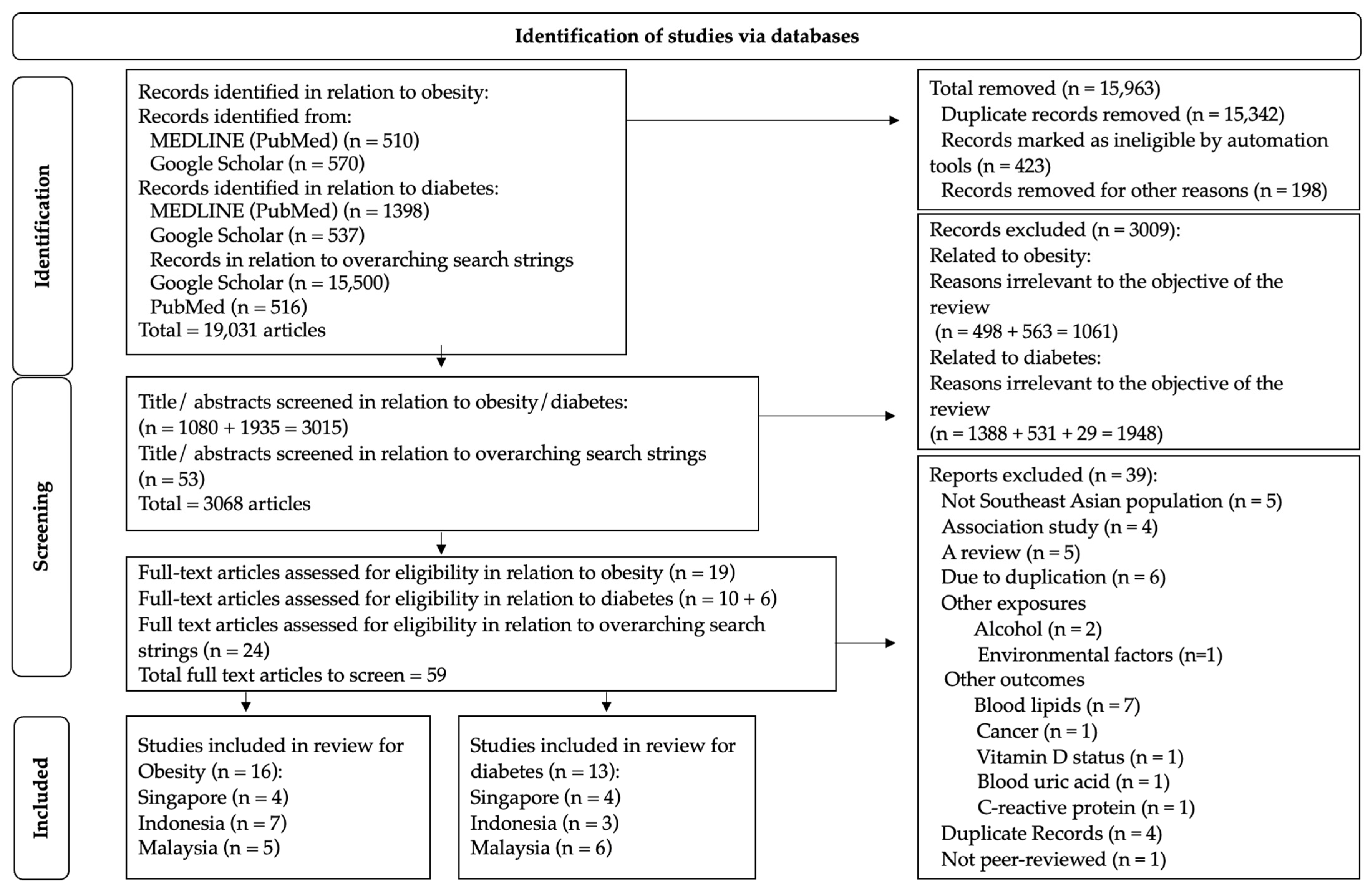
2.1. Study Identification and Source Strategy
2.2. Data Extraction
2.3. Study Selection: Inclusion and Exclusion Criteria
2.4. Data Items and Effect Measures
2.5. Risk of Bias and Certainty Assessment
3. Results and Discussion
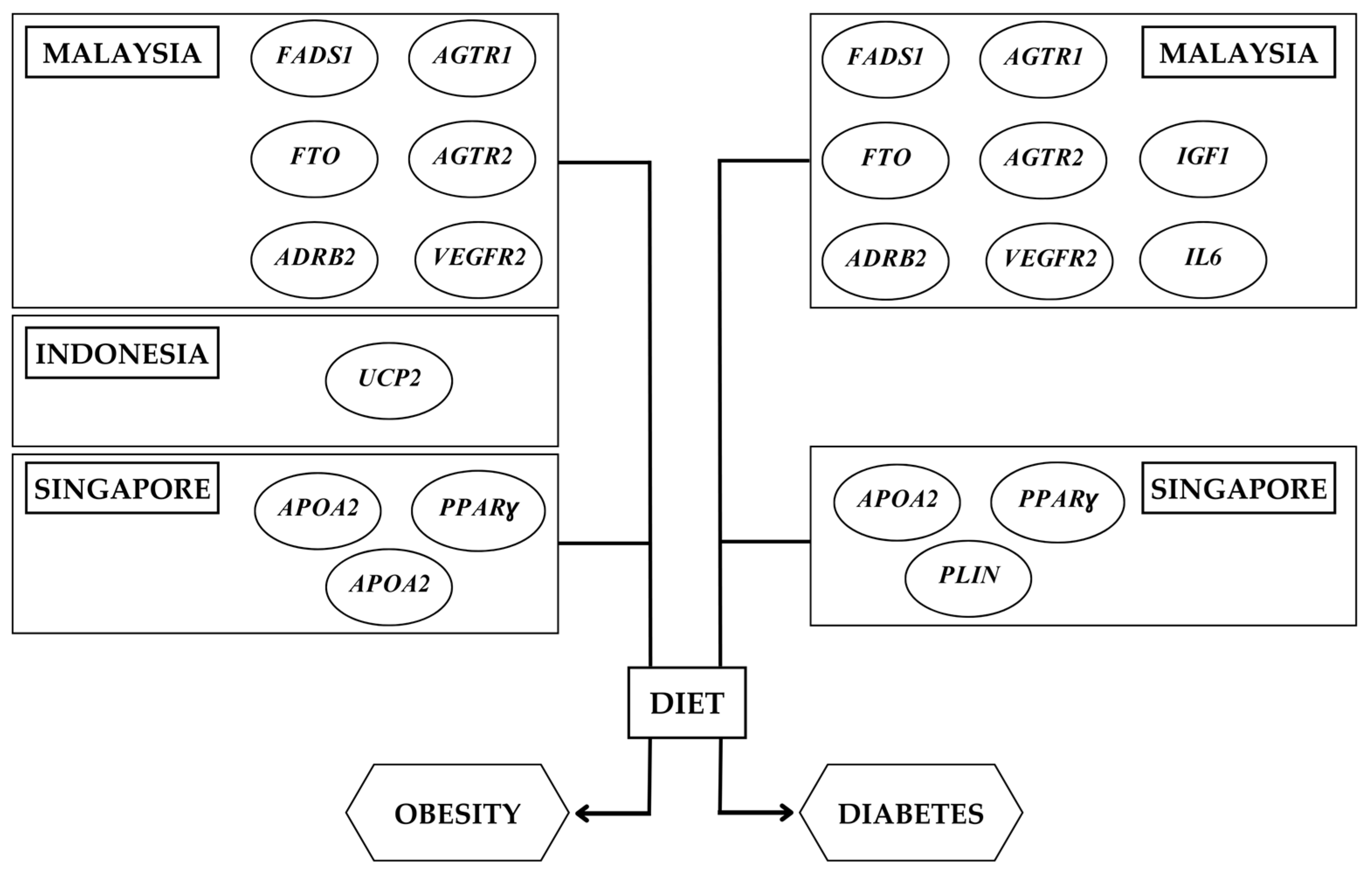
3.1. Nutrigenetics Studies in Southeast Asia
3.2. Gene–Diet Interactions on Obesity-Related Outcomes in the Southeast Asian Population
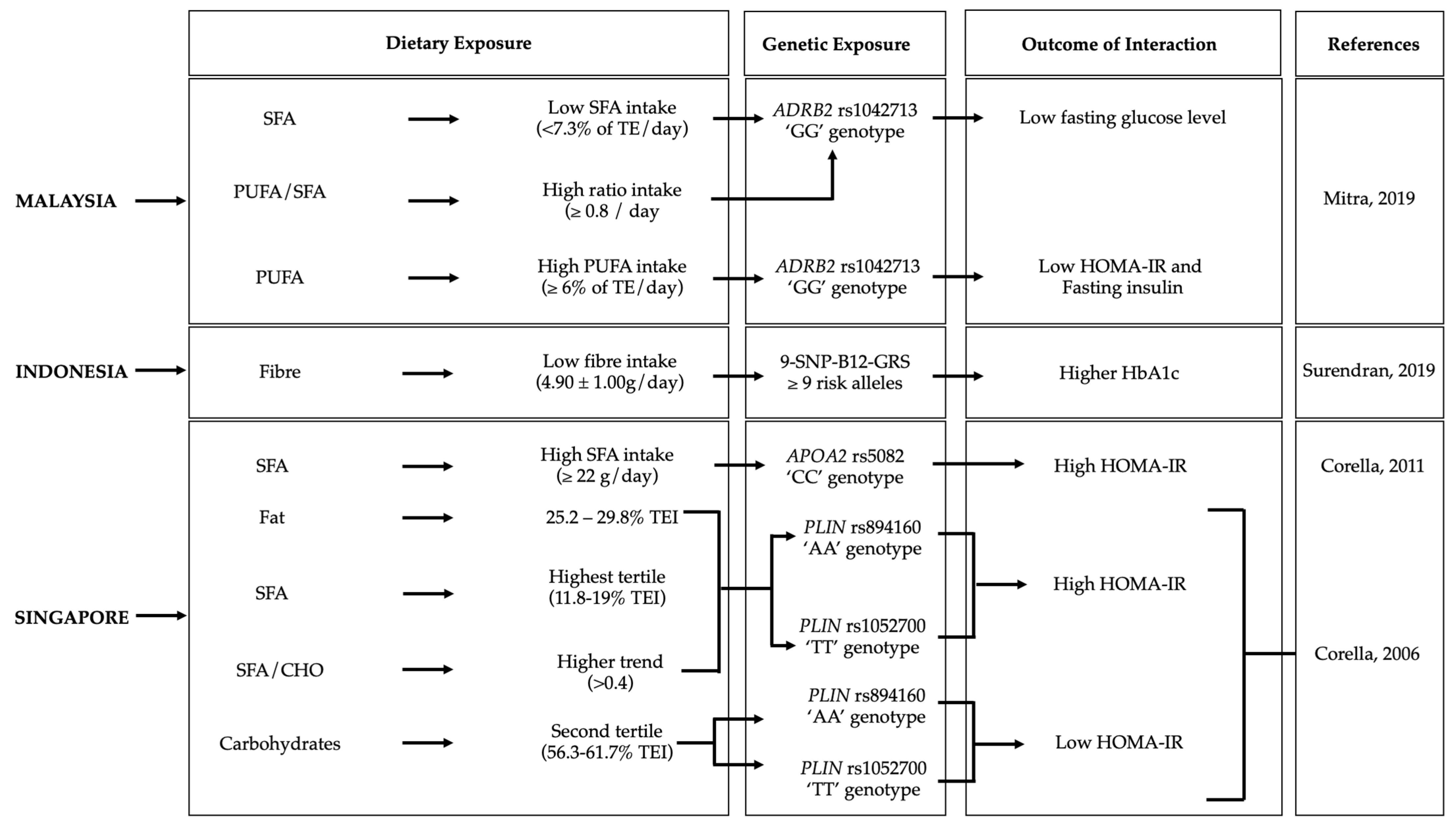
3.2.1. Malaysia
3.2.2. Indonesia
3.2.3. Singapore
3.3. Gene–Diet Interactions on Diabetes-Related Outcomes in the Southeast Asian Population
3.3.1. Malaysia
3.3.2. Indonesia
3.3.3. Singapore
4. Precision Nutrition Approach for the Southeast Asian Population
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Cesare, M.; Sorić, M.; Bovet, P.; Miranda, J.J.; Bhutta, Z.; Stevens, G.A.; Laxmaiah, A.; Kengne, A.P.; Bentham, J. The epidemiological burden of obesity in childhood: A worldwide epidemic requiring urgent action. BMC Med. 2019, 17, 212. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S. GeNuIne (gene-nutrient interactions) Collaboration: Towards implementing multi-ethnic population-based nutrigenetic studies of vitamin B(12) and D deficiencies and metabolic diseases. Proc. Nutr. Soc. 2021, 80, 1–11. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Obesity. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 3 November 2022).
- World Health Organisation. Diabetes. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 3 November 2022).
- World Health Organisation. Obesity and Overweight in South East Asia. Available online: https://www.who.int/southeastasia/health-topics/obesity (accessed on 3 November 2022).
- Biswas, T.; Townsend, N.; Magalhaes, R.J.S.; Islam, M.S.; Hasan, M.M.; Mamun, A. Current Progress and Future Directions in the Double Burden of Malnutrition among Women in South and Southeast Asian Countries. Curr. Dev. Nutr. 2019, 3, nzz026. [Google Scholar] [CrossRef] [PubMed]
- Association of Southeast Asian Nations: ASEAN Food and Nutrition Security Report 2021, Volume 1. Available online: https://asean.org/book/asean-food-and-nutrition-security-report-2021-volume-1-the-asean-secretariat-jakarta/ (accessed on 4 November 2022).
- ASEAN Food and Nutrition Security Report 2021 Volume 2—Food and Nutrition Security Country Profiles. Available online: https://asean.org/book/asean-food-and-nutrition-security-report-2021-volume-2-food-and-nutrition-security-country-profiles/ (accessed on 8 November 2022).
- Phillips, C.M. Nutrigenetics and metabolic disease: Current status and implications for personalised nutrition. Nutrients 2013, 5, 32–57. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S.; Franks, P.W.; Barroso, I.; Brage, S.; Ekelund, U.; Wareham, N.J.; Loos, R.J. Habitual energy expenditure modifies the association between NOS3 gene polymorphisms and blood pressure. Am. J. Hypertens. 2008, 21, 297–302. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Li, S.; Zhao, J.H.; Luan, J.; Bingham, S.A.; Khaw, K.T.; Ekelund, U.; Wareham, N.J.; Loos, R.J. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am. J. Clin. Nutr. 2009, 90, 425–428. [Google Scholar] [CrossRef]
- Alathari, B.E.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Sudji, I.R.; Lovegrove, J.A.; Lipoeto, N.I.; Vimaleswaran, K.S. Interaction between Vitamin D-Related Genetic Risk Score and Carbohydrate Intake on Body Fat Composition: A Study in Southeast Asian Minangkabau Women. Nutrients 2021, 13, 326. [Google Scholar] [CrossRef]
- Bodhini, D.; Gaal, S.; Shatwan, I.; Ramya, K.; Ellahi, B.; Surendran, S.; Sudha, V.; Anjana, M.R.; Mohan, V.; Lovegrove, J.A.; et al. Interaction between TCF7L2 polymorphism and dietary fat intake on high density lipoprotein cholesterol. PLoS ONE 2017, 12, e0188382. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Bodhini, D.; Lakshmipriya, N.; Ramya, K.; Anjana, R.M.; Sudha, V.; Lovegrove, J.A.; Kinra, S.; Mohan, V.; Radha, V. Interaction between FTO gene variants and lifestyle factors on metabolic traits in an Asian Indian population. Nutr. Metab. 2016, 13, 39. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Cavadino, A.; Hyppönen, E. APOA5 genotype influences the association between 25-hydroxyvitamin D and high density lipoprotein cholesterol. Atherosclerosis 2013, 228, 188–192. [Google Scholar] [CrossRef]
- Bishwajit, G. Nutrition transition in South Asia: The emergence of non-communicable chronic diseases. F1000Res 2015, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Periasamy, M. Genetic and Environmental Factors Contributing to Visceral Adiposity in Asian Populations. Endocrinol Metab 2020, 35, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Wuni, R.; Adela Nathania, E.; Ayyappa, A.K.; Lakshmipriya, N.; Ramya, K.; Gayathri, R.; Geetha, G.; Anjana, R.M.; Kuhnle, G.G.C.; Radha, V.; et al. Impact of Lipid Genetic Risk Score and Saturated Fatty Acid Intake on Central Obesity in an Asian Indian Population. Nutrients 2022, 14, 2713. [Google Scholar] [CrossRef] [PubMed]
- Wuni, R.; Kuhnle, G.G.C.; Wynn-Jones, A.A.; Vimaleswaran, K.S. A Nutrigenetic Update on CETP Gene-Diet Interactions on Lipid-Related Outcomes. Curr. Atheroscler. Rep. 2022, 24, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S. A nutrigenetics approach to study the impact of genetic and lifestyle factors on cardiometabolic traits in various ethnic groups: Findings from the GeNuIne Collaboration. Proc. Nutr. Soc. 2020, 79, 194–204. [Google Scholar] [CrossRef]
- Alsulami, S.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; Vimaleswaran, K.S. Interaction between the genetic risk score and dietary protein intake on cardiometabolic traits in Southeast Asian. Genes Nutr. 2020, 15, 19. [Google Scholar] [CrossRef]
- Dhanapal, A.; Wuni, R.; Ventura, E.F.; Chiet, T.K.; Cheah, E.S.G.; Loganathan, A.; Quen, P.L.; Appukutty, M.; Noh, M.F.M.; Givens, I.; et al. Implementation of Nutrigenetics and Nutrigenomics Research and Training Activities for Developing Precision Nutrition Strategies in Malaysia. Nutrients 2022, 14, 5108. [Google Scholar] [CrossRef]
- Thiruvenkataswamy, C.S.; Appukutty, M.; Vimaleswaran, K.S. Role of precision nutrition in improving military performance. Pers. Med. 2022, 19, 167–170. [Google Scholar] [CrossRef]
- Ching, Y.K.; Chin, Y.S.; Appukutty, M.; Ramanchadran, V.; Yu, C.Y.; Ang, G.Y.; Gan, W.Y.; Chan, Y.M.; Teh, L.K.; Salleh, M.Z. Interaction of Dietary Linoleic Acid and α-Linolenic Acids with rs174547 in FADS1 Gene on Metabolic Syndrome Components among Vegetarians. Nutrients 2019, 11, 1686. [Google Scholar] [CrossRef]
- Lee, S.S.; Ling, K.H.; Tusimin, M.; Subramaniam, R.; Rahim, K.F.; Loh, S.P. Interplay between Maternal and Neonatal Vitamin D Deficiency and Vitamin-D-Related Gene Polymorphism with Neonatal Birth Anthropometry. Nutrients 2022, 14, 564. [Google Scholar] [CrossRef]
- Tan, P.Y.; Mitra, S.R. The Combined Effect of Polygenic Risk from FTO and ADRB2 Gene Variants, Odds of Obesity, and Post-Hipcref Diet Differences. Lifestyle Genom. 2020, 13, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Yap, R.W.K.; Shidoji, Y.; Yap, W.S.; Masaki, M. Association and Interaction Effect of AGTR1 and AGTR2 Gene Polymorphisms with Dietary Pattern on Metabolic Risk Factors of Cardiovascular Disease in Malaysian Adults. Nutrients 2017, 9, 853. [Google Scholar] [CrossRef] [PubMed]
- Yap, R.W.; Shidoji, Y.; Hon, W.M.; Masaki, M. Interaction between VEGF receptor-2 gene polymorphisms and dietary patterns on blood glucose and lipid levels in Chinese Malaysian adults. J. Nutr. Nutr. 2011, 4, 309–321. [Google Scholar] [CrossRef]
- Muhammad, H.F.L.; Sulistyoningrum, D.C.; Huriyati, E.; Lee, Y.Y.; Muda, W. The interaction between energy intake, physical activity and UCP2 -866G/A gene variation on weight gain and changes in adiposity: An Indonesian Nutrigenetic Cohort (INDOGENIC). Br. J. Nutr. 2021, 125, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Huriyati, E.; Luglio, H.F.; Ratrikaningtyas, P.D.; Tsani, A.F.; Sadewa, A.H.; Juffrie, M. Dyslipidemia, insulin resistance and dietary fat intake in obese and normal weight adolescents: The role of uncoupling protein 2 -866G/A gene polymorphism. TInt. J. Mol. Epidemiol. Genet. 2016, 7, 67–73. [Google Scholar]
- Muhammad, H.F.L.; Sulistyoningrum, D.C.; Huriyati, E.; Lee, Y.Y.; Manan Wan Muda, W.A. The Interaction between Coffee: Caffeine Consumption, UCP2 Gene Variation, and Adiposity in Adults-A Cross-Sectional Study. J. Nutr. Metab. 2019, 2019, 9606054. [Google Scholar] [CrossRef]
- Surendran, S.; Aji, A.S.; Ariyasra, U.; Sari, S.R.; Malik, S.G.; Tasrif, N.; Yani, F.F.; Lovegrove, J.A.; Sudji, I.R.; Lipoeto, N.I.; et al. A nutrigenetic approach for investigating the relationship between vitamin B12 status and metabolic traits in Indonesian women. J. Diabetes Metab. Disord. 2019, 18, 389–399. [Google Scholar] [CrossRef]
- Aji, A.S.; Lipoeto, N.I.; Yusrawati, Y.; Malik, S.G.; Kusmayanti, N.A.; Susanto, I.; Nurunniyah, S.; Alfiana, R.D.; Wahyuningsih, W.; Majidah, N.M.; et al. Impact of maternal dietary carbohydrate intake and vitamin D-related genetic risk score on birth length: The Vitamin D Pregnant Mother (VDPM) cohort study. BMC Pregnancy Childbirth 2022, 22, 690. [Google Scholar] [CrossRef]
- Chang, X.; Dorajoo, R.; Sun, Y.; Han, Y.; Wang, L.; Khor, C.C.; Sim, X.; Tai, E.S.; Liu, J.; Yuan, J.M.; et al. Gene-diet interaction effects on BMI levels in the Singapore Chinese population. Nutr. J. 2018, 17, 31. [Google Scholar] [CrossRef]
- Huang, T.; Wang, T.; Heianza, Y.; Wiggs, J.; Sun, D.; Choi, H.K.; Chai, J.F.; Sim, X.; Khor, C.C.; Friedlander, Y.; et al. Fish and marine fatty acids intakes, the FADS genotypes and long-term weight gain: A prospective cohort study. BMJ Open 2019, 9, e022877. [Google Scholar] [CrossRef]
- Corella, D.; Tai, E.S.; Sorlí, J.V.; Chew, S.K.; Coltell, O.; Sotos-Prieto, M.; García-Rios, A.; Estruch, R.; Ordovas, J.M. Association between the APOA2 promoter polymorphism and body weight in Mediterranean and Asian populations: Replication of a gene-saturated fat interaction. Int. J. Obes. 2011, 35, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Tai, E.S.; Corella, D.; Deurenberg-Yap, M.; Adiconis, X.; Chew, S.K.; Tan, C.E.; Ordovas, J.M. Differential effects of the C1431T and Pro12Ala PPARgamma gene variants on plasma lipids and diabetes risk in an Asian population. J. Lipid Res. 2004, 45, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Chan, Y.M.; Ramachandran, V.; Shariff, Z.M.; Chin, Y.S.; Arumugam, M. Dietary Acid Load and Its Interaction with IGF1 (rs35767 and rs7136446) and IL6 (rs1800796) Polymorphisms on Metabolic Traits among Postmenopausal Women. Nutrients 2021, 13, 2161. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.R.; Tan, P.Y.; Amini, F. Association of ADRB2 rs1042713 with Obesity and Obesity-Related Phenotypes and Its Interaction with Dietary Fat in Modulating Glycaemic Indices in Malaysian Adults. J. Nutr. Metab. 2019, 2019, 8718795. [Google Scholar] [PubMed]
- Li, H.; Khor, C.C.; Fan, J.; Lv, J.; Yu, C.; Guo, Y.; Bian, Z.; Yang, L.; Millwood, I.Y.; Walters, R.G.; et al. Genetic risk, adherence to a healthy lifestyle, and type 2 diabetes risk among 550,000 Chinese adults: Results from 2 independent Asian cohorts. Am. J. Clin. Nutr. 2020, 111, 698–707. [Google Scholar] [PubMed]
- Corella, D.; Qi, L.; Tai, E.S.; Deurenberg-Yap, M.; Tan, C.E.; Chew, S.K.; Ordovas, J.M. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high-saturated fat, low-carbohydrate diet. Diabetes Care 2006, 29, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Downes, M.J.; Brennan, M.L.; Williams, H.C.; Dean, R.S. Development of a critical appraisal tool to assess the quality of cross-sectional studies (AXIS). BMJ Open 2016, 6, e011458. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef]
- RoB 2: A Revised Cochrane Risk-Of-Bias Tool for Randomized Trials|Cochrane Bias. Available online: https://methods.cochrane.org/bias/resources/rob-2-revised-cochrane-risk-bias-tool-randomized-trials (accessed on 24 October 2022).
- Liu, S.J.; Zhi, H.; Chen, P.Z.; Chen, W.; Lu, F.; Ma, G.S.; Dai, J.C.; Shen, C.; Liu, N.F.; Hu, Z.B.; et al. Fatty acid desaturase 1 polymorphisms are associated with coronary heart disease in a Chinese population. Chin. Med. J. 2012, 125, 801–806. [Google Scholar]
- Agarwal, S.; Kovilam, O.; Agrawal, D.K. Vitamin D and its impact on maternal-fetal outcomes in pregnancy: A critical review. Crit. Rev. Food Sci. Nutr. 2018, 58, 755–769. [Google Scholar] [CrossRef]
- Cohen, J.B.; Stephens-Shields, A.J.; Denburg, M.R.; Anderson, A.H.; Townsend, R.R.; Reese, P.P. Obesity, Renin-Angiotensin System Blockade and Risk of Adverse Renal Outcomes: A Population-Based Cohort Study. Am. J. Nephrol. 2016, 43, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Pahlavani, M.; Kalupahana, N.S.; Ramalingam, L.; Moustaid-Moussa, N. Regulation and Functions of the Renin-Angiotensin System in White and Brown Adipose Tissue. Compr. Physiol. 2017, 7, 1137–1150. [Google Scholar] [PubMed]
- Tam, J.; Duda, D.G.; Perentes, J.Y.; Quadri, R.S.; Fukumura, D.; Jain, R.K. Blockade of VEGFR2 and not VEGFR1 can limit diet-induced fat tissue expansion: Role of local versus bone marrow-derived endothelial cells. PLoS ONE 2009, 4, e4974. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, K.A.; Barroso, I. The genetics of obesity: FTO leads the way. Trend Genet. 2010, 26, 266–274. [Google Scholar] [CrossRef]
- Abbas, M.A. Physiological functions of Vitamin D in adipose tissue. J. Steroid Biochem. Mol. Biol. 2017, 165, 369–381. [Google Scholar] [CrossRef]
- Sirotkin, A.V.; Kolesárová, A. The anti-obesity and health-promoting effects of tea and coffee. Physiol. Res. 2021, 70, 161–168. [Google Scholar] [CrossRef]
- Pan, M.H.; Tung, Y.C.; Yang, G.; Li, S.; Ho, C.T. Molecular mechanisms of the anti-obesity effect of bioactive compounds in tea and coffee. Food Funct. 2016, 7, 4481–4491. [Google Scholar]
- Li, J.; Jiang, R.; Cong, X.; Zhao, Y. UCP2 gene polymorphisms in obesity and diabetes, and the role of UCP2 in cancer. FEBS Lett. 2019, 593, 2525–2534. [Google Scholar] [CrossRef]
- Allister, E.M.; Robson-Doucette, C.A.; Prentice, K.J.; Hardy, A.B.; Sultan, S.; Gaisano, H.Y.; Kong, D.; Gilon, P.; Herrera, P.L.; Lowell, B.B.; et al. UCP2 regulates the glucagon response to fasting and starvation. Diabetes 2013, 62, 1623–1633. [Google Scholar] [CrossRef]
- Evans, R.M.; Barish, G.D.; Wang, Y.X. PPARs and the complex journey to obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef]
- Murff, H.J.; Edwards, T.L. Endogenous Production of Long-Chain Polyunsaturated Fatty Acids and Metabolic Disease Risk. Curr. Cardiovasc. Risk Rep. 2014, 8, 1–9. [Google Scholar] [CrossRef]
- Zaki, M.E.; Amr, K.S.; Abdel-Hamid, M. APOA2 Polymorphism in Relation to Obesity and Lipid Metabolism. Cholesterol 2013, 2013, 289481. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, J.; Lin, H.; Huo, X.; Zhu, Q.; Zhang, M. Relationship between fat mass and obesity-associated gene expression and type 2 diabetes mellitus severity. Exp. Ther. Med. 2018, 15, 2917–2921. [Google Scholar] [CrossRef]
- Grant, S.F.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Manolescu, A.; Sainz, J.; Helgason, A.; Stefansson, H.; Emilsson, V.; Helgadottir, A.; et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006, 38, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Ceasrine, A.M.; Lin, E.E.; Lumelsky, D.N.; Iyer, R.; Kuruvilla, R. Adrb2 controls glucose homeostasis by developmental regulation of pancreatic islet vasculature. Elife 2018, 7, e39689. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Niu, F.; Liu, D.; Sun, J.; Zhang, X.; Zhang, J.; Guo, S. Association between AGTR1 A1166C polymorphism and the susceptibility to diabetic nephropathy: Evidence from a meta-analysis. Medicine 2018, 97, e07689. [Google Scholar] [CrossRef]
- Chou, E.; Suzuma, I.; Way, K.J.; Opland, D.; Clermont, A.C.; Naruse, K.; Suzuma, K.; Bowling, N.L.; Vlahos, C.J.; Aiello, L.P.; et al. Decreased cardiac expression of vascular endothelial growth factor and its receptors in insulin-resistant and diabetic States: A possible explanation for impaired collateral formation in cardiac tissue. Circulation 2002, 105, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Møller, N.; Jørgensen, J.O. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev 2009, 30, 152–177. [Google Scholar] [CrossRef]
- Rehman, K.; Akash, M.S.H.; Liaqat, A.; Kamal, S.; Qadir, M.I.; Rasul, A. Role of Interleukin-6 in Development of Insulin Resistance and Type 2 Diabetes Mellitus. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 229–236. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Wang, B.; Song, Y.; Lin, T.; Zhou, Z.; Wang, Z.; Wei, Y.; Guo, H.; Chen, P.; et al. Vitamin B(12) and risk of diabetes: New insight from cross-sectional and longitudinal analyses of the China Stroke Primary Prevention Trial (CSPPT). BMJ Open Diabetes Res. Care 2020, 8, e001423. [Google Scholar] [CrossRef]
- Duesing, K.; Charpentier, G.; Marre, M.; Tichet, J.; Hercberg, S.; Balkau, B.; Froguel, P.; Gibson, F. Evaluating the association of common APOA2 variants with type 2 diabetes. BMC Med. Genet. 2009, 10, 13. [Google Scholar]
- Castellani, L.W.; Goto, A.M.; Lusis, A.J. Studies with apolipoprotein A-II transgenic mice indicate a role for HDLs in adiposity and insulin resistance. Diabetes 2001, 50, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, S.M.; Lazar, M.A. Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol. Sci. 2004, 25, 331–336. [Google Scholar] [CrossRef]
- Dumont, J.; Goumidi, L.; Grenier-Boley, B.; Cottel, D.; Marécaux, N.; Montaye, M.; Wagner, A.; Arveiler, D.; Simon, C.; Ferrières, J.; et al. Dietary linoleic acid interacts with FADS1 genetic variability to modulate HDL-cholesterol and obesity-related traits. Clin. Nutr. 2018, 37, 1683–1689. [Google Scholar] [CrossRef]
- Chen, Y.; Estampador, A.C.; Keller, M.; Poveda, A.; Dalla-Riva, J.; Johansson, I.; Renström, F.; Kurbasic, A.; Franks, P.W.; Varga, T.V. The combined effects of FADS gene variation and dietary fats in obesity-related traits in a population from the far north of Sweden: The GLACIER Study. Int. J. Obes. 2019, 43, 808–820. [Google Scholar] [CrossRef]
- Qi, L.; Zhang, C.; Greenberg, A.; Hu, F.B. Common variations in perilipin gene, central obesity, and risk of type 2 diabetes in US women. Obesity 2008, 16, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Li, C.; Xie, J.; Xu, G.; Li, Y.; Liu, J.; Chen, B.; Pan, J.; Shen, M.; Yang, L.; et al. Association between three genetic variants of the Perilipin Gene (PLIN) and glucose metabolism: Results from a replication study among Chinese adults and a meta-analysis. Endocr. Res. 2013, 38, 263–279. [Google Scholar] [CrossRef]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Choi, M.S.; Curi, R.; de Luis, D.A.; Gil, Á.; et al. Guide and Position of the International Society of Nutrigenetics/Nutrigenomics on Personalised Nutrition: Part 1Fields of Precision Nutrition. J. Nutr. Nutr. 2016, 9, 12–27. [Google Scholar] [CrossRef]
- Maria Catherine, B. Otero LFB: Role of Nutrigenomics in Modern-Day Healthcare and Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]

| Gene | Genetic Variation | Study Design | n (Men/Women) | Ethnicity | Age (Years) | Dietary Factors | Outcomes | Pinteraction | Interpretation | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Malaysia | ||||||||||
| FADS1 | rs174547 | C-S | 200 (69/131) | Chinese Indians | >18 | Linoleic acid (7.9 ± 3.6 g/day) | WC | 0.177 | No significant interaction; vegetarians with TT genotype had higher odds of MetS, larger WC, and low HDL-c | [24] |
| α-Linolenic Aaid (0.4–0.8 g/day) | 0.258 | |||||||||
| Maternal VDR | rs2228570 | C-S | 217 (107/110) | Malay Chinese Indians Kadazan Bajau Suluk Mixed Ethnic | 28.9 ± 4.2 | Maternal 25OHD (< or >30 nmol/L), infant 25OHD (< or >30 nmol/L) | Infant birth weight | 0.018 | Significant interaction. Inverse association of maternal Vit D deficiency with neonatal birth anthropometry; neonatal G allele carriers associated with higher birth weight | [25] |
| Maternal GC | rs7041 | |||||||||
| Maternal GC | rs4588 | |||||||||
| Cord VDR | rs2228570 | |||||||||
| Cord GC | rs7041 | |||||||||
| Cord GC | rs4588 | |||||||||
| FTO | rs9930501 | RCT | 103 (16/87) | Malay Chinese Indians | >18 | Hipcref (high-protein calorie-restricted) diet | BMI | 0.125 | No significant interactions | [26] |
| rs9930506 | Body weight | 0.058 | ||||||||
| rs9932754 | WC | 0.224 | ||||||||
| ADRB2 | rs1042713 | WHR | 0.369 | |||||||
| Fat mass | 0.234 | |||||||||
| rs1042714 | BFP | 0.468 | ||||||||
| Muscle mass | 0.068 | |||||||||
| AGTR1 | rs5186 | C-S | 507 (154/353) | Malay Chinese Indian | 30–65 | Vegetables, fruits, soy diet (VFSD) in Malay | BMI | 0.994 | No significant interactions; Malay and Chinese showed high risk for lipids with gene–diet interactions but not Indians | [27] |
| Rice, egg and fish diet (REFD) in Chinese | 0.66 | |||||||||
| AGTR2 | rs1403543 | VFSD in Chinese females | 0.053 | |||||||
| VEGFR2 | Rs1870377 | P-C | 179 | Chinese | 30–65 | Meat, rice, and noodles diet | BMI | 0.408 | No significant interactions | [28] |
| Indonesia | ||||||||||
| UCP2 | rs659366 (−866G/A) AA + GA Genotype | P-C | 203 | Indonesian | 20–56 | Total energy intake (kcal) [2570 ± 1066 (baseline) to 2120 ± 1042 (after 2 years)] | Changes in body weight (kg) | 0.263 | No significant interactions | [29] |
| Changes in % body fat | 0.303 | |||||||||
| Changes in WHR | 0.464 | |||||||||
| Fat intake % [23.9 ± 11.2] | Changes in body weight (kg) | 0.896 | ||||||||
| Changes in % body fat | 0.965 | |||||||||
| Changes in WHR | 0.996 | |||||||||
| Carbohydrate intake % [63.9 ± 11.2 (baseline) to 64.2 ± 10.4 (after 2 years)] | Changes in body weight (kg) | 0.433 | ||||||||
| Changes in % body fat | 0.839 | |||||||||
| Changes in WHR | 0.665 | |||||||||
| Protein intake % [12·3 ± 3.4 (baseline) to 12.4 ± 3.7 (after 2 years)] | Changes in body weight (kg) | 0.076 | ||||||||
| Changes in % body fat | 0.360 | |||||||||
| Changes in WHR | 0.355 | |||||||||
| Physical activity (MET-min/week) | Changes in body weight (kg) | 0.251 | ||||||||
| Changes in % body fat | 0.979 | |||||||||
| Changes in WHR | 0.684 | |||||||||
| UCP2 | rs659366 (−866G/A) GG Genotype | P-C | 120 | Indonesian | 20–56 | Total energy intake (kcal) [2570 ± 1066 (baseline) to 2120 ±1042 (after 2 years)] | Changes in body weight (kg) | 0.016 | Significant interaction between UCP2 gene variation and total energy intake on body weight change and BFP Significant interaction between UCP2 gene variation and physical activity on WHR | |
| Changes in % body fat | 0.034 | |||||||||
| Changes in WHR | 0.070 | |||||||||
| Fat intake % [23.9 ±11.2] | Changes in body weight (kg) | 0.682 | ||||||||
| Changes in % body fat | 0.974 | |||||||||
| Changes in WHR | 0.753 | |||||||||
| Carbohydrate intake % [63.9 ± 11.2 (baseline) to 64.2 ±10.4 (after 2 years)] | Changes in body weight (kg) | 0.580 | ||||||||
| Changes in % body fat | 0.771 | |||||||||
| Changes in WHR | 0.826 | |||||||||
| Protein intake % [12·3 ± 3.4 (baseline) to 12.4 ± 3.7 (after 2 years)] | Changes in body weight (kg) | 0.830 | ||||||||
| Changes in % body fat | 0.913 | |||||||||
| Changes of WHR | 0.103 | |||||||||
| Physical activity (MET-min/week) | Changes in body weight (kg) | 0.666 | ||||||||
| Changes in % body fat | 0.653 | |||||||||
| Changes in WHR | 0.040 | |||||||||
| GRS | Vitamin D GRS [DHCR7, CYP2R1, CYP24A1, GC, CASR] | C-S | 110 | Minangkabau women | 25–60 | Carbohydrate [(235.2 g ± 73.5)] | BFP | 0.049 | A significant interaction between Vitamin D GRS and carbohydrate intake on logBFP; carriers of more than 2 risk alleles and consumed high carbohydrate amounts had significantly high logBFP | [12] |
| Metabolic GRS [FTO, TCF7L2, MC4R, KCNQ1, CDKN2A/B] | Carbohydrate [(235.2 g ± 73.5)] and protein [77.87 g ± 220.5] | BMI WC BFP | 0.997 | No significant interactions | ||||||
| UCP2 | rs659366 (−866G/A) | C-C | 261 (145/116) | Indonesian | 15–21 | Fat intake % [(>24.14 ± 5.95] | Obesity | 0.006 | A significant interaction between UCP2 rs659366 (−866G/A) and fat intake on obesity | [30] |
| UCP2 | rs659366 (−866 AA + GA) | C-S | 455 (223/232) | Indonesian | 19–56 | Coffee intake ml (34.5 ± 89.6) | BMI | 0.214 | Significant interaction between UCP2 rs659366 (−866 AA + GA) and coffee intake on body fat | [31] |
| Body fat | 0.015 | |||||||||
| WC | 0.302 | |||||||||
| Hip circumference | 0.253 | |||||||||
| rs659366 (−866 GG) | Coffee intake ml (37.1 ± 106.4) | BMI | 0.231 | |||||||
| Body fat | 0.313 | |||||||||
| WC | 0.510 | |||||||||
| Hip circumference | 0.421 | |||||||||
| GRS | 9-SNP-B12-GRS | C-S | 117 | Minangkabau women | 25–60 | Fat (59.00 g ± 33.10) | BMI | 0.933 | A significant interaction between B12 GRS and protein energy (%) on BFP | [32] |
| WC | 0.444 | |||||||||
| BFP | 0.275 | |||||||||
| Carbohydrate (233 g ± 71) | BMI | 0.685 | ||||||||
| WC | 0.875 | |||||||||
| BFP | 0.064 | |||||||||
| Protein energy (76.90 g ± 36.50) | BMI | 0.993 | ||||||||
| WC | 0.395 | |||||||||
| BFP | 0.034 | |||||||||
| Fiber energy (8.80 g ± 4.50) | BMI | 0.155 | ||||||||
| WC | 0.547 | |||||||||
| BFP | 0.697 | |||||||||
| 9-SNP-Metabolic-GRS | Fat (59.00 g ± 33.10) | BMI | 0.422 | A significant interaction between metabolic GRS and protein energy (%) on WC | ||||||
| WC | 0.812 | |||||||||
| BFP | 0.775 | |||||||||
| Carbohydrate ((233 g ± 71) | BMI | 0.230 | ||||||||
| WC | 0.072 | |||||||||
| BFP | 0.844 | |||||||||
| Protein energy (76.90 g ± 36.50) | BMI | 0.110 | ||||||||
| WC | 0.032 | |||||||||
| BFP | 0.568 | |||||||||
| Fiber energy (8.80 g ± 4.50) | BMI | 0.273 | ||||||||
| WC | 0.648 | |||||||||
| BFP | 0.423 | |||||||||
| GRS | 6SNP-Vitamin D-GRS (≤3) | P-C | 183 | Minangkabau women | 29.6 ± 5.56 | Maternal carbohydrate intake during third trimester | Infant birth weight | 0.611 | Significant interactions between VDR GRS and carbohydrate intake on new-born birth length Pregnant women with a high genetic risk of vitamin D deficiency with high carbohydrate intake gave birth to babies with lower birth lengths | [33] |
| Infant birth length | 0.065 | |||||||||
| 6SNP-Vitamin D-GRS (≥4) | Infant birth weight | 0.872 | ||||||||
| Infant birth length | 0.073 | |||||||||
| 4-SNP-GRS[DHCR7, GC, CYP24A1 and CYP2R1] (<3) | Infant birth weight | 0.841 | ||||||||
| Infant birth length | 0.256 | |||||||||
| 4-SNP-GRS[DHCR7, GC, CYP24A1 and CYP2R1] (≥3) | Infant birth weight | 0.795 | ||||||||
| Infant birth length | 0.079 | |||||||||
| 2-SNP-VDR-GRS (<2) | Infant birth weight | 0.810 | ||||||||
| Infant birth length | 0.032 | |||||||||
| 2-SNP-VDR-GRS (≥2) | Infant birth weight | 0.775 | ||||||||
| Infant birth length | 0.099 | |||||||||
| GRS | 15-SNP-cardiometabolic disease related traits-GRS | C-S | 110 | Minangkabau women | 25–60 | Carbohydrate (53.97 ± 9.44) | BMI | 0.961 | Significant interaction between GRS and protein intake on obesity-related outcome | [21] |
| WC | 0.224 | |||||||||
| Protein (13.51 ± 1.18% TEI) | BMI | 0.282 | ||||||||
| WC | 0.002 | |||||||||
| Fat (28.95 ± 7.99) | BMI | 0.721 | ||||||||
| WC | 0.577 | |||||||||
| BMI | 0.876 | |||||||||
| Fiber g (8.78 ± 4.29) | WC | 0.614 | ||||||||
| Singapore | ||||||||||
| CCDC171 | rs4740619 | C-S P-C | 7817 | Chinese | 24–95 | Cholesterol (lowest consumption group 184.35 ± 116.00 to highest consumption group 260.61 ± 150.56) | BMI | 0.043 | Significant interaction was observed; CCDC171 rs4740619 interaction with cholesterol showed increased BMI level in subjects | [34] |
| FADS | rs174570 | P-C | 5264 | Chinese | 30–55 | Total fish (0.16 servings per day ±0.07) Food sourced EPA + DHA (0.33 g/d ±0.20) | BMI | 0.035 | Significant interaction was observed.; long-term BMI changes in people with high fish/n-3 PUFA intake carrying signature allele show increased weight gain and risk of obesity | [35] |
| APOA2 | rs5082(−265T > C) | C-S | 3605 (1714/1891) | Chinese Malay Indian | 18–69 | SFA intake (22 g) | BMI | 0.758 | No significant interactions | [36] |
| PPAR-ϒ | rs1801282(Pro12Ala) | C-S | 4038 (1869/2169) | Chinese Malay Indian | 18–69 | PUFA/SFA | BMI | 0.873 | No significant interactions | [37] |
| rs3856806(C1431T) | 0.472 | |||||||||
| Genes | Genetic Variations | Study Design | n (Male/Female) | Ethnicity | Age (Years) | Dietary Factors | Outcomes | Pinteraction | Interpretation | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Malaysia | ||||||||||
| FTO | rs9930501 | RCT | 103 (16/87) | Malay Chinese Indians | >18 | Hipcref a (high-protein calorie-restricted) control diet b | Fasting glucose | 0.381 | Significant interaction observed;participants showed a greater reduction in hsCRP levels with the Hipcref diet compared to normal diet | [26] |
| rs9930506 | Fasting insulin | 0.121 | ||||||||
| rs9932754 | ||||||||||
| ADRB2 | rs1042713 | HOMA-IR | 0.122 | |||||||
| rs1042714 | hs-CRP | 0.048 | ||||||||
| AGTR1 | rs5186 | C-S | 507 (154/353) | Malay Chinese Indian | 30–65 | Vegetables, fruits, and soy diet (VFSD) in Malays | HbA1C | 0.537 | No significant interactions | [27] |
| Rice, egg, and fish diet (REFD) in Chinese | 0.844 | |||||||||
| AGTR2 | rs1403543 | VFSD in Chinese females | 0.989 | |||||||
| VEGFR2 | rs1870377 | C-S | 179 | Chinese | 30–65 | Meat, rice, and noodles diet | Blood glucose | p > 0.05 | No significant interactions | [28] |
| HbA1c | p > 0.05 | |||||||||
| FADS1 | rs174547 | C-S | 200 (69/131) | Chinese Indians | >18 | Linoleic acid (7.9 ± 3.6 g/day) | Log FBG | 0.807 | No significant interactions | [24] |
| α-Linolenic acid (0.4–0.8 g/day) | 0.293 | |||||||||
| IGF1 | rs35767 | C-S | 211 | Chinese | 66.7 ± 6 | DAL (using PRAL) | FBG | NS | No significant interactions; study shows association between DAL and high FBG, indicating a potential risk factor for diabetes | [38] |
| IGF1 | rs7136446 | |||||||||
| IL6 | rs1800796 | |||||||||
| ADRB2 | rs1042713 | C-S | 126 | Malaysian Chinese Indians | 18–74 | Saturated fat intake (<7.3% of total energy/day) PUFA intake (≥0.8/day) PUFA:SFA ratio (≥6% of TE/ day) | FBG | 0.011 | Significant gene diet interactions; G allele carriers of ADRB2 rs1042713 are associated with increased odds of insulin resistance | [39] |
| HOMA-IR | 0.026 | |||||||||
| Fasting insulin | 0.036 | |||||||||
| Indonesia | ||||||||||
| GRS | 9-SNP-B12-GRS | C-S | 117 | Minangkabau women | 25–60 | Fat (59.00 ± 33.10) | log HbA1c | 0.175 | Significant interaction between B12 GRS and fiber intake on HbA1c levels; individuals with ≥9 risk alleles who consumed low fiber diet had significantly higher HbA1c levels | [32] |
| log FBG | 0.374 | |||||||||
| log fasting serum insulin | 0.757 | |||||||||
| Carbohydrate (233 ± 71) | log HbA1c | 0.091 | ||||||||
| log FBG | 0.260 | |||||||||
| log fasting serum insulin | 0.341 | |||||||||
| Protein (76.90 ± 36.50) | log HbA1c | 0.150 | ||||||||
| log FBG | 0.368 | |||||||||
| log fasting serum insulin | 0.073 | |||||||||
| Fiber intake (8.80 ± 4.50) | log HbA1c | 0.042 | ||||||||
| log FBG | 0.380 | |||||||||
| log fasting serum insulin | 0.215 | |||||||||
| 9-SNP-Metabolic-GRS | Fat (59.00 ± 33.10) | log HbA1c | 0.298 | |||||||
| log FBG | 0.634 | |||||||||
| log fasting serum insulin | 0.108 | |||||||||
| Carbohydrate (233 ± 71) | log HbA1c | 0.166 | ||||||||
| log FBG | 0.771 | |||||||||
| log fasting serum insulin | 0.104 | |||||||||
| Protein (76.90 ± 36.50) | log HbA1c | 0.155 | ||||||||
| log FBG | 0.929 | |||||||||
| log fasting serum insulin | 0.890 | |||||||||
| Fiber intake (g/d) (4.90 ± 1.00 g/day) | log HbA1c | 0.851 | ||||||||
| log FBG | 0.215 | |||||||||
| log fasting serum insulin | 0.947 | |||||||||
| GRS | Metabolic GRS [FTO, TCF7L2, MC4R, KCNQ1, CDKN2A/B] | C-S | 110 | Minangkabau women | 25–60 | Carbohydrate (g/d) (233.7 ± 75.1 g) | Glucose | 0.360 | No significant interactions | [12] |
| HbA1c | 0.780 | |||||||||
| Fasting insulin | 0.630 | |||||||||
| Protein (g/d) (77.2 ± 41.7 g) | Glucose | 0.560 | ||||||||
| HbA1c | 0.680 | |||||||||
| Fasting insulin | 0.220 | |||||||||
| Fat (g/d) (61.2 ± 36.1 g) | Glucose | 0.700 | ||||||||
| HbA1c | 0.780 | |||||||||
| Fasting insulin | 0.440 | |||||||||
| Fiber (g/d) (8.6 ± 4.3 g) | Glucose | 0.830 | ||||||||
| HbA1c | 0.530 | |||||||||
| Fasting insulin | 0.440 | |||||||||
| GRS | 15-SNP-cardiometabolic disease related traits-GRS | C-S | 110 | Minangkabau women | 25–60 | Carbohydrates % (53.97 ± 9.44) | logGlucose | 0.882 | No significant interactions | [21] |
| logInsulin | 0.336 | |||||||||
| logHbA1c | 0.766 | |||||||||
| Protein % (16.93 ± 3.32) | logGlucose | 0.751 | ||||||||
| logInsulin | 0.341 | |||||||||
| logHbA1c | 0.638 | |||||||||
| Fat % (28.95 ± 7.99) | logGlucose | 0.732 | ||||||||
| logInsulin | 0.480 | |||||||||
| logHbA1c | 0.935 | |||||||||
| Fiber g (8.78 ± 4.29) | logGlucose | 0.833 | ||||||||
| logInsulin | 0.216 | |||||||||
| logHbA1c | 0.162 | |||||||||
| Singapore | ||||||||||
| GRS | DM 37-SNP GRS | P-C | 38,434 | Chinese | 30–79 | Alcohol (men: 10–25 g/d; women: 5–15 g/d) | Diabetes risk | NS | No significant interaction; but a healthy lifestyle and any genetic risk category was associated with a significantly lower risk of diabetes | [40] |
| VFSD pattern | ||||||||||
| Meat—dim sum pattern | ||||||||||
| PLIN | rs894160(11482G > A) | C-S | 4107 | Chinese Malay Indian | 18–69 | Total fat % (25.2–29.8 TEI) | Fasting glucose | 0.425 | Significant interaction between PLIN variants and dietary factors on diabetes related outcomes | [41] |
| Fasting insulin | 0.01 | |||||||||
| HOMA-IR | 0.007 | |||||||||
| SFA % (9.4–11.8) | Fasting glucose | 0.004 | ||||||||
| Fasting insulin | 0.004 | |||||||||
| HOMA-IR | 0.003 | |||||||||
| Carbohydrates % (56.3–61.7) | Fasting glucose | 0.145 | ||||||||
| Fasting insulin | 0.007 | |||||||||
| HOMA-IR | 0.004 | |||||||||
| rs1052700(14995A > T) | Total fat % (25.2–29.8 TEI) | Fasting glucose | 0.448 | |||||||
| Fasting insulin | 0.014 | |||||||||
| HOMA-IR | 0.012 | |||||||||
| SFA % (9.4–11.8) | Fasting glucose | 0.009 | ||||||||
| Fasting insulin | 0.014 | |||||||||
| HOMA-IR | 0.005 | |||||||||
| Carbohydrates % (56.3–61.7) | Fasting glucose | 0.293 | ||||||||
| Fasting insulin | 0.008 | |||||||||
| HOMA-IR | 0.012 | |||||||||
| PPAR-ϒ | rs1801282(Pro12Ala) | C-S | 4038 1869/2169) | Chinese Malay Indian | 18–69 | PUFA/SFA | Insulin | 0.089 | No significant interactions | [37] |
| rs3856806(C1431T) | 0.175 | |||||||||
| APOA2 | rs5082(−265T > C) | C-S | 3605 (1714/1891) | Chinese Malay Indian | 18–69 | SFA intake (22 g) | HOMA-IR | 0.026 | Significant interaction between APOA2 rs5082(−265T > C) and SFA intake on HOMA-IR | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sekar, P.; Ventura, E.F.; Dhanapal, A.C.T.A.; Cheah, E.S.G.; Loganathan, A.; Quen, P.L.; Appukutty, M.; Taslim, N.A.; Hardinsyah, H.; Md Noh, M.F.; et al. Gene–Diet Interactions on Metabolic Disease-Related Outcomes in Southeast Asian Populations: A Systematic Review. Nutrients 2023, 15, 2948. https://doi.org/10.3390/nu15132948
Sekar P, Ventura EF, Dhanapal ACTA, Cheah ESG, Loganathan A, Quen PL, Appukutty M, Taslim NA, Hardinsyah H, Md Noh MF, et al. Gene–Diet Interactions on Metabolic Disease-Related Outcomes in Southeast Asian Populations: A Systematic Review. Nutrients. 2023; 15(13):2948. https://doi.org/10.3390/nu15132948
Chicago/Turabian StyleSekar, Padmini, Eduard Flores Ventura, Anto Cordelia T. A. Dhanapal, Eddy Seong Guan Cheah, Annaletchumy Loganathan, Phoon Lee Quen, Mahenderan Appukutty, Nurpudji Astuti Taslim, Hardinsyah Hardinsyah, Mohd Fairulnizal Md Noh, and et al. 2023. "Gene–Diet Interactions on Metabolic Disease-Related Outcomes in Southeast Asian Populations: A Systematic Review" Nutrients 15, no. 13: 2948. https://doi.org/10.3390/nu15132948
APA StyleSekar, P., Ventura, E. F., Dhanapal, A. C. T. A., Cheah, E. S. G., Loganathan, A., Quen, P. L., Appukutty, M., Taslim, N. A., Hardinsyah, H., Md Noh, M. F., Lovegrove, J. A., Givens, I., & Vimaleswaran, K. S. (2023). Gene–Diet Interactions on Metabolic Disease-Related Outcomes in Southeast Asian Populations: A Systematic Review. Nutrients, 15(13), 2948. https://doi.org/10.3390/nu15132948








