Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems
Abstract
:1. Introduction
1.1. Automated Insulin Delivery (AID) Technologies
1.2. Motivation to Study Glucose Variability
1.3. Paper Contributions and Organisation
- Methods and techniques for data cleaning and glycemic variability analysis for large-scale diabetes data, i.e., OpenAPS Data Commons, originating from open-source AID technologies. We further calculate standard clinical metrics to analyse the glucose variability outcomes and add to previous demonstrations of the effectiveness of open-source AID technologies.
- The application of a machine-learning-based hierarchical clustering algorithm in order to understand distinct patterns across glucose profiles of insulin-requiring individuals. We discovered that there are no obvious sub-populations in the open-source AID user community that are being underserved.
- The first in-depth timeseries analysis for glucose variability outcomes and data-driven comparative analysis of the outcomes in open-source AID users based on self-reported population genders. We demonstrate some gender-wise differences in glucose mean and distribution during the times of a day, days of a week and month, and months of a year.
2. Related Work
2.1. Popular Metrics for Assessing Glucose Variability
2.2. Software Tools for Automated Variability Analysis of Continuous Glucose Monitoring (CGM) Data
2.3. Comprehensive Review of Efficiency and Performance of Glucose Variability Metrics
3. Materials and Methods
3.1. Diabetes Data Collection and Anonymisation Highlights
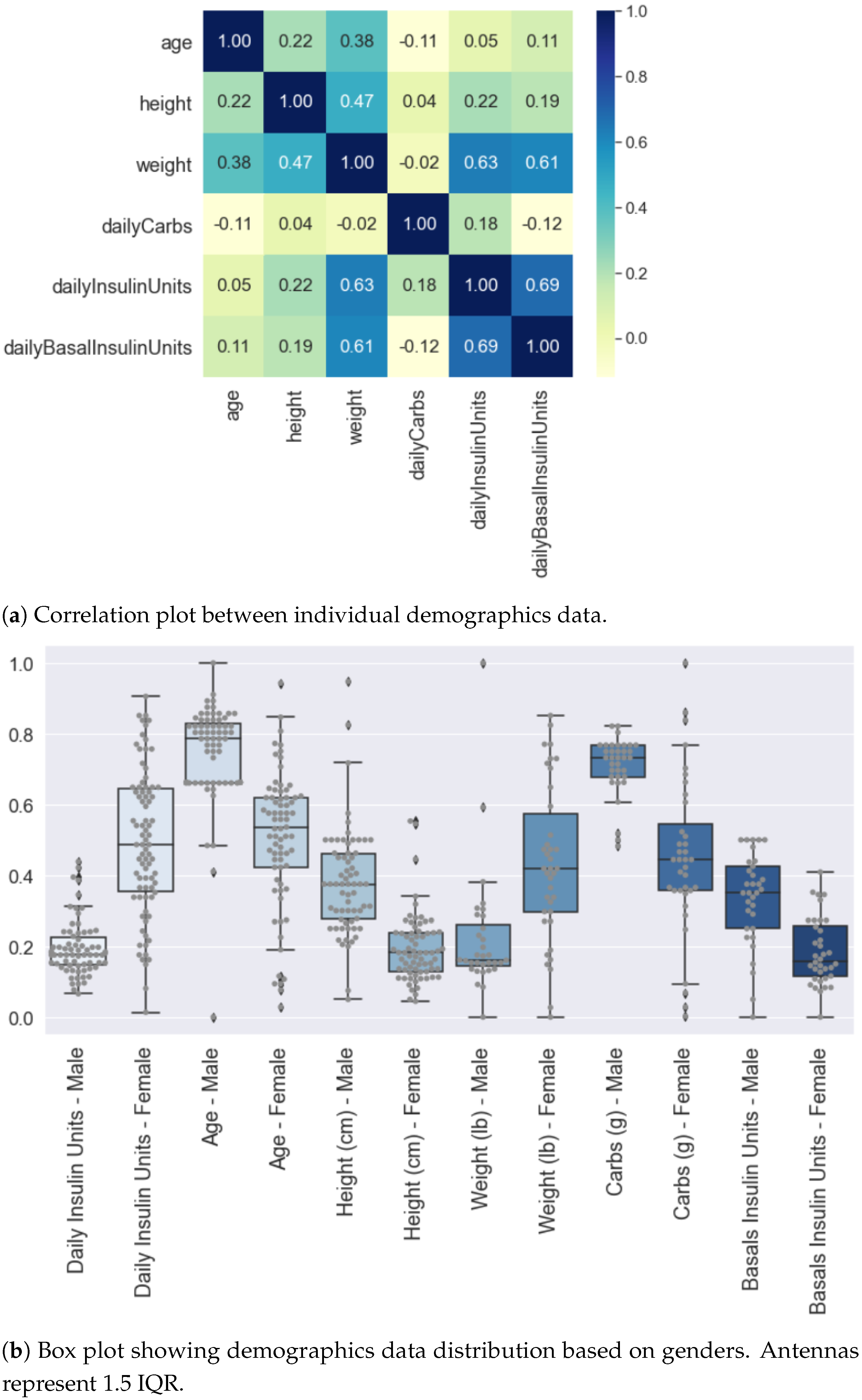
Demographics Data Collection
3.2. Glucose and Demographics Data Cleaning
- File formatting: We use glucose entries files in .csv format to ease data visualisation support using spreadsheets. Since the originating source contains a large volume of .json data, there are multiple .csv files for each individual. The files were previously converted to .csv using the Unzip-Zip-CSVify-OpenHumans-data.sh script (this is an open-source script along with other open-source tools used for processing large complex data such as the OpenAPS Data Commons data coming from the Open Humans platform, https://github.com/danamlewis/OpenHumansDataTools, accessed on 25 April 2022).
- Unified datastore: We pull glucose entries data in .csv format for all individuals to a common directory representing a unified data store.
- Timestamp cleaning and consistency: Each glucose entry file contains inconsistent timestamps. The cleanest timestamps were appended with letters such as T and Z represented in the following example format: “2018-08-04T23:58:50Z”. These were cleaned simply by trimming the alphabet letter. Multiple types of timestamp formats represented in different time zones—GMT, PDT, CES, CST, etc.—were found in single glucose entries files. We cleaned such instances programmatically by employing “regex” functions exposed by the Python Pandas package and lambda functions. Furthermore, we noticed an overlap of different formats of timestamps between the data rows. Some timestamp values were accompanied by the abbreviations for the day of the week such as “Mon”, “Tue”, “Wed”, etc., alongside the year and date. Although a lot of the inconsistent timestamps were cleaned programmatically, some of them required manual labour efforts as well. After all the pre-processing, we converted the timestamps to a consistent date–time format.
- Glucose entries cleaning and consistency: To maintain the consistency of glucose entries, we performed the following steps during the data cleaning phase:
- We noticed some glucose data samples contain text such as “null”. Data rows with “null” were removed in each glucose entry file.
- Multiple .csv files for the same individual were merged, and data were organised in the increasing order of the timestamps in dataframe columns.
- All the duplicate timeseries values (if any) were removed, and only the first entry for duplicate timestamps was kept.
- All infinite numbers represented as “inf” were replaced with “NaN”, and all rows with “NaN” were dropped.
- Based on CGM device knowledge, decisions were made to remove data values that represent error terms, as they should not be included in calculations for glucose values (units mg/dL). This includes removing every data point less than 39 and greater than 1000. Any data point greater than 400 and less than 1000 was replaced with 400. No further interpolations were performed to cover the error terms.
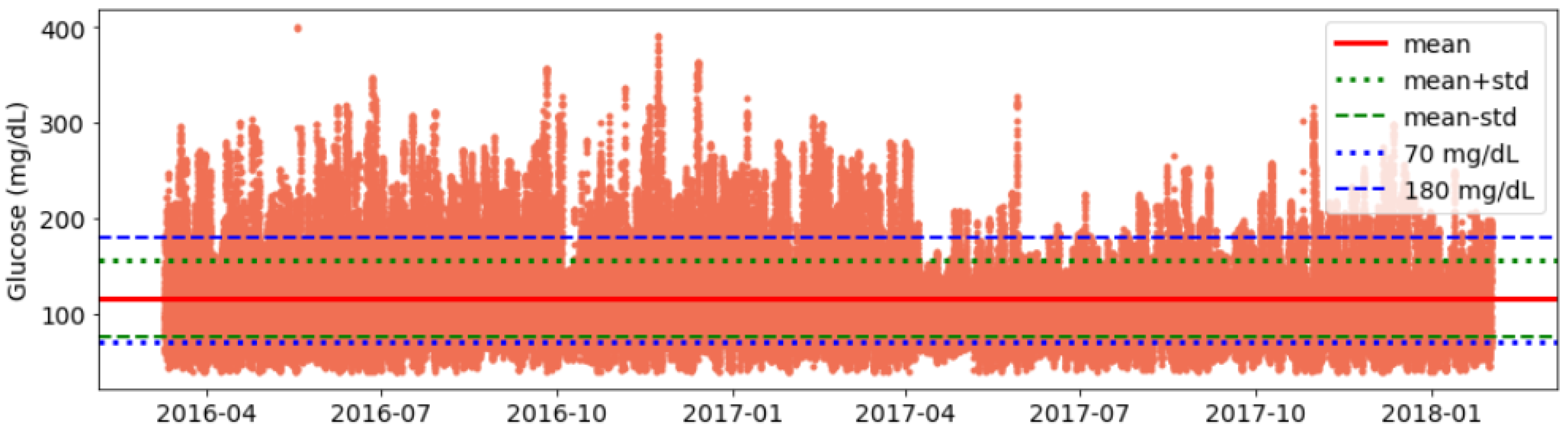
- Validation of data consistency: Finally, we plot glucose data and visualise it for each individual to identify any anomalies or inconsistencies in timestamps and corrected them (See Figure A1).
3.3. Glucose Analysis Metrics
- Count—Total CGM points to calculate the amount of data.
- Mean—Average of CGM data.
- Min—Minimum CGM data value.
- Max—Maximum CGM data value.
- Q1 or 25%—First quartile.
- Q2 or 50%—Second quartile.
- Q3 or 75%—Third quartile.
- IQR—Interquartile range.
- SD—Interday standard deviation of CGM data.
- CV—Interday coefficient of variation.
- TOR < 70—Time outside range, i.e., hypoglycemia. Total glucose data points less than 70 mg/dL in percentage.
- TIR—Time inside range, i.e., total glucose data points within target range between 70 mg/dL and 180 mg/dL in percentage.
- TOR > 180—Time outside range, i.e., hyperglycemia. Total glucose data points greater than 180 mg/dL in percentage.
- POR—Total percentage of time outside range (range in standard deviations from mean).
- J_index—Glycemic variability.
- LBGI—Glycemic variability metric to calculate low blood glucose index.
- HBGI—Glycemic variability metric to calculate high blood glucose index.
- GMI—Glycemic management indicator.
3.4. Summary of Glucose and Demographics Data
4. Results
4.1. Demographics Data Analysis
4.2. Glucose Data Analysis
- It can be seen that the interday glucose standard deviation is 49.75 mg/dL with a minimum and maximum of 14.71 mg/dL and 77.33 mg/dL, respectively.
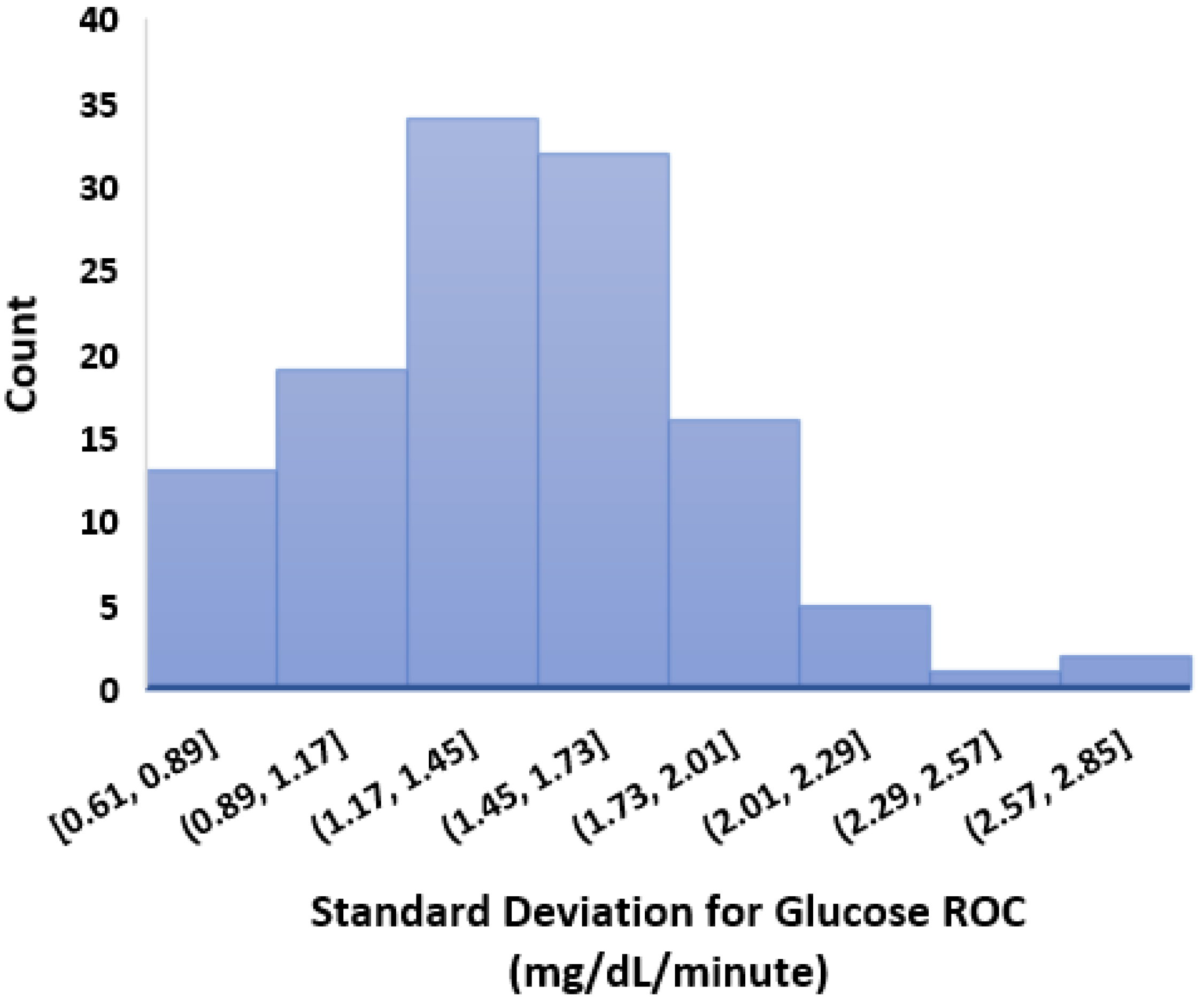
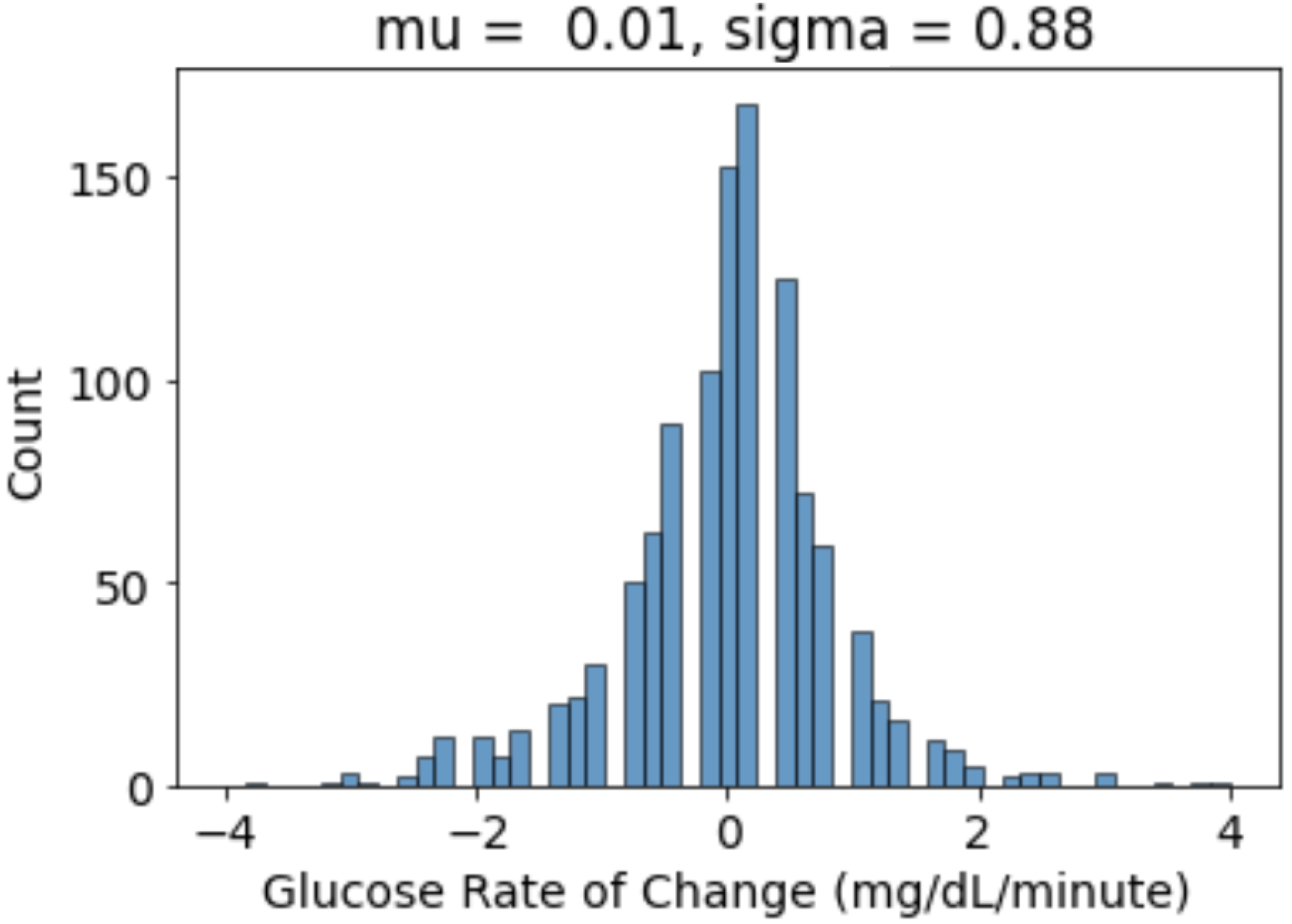
- We calculated the glucose rate of change for each individual in our dataset using the formula . We analysed visually and statistically the glucose rate of change (ROC) for each insulin-requiring individual (see Figure 4). The standard deviation of glucose ROC has a minimum, average, and maximum of {0.61, 1.42, 2.69} mg/dL per minute, respectively. The Shapiro–Wilk test (statistical test for normality) performed on SD of glucose ROC data yielded p = 0.205 (i.e., >0.05) indicating that the data follow a normal distribution. According to rule of distribution statistics, 99.7% of the data lies between 0.29 and 2.46.
- We further analysed the distribution of standard deviation of rate of change (ROC) for individual glucose profiles in the dataset (see Figure A2). The minimum, average, and maximum of SD ROC calculated separately for males and females is {0.60, 1.36, 2.6} and {0.7, 1.4, 2} mg/dL per minute, respectively.
- The average interday coefficient of variation (CV) is 35.43. A total of 25% of the insulin-requiring individuals have CV less than 32.42 and greater than 38.47, whereas the interquartile range is 6.
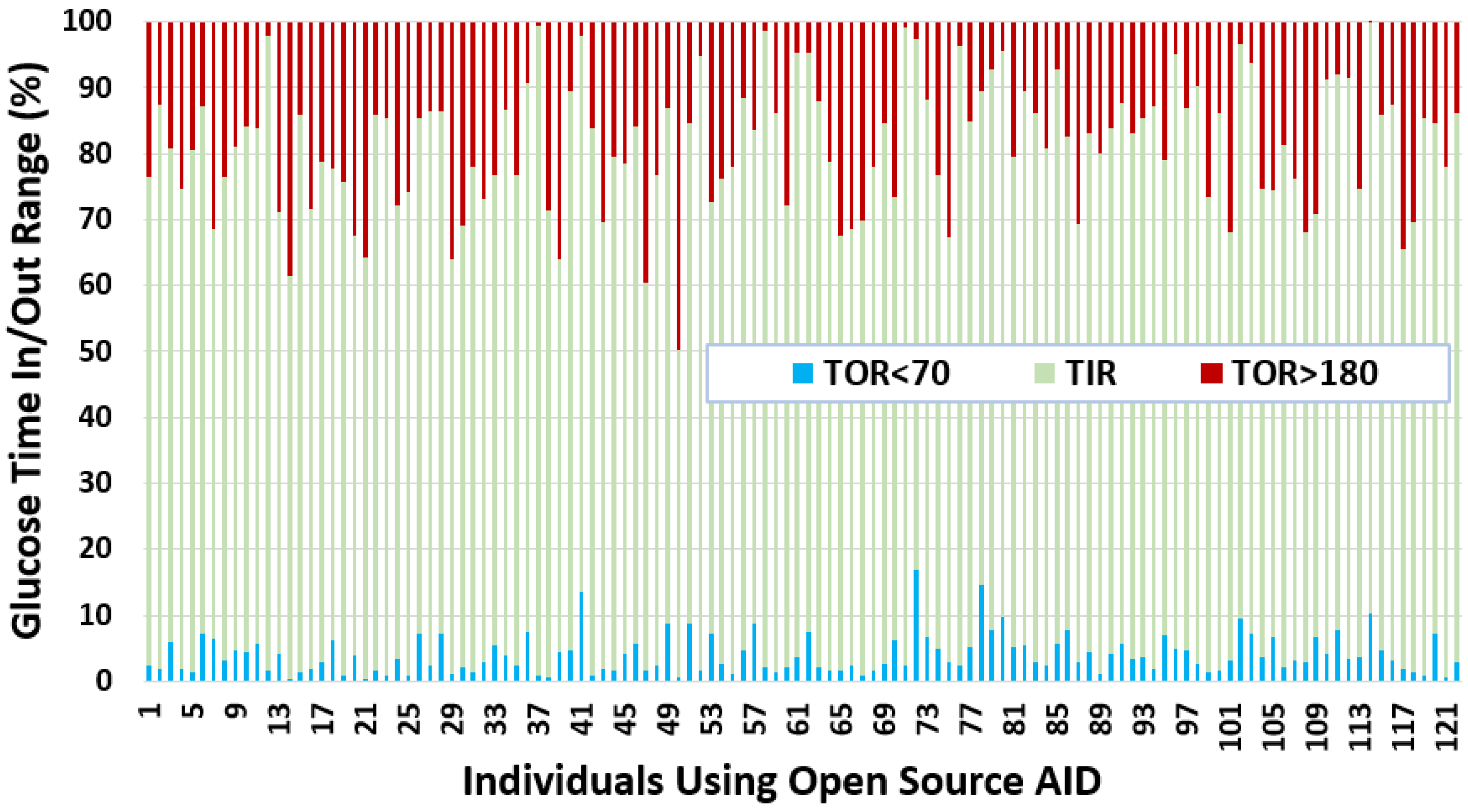
- We observe that the average TIR is 77.27% for people using DIY technologies. Furthermore, less than 25% of the individuals have TIR less than 71%. However, over 25% of the insulin-requiring individuals achieved a TIR higher than 84%. The minimum, average, and maximum for TOR < 70 and TOR > 180 is {0.23%, 4%, 16.97%} and {0.05%, 18.74%, 49.67%}, respectively.
- The minimum, average, and maximum for TOR < 70 and TOR > 180 highly correlate to LBGI and HBGI, respectively.
- The mean J_index and GMI for insulin-requiring individuals in our dataset is 36.42 and 6.63, respectively.
| Min | Max | Average | Q1 | Q2 | Q3 | IQR | |
|---|---|---|---|---|---|---|---|
| Interday SD (mg/dL) | 14.71 | 77.33 | 49.75 | 43.13 | 49.34 | 58.10 | 14.97 |
| Glucose ROC SD | 0.61 | 2.69 | 1.42 | 1.15 | 1.41 | 1.66 | 0.51 |
| Interday CV (%) | 16.86 | 44.94 | 35.43 | 32.42 | 35.87 | 38.47 | 6.05 |
| TIR (%) | 49.75 | 98.45 | 77.26 | 71.18 | 77.91 | 84.07 | 12.89 |
| TOR < 70 (%) | 0.23 | 16.97 | 4.01 | 1.79 | 3.22 | 5.61 | 3.83 |
| TOR > 180 (%) | 0.05 | 49.67 | 18.74 | 12.72 | 17.14 | 25.52 | 12.81 |
| LGBI | 0.13 | 3.82 | 1.09 | 0.64 | 0.95 | 1.42 | 0.78 |
| HBGI | 0.03 | 13.25 | 4.36 | 2.83 | 3.95 | 5.89 | 3.06 |
| J_index | 10.39 | 73.93 | 36.42 | 29.71 | 35.49 | 43.84 | 14.14 |
| GMI | 5.40 | 7.96 | 6.63 | 6.38 | 6.63 | 6.92 | 0.53 |
4.3. Clustering Glucose Profiles
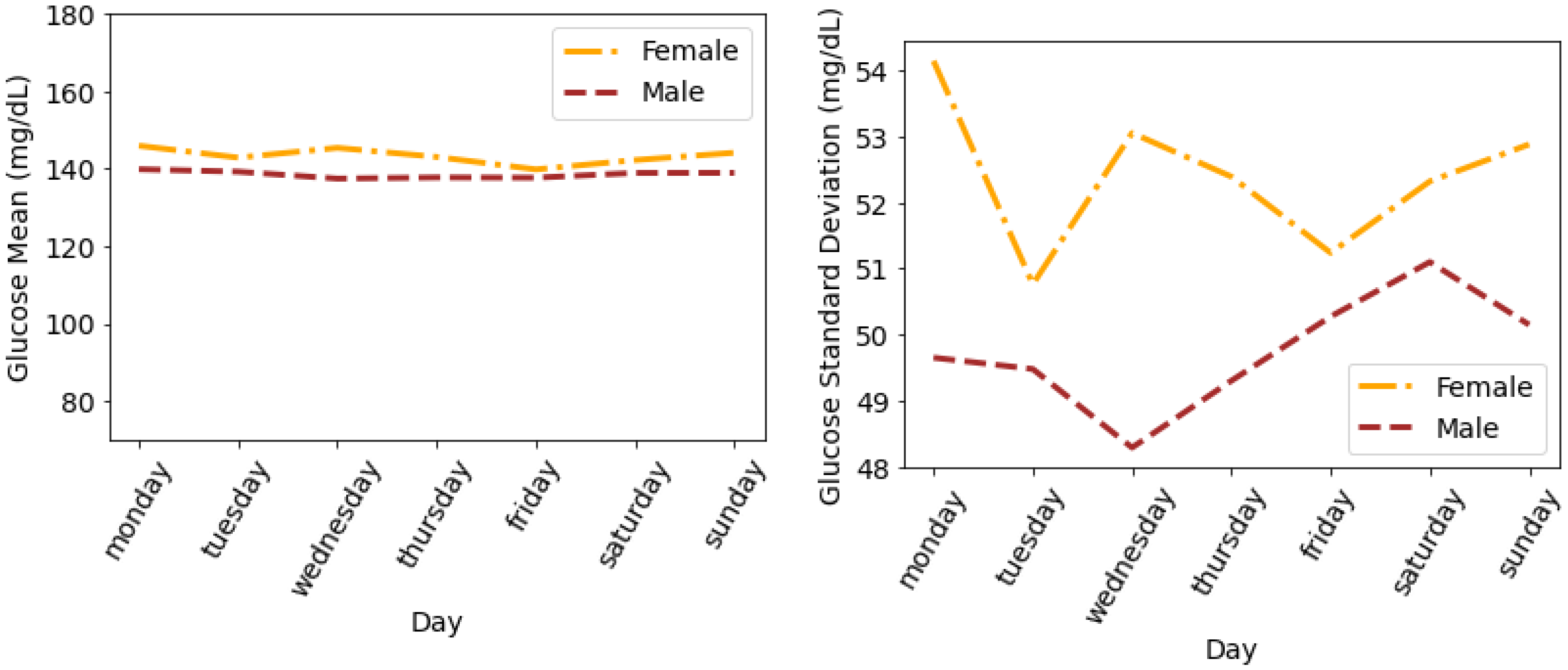
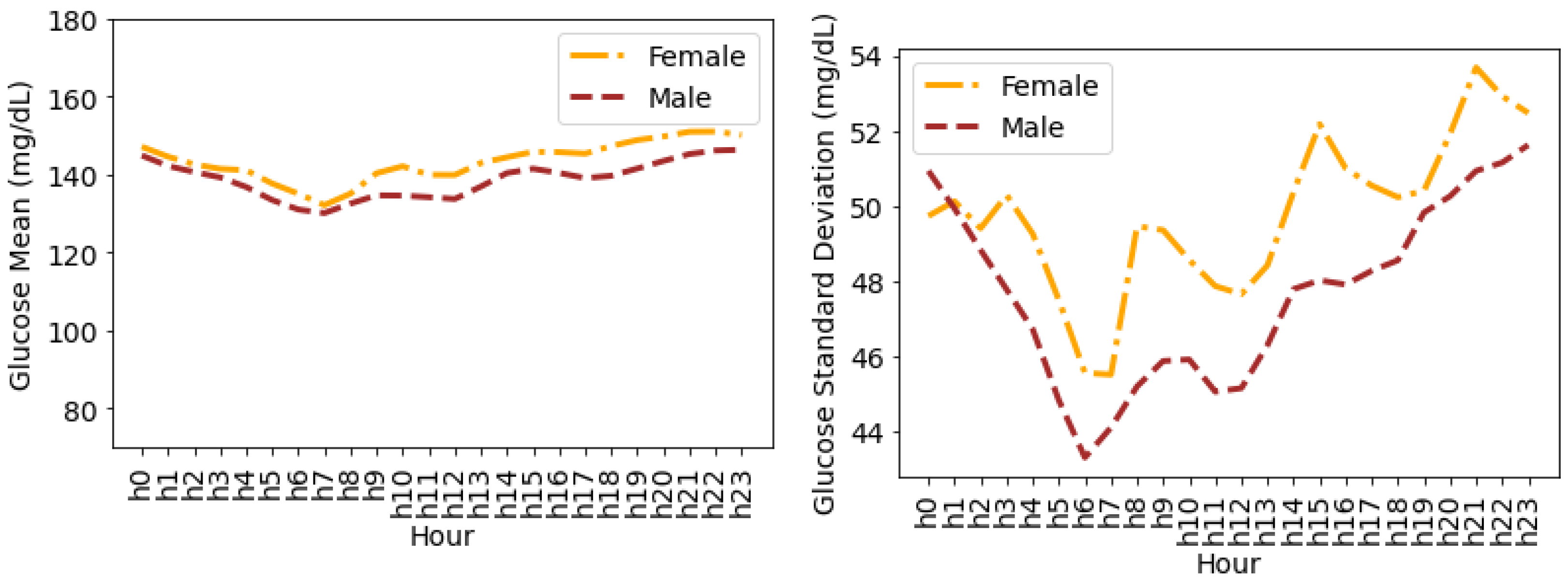
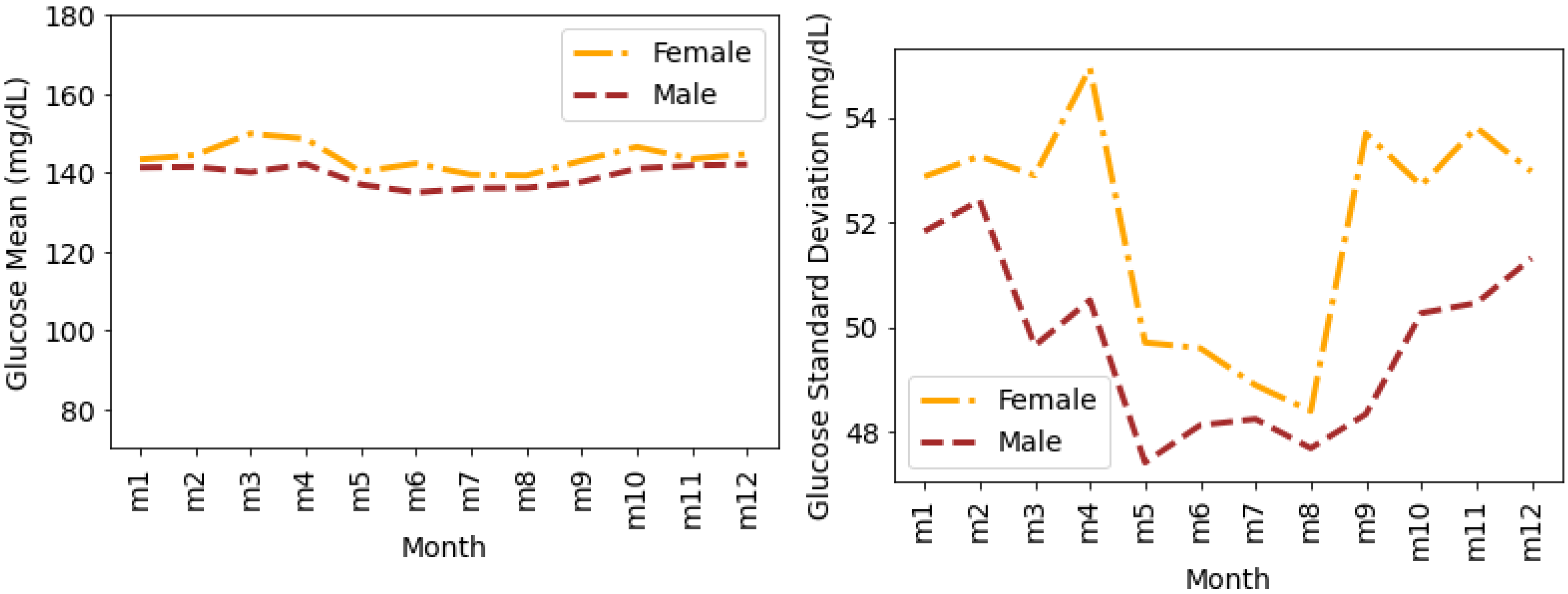
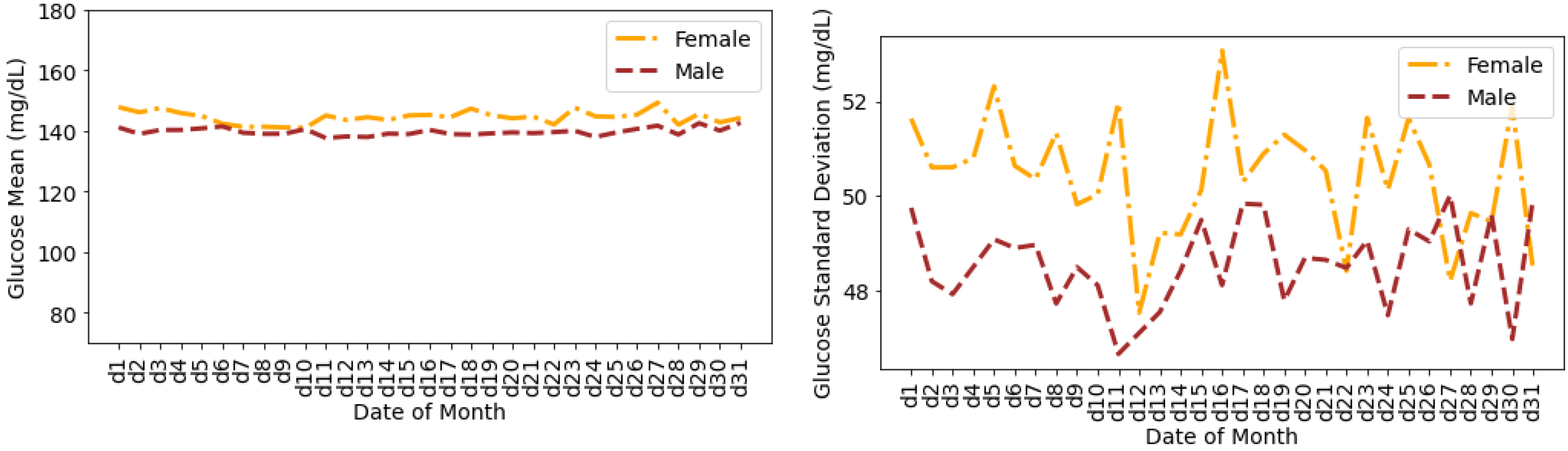
4.4. Glucose Variability Analysis Based on Gender
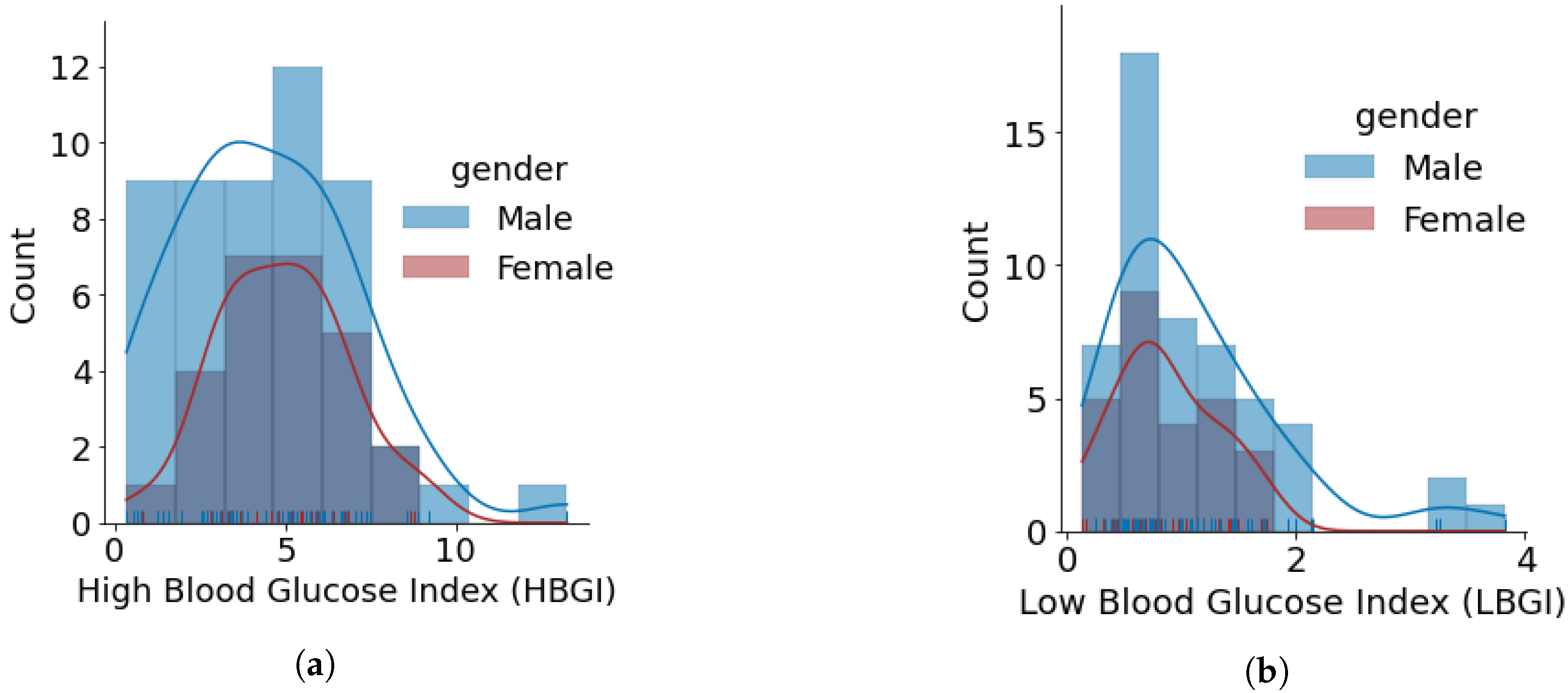
- The minimum, average, and maximum LBGI for females and males are {0.13, 0.87, 1.74} and {0.24, 1.11, 3.82}, respectively. Similarly, the minimum, average, and maximum HBGI for females and males is {0.80, 4.90, 8.75} and {0.33, 4.44, 13.24}, respectively. It can be seen that LBGI and HBGI distributions for males and females are positively skewed. The Shapiro–Wilk test on LBGI and HBGI yielded p < 0.05, confirming that the data do not belong to a normal distribution.
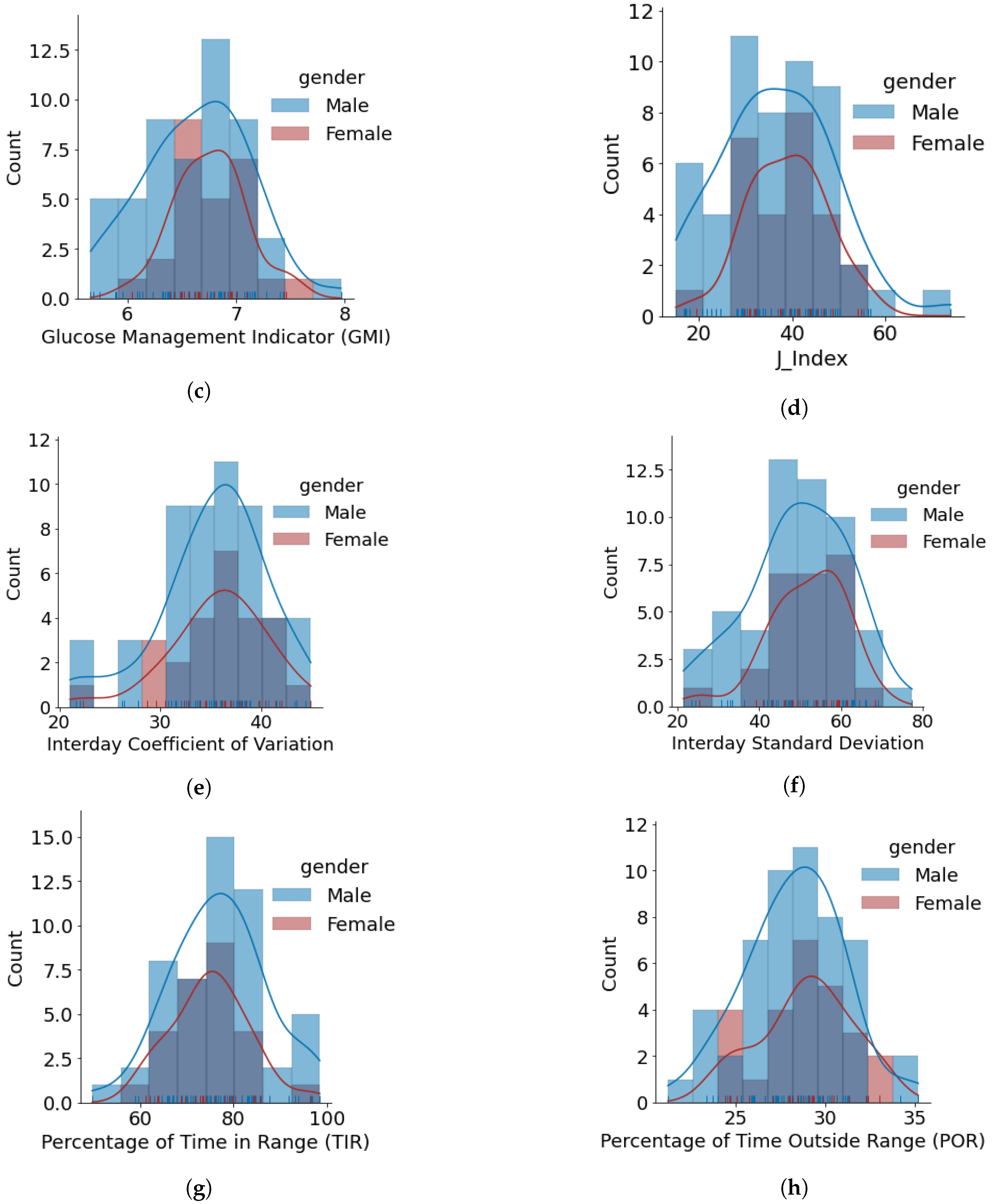
- The minimum, average, and maximum GMI for females and males is {6.04, 6.76, 7.46} and {5.66, 6.64, 7.96}, respectively. We do not observe any differences in GMI distribution patterns for males and females. However, the Shapiro–Wilk test yielded p > 0.05, indicating that the GMI follow a normal distribution.
- J_index distributions show a slight positive skew with higher average statistics for females with 38.99 as compared to males with 36.63. p > 0.05 is obtained from the Shapiro–Wilk test, indicating that J_index follows a normal distribution.
- There are no distinct differences between females and males for both CV and SD. The average CV in percentage and SD in mg/dL are {35.80, 51.93} for females and {35.23, 49.62} for males. Furthermore, there is a negative skew for both males and females in CV distributions. Using the Shapiro–Wilk test, p < 0.05 for CV confirms its non-uniform distribution. The SD distributions are more uniform (p > 0.05).
- Average TIR for females and males is 75.13 and 76.90. The distributions for both females and males are uniform (p > 0.05) and exhibit similar patterns. For our datasets, more than 75% of males and females achieve TIR of over 70%. The maximum TIR achieved for females and males in 96.25% and 98.45%
- The POR distributions for both genders are uniform (p > 0.05), where average POR for females is 28.92% and males is 28.26%.
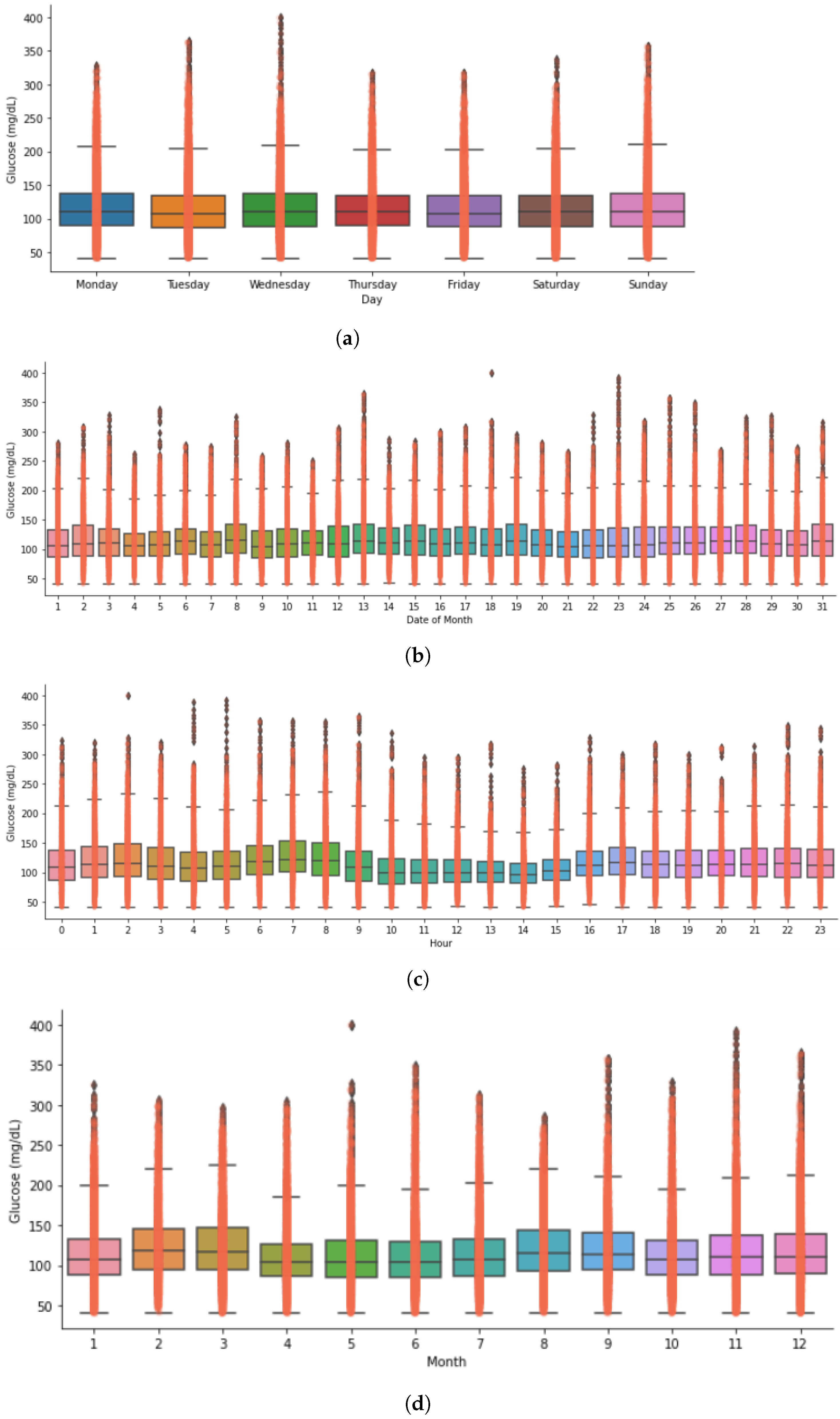
Timeseries Analysis for Glucose Data based on Gender
5. Discussion
Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OPEN | Outcomes of Patients’ Evidence with Novel, Do-it-Yourself Artificial Pancreas Technology |
| OpenAPS | Open-Source Artificial Pancreas System |
| AID | Automated Insulin Delivery |
| APS | Artificial Pancreas System |
| HCL | Hybrid Closed Loop |
| T1D | Type 1 Diabetes |
| CGM | Continuous Glucose Monitoring |
| PwD | People With Diabetes (any type) |
| HbA1c | Hemoglobin A1c |
| TIR | Time In Range |
| GV | Glucose Variability |
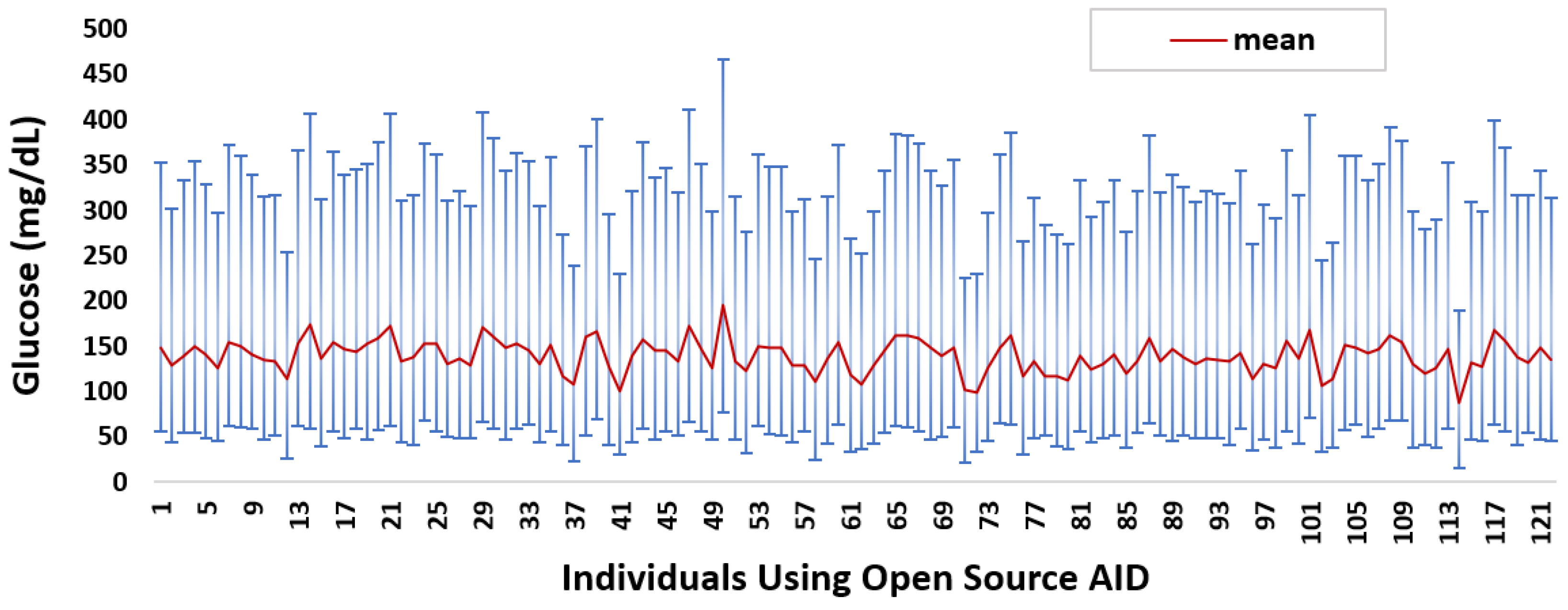
Appendix A. Individual-Level Glucose Data Analysis
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C.; et al. IDF diabetes atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- Garg, S.K.; Rewers, A.H.; Akturk, H.K. Ever-Increasing Insulin-Requiring Patients Globally. Diabetes Technol. Ther. 2018, 20, S2-1–S2-4. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D. Setting expectations for successful artificial pancreas/hybrid closed loop/automated insulin delivery adoption. J. Diabetes Sci. Technol. 2018, 12, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D. How it started, how it is going: The future of artificial pancreas systems (automated insulin delivery systems). J. Diabetes Sci. Technol. 2021, 15, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.; Leibrand, S.; Community, O. Real-world use of open source artificial pancreas systems. J. Diabetes Sci. Technol. 2016, 10, 1411. [Google Scholar] [CrossRef]
- Lewis, D.M. Do-it-yourself artificial pancreas system and the OpenAPS movement. Endocrinol. Metab. Clin. 2020, 49, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Knoll, C.; Peacock, S.; Wäldchen, M.; Cooper, D.; Aulakh, S.K.; Raile, K.; Hussain, S.; Braune, K. Real-world evidence on clinical outcomes of people with type 1 diabetes using open-source and commercial automated insulin dosing systems: A systematic review. Diabet. Med. 2021, 39, e14741. [Google Scholar] [CrossRef]
- Patel, R.; Crabtree, T.; Taylor, N.; Langeland, L.; Gazis, T.; Mendis, B.; Wilmot, E.; Idris, I. Safety and effectiveness of Do-It-Yourself Artificial Pancreas System (DIYAPS) compared with continuous subcutaneous insulin infusions (CSII) in combination with Free Style Libre (FSL) in people with Type 1 diabetes. Diabet. Med. 2022, 39, e14793. [Google Scholar] [CrossRef]
- Rodbard, D. Glucose variability: A review of clinical applications and research developments. Diabetes Technol. Ther. 2018, 20, S2–S5. [Google Scholar] [CrossRef]
- Ceriello, A. Glucose variability and diabetic complications: Is it time to treat? Diabetes Care 2020, 43, 1169–1171. [Google Scholar] [CrossRef]
- OpenAPS. Available online: https://OpenAPS.org (accessed on 25 April 2022).
- Lewis, D.M.; Swain, R.S.; Donner, T.W. Improvements in A1C and Time-in-Range in DIY Closed-Loop (OpenAPS) Users. Diabetes 2020, 67, 352-OR. [Google Scholar] [CrossRef]
- Zabinsky, J.; Howell, H.; Ghezavati, A.; Lewis, D.M.; Nguyen, A.; Wong, J.C. 988-P: Do-it-yourself artificial pancreas systems for type 1 diabetes reduce hyperglycemia without increasing hypoglycemia. Diabetes 2020, 69, 988-P. [Google Scholar] [CrossRef]
- Melmer, A.; Züger, T.; Lewis, D.M.; Leibrand, S.; Stettler, C.; Laimer, M. Glycaemic control in individuals with type 1 diabetes using an open source artificial pancreas system (OpenAPS). Diabetes Obes. Metab. 2019, 21, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Luo, S.; Zheng, X.; Bi, Y.; Xu, W.; Yan, J.; Yang, D.; Weng, J. Use of a do-it-yourself artificial pancreas system is associated with better glucose management and higher quality of life among adults with type 1 diabetes. Ther. Adv. Endocrinol. Metab. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Volkova, A.R.; Chernaya, M.; Vlasova, K.A. Experience of using insulin therapy with the closed loop method among patients with type 1 diabetes mellitus in Russia. In Proceedings of the Endocrine Abstracts, Virtual, UK, 5–9 September 2020; Volume 70. [Google Scholar]
- Gawrecki, A.; Zozulinska-Ziolkiewicz, D.; Michalak, M.A.; Adamska, A.; Michalak, M.; Frackowiak, U.; Flotynska, J.; Pietrzak, M.; Czapla, S.; Gehr, B.; et al. Safety and glycemic outcomes of do-it-yourself AndroidAPS hybrid closed-loop system in adults with type 1 diabetes. PLoS ONE 2021, 16, e0248965. [Google Scholar] [CrossRef] [PubMed]
- Jeyaventhan, R.; Gallen, G.; Choudhary, P.; Hussain, S. A Real-World Study of User Characteristics, Safety and Efficacy of Open-Source Closed-Loop Systems and Medtronic 670G. Diabetes Obes. Metab. 2021, 23, 1989–1994. [Google Scholar] [CrossRef] [PubMed]
- Greshake Tzovaras, B.; Angrist, M.; Arvai, K.; Dulaney, M.; Estrada-Galiñanes, V.; Gunderson, B.; Head, T.; Lewis, D.; Nov, O.; Shaer, O.; et al. Open Humans: A platform for participant-centered research and personal data exploration. GigaScience 2019, 8, giz076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, A.; Salinas, J.; Crump, C. Computational modeling approaches to characterize risk and achieve safe, effective, and trusted designs in the development of artificial intelligence and autonomous closed-loop medical systems. In Proceedings of the Artificial Intelligence and Machine Learning for Multi-Domain Operations Applications III, Virtual, FL, USA, 12–17 April 2021; Volume 11746, p. 1174623. [Google Scholar]
- Rodríguez-Rodríguez, I.; Chatzigiannakis, I.; Rodríguez, J.V.; Maranghi, M.; Gentili, M.; Zamora-Izquierdo, M. Utility of big data in predicting short-term blood glucose levels in type 1 diabetes mellitus through machine learning techniques. Sensors 2019, 19, 4482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosquera-Lopez, C.; Dodier, R.; Tyler, N.; Resalat, N.; Jacobs, P. Leveraging a Big Dataset to Develop a Recurrent Neural Network to Predict Adverse Glycemic Events in Type 1 Diabetes. IEEE J. Biomed. Health Inform. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Mosquera-Lopez, C.; Jacobs, P.G. Incorporating glucose variability into glucose forecasting accuracy assessment using the new glucose variability impact index and the prediction consistency index: An LSTM case example. J. Diabetes Sci. Technol. 2021, 16, 7–18. [Google Scholar] [CrossRef]
- Celebrating 10,000 donations to the Tidepool Big Data Donation Project. Available online: https://www.tidepool.org/blog/celebrating-10000-donations (accessed on 25 April 2022).
- Tidepool: Big Data Donation Project. Available online: https://www.tidepool.org/bigdata/ (accessed on 25 April 2022).
- Kovatchev, B.; Cobelli, C. Glucose variability: Timing, risk analysis, and relationship to hypoglycemia in diabetes. Diabetes Care 2016, 39, 502–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeVries, J.H. Glucose variability: Where it is important and how to measure it. Diabetes 2013, 62, 1405–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, A. Programming Scripts for Demographics and Glucose Variability Analysis for OpenAPS Data Commons Dataset. 2022. Available online: https://github.com/danamlewis/OpenHumansDataTools/tree/master/bin/GV-demographics-scripts (accessed on 25 April 2022).
- Kovatchev, B.P. Metrics for glycemic control—From HbA 1c to continuous glucose monitoring. Nat. Rev. Endocrinol. 2017, 13, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Kovatchev, B.P.; Otto, E.; Cox, D.; Gonder-Frederick, L.; Clarke, W. Evaluation of a new measure of blood glucose variability in diabetes. Diabetes Care 2006, 29, 2433–2438. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, C.; Donath, S.; Vidmar, S.; Werther, G.; Cameron, F. A novel approach to continuous glucose analysis utilizing glycemic variation. Diabetes Technol. Ther. 2005, 7, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Service, F.J.; Molnar, G.D.; Rosevear, J.W.; Ackerman, E.; Gatewood, L.C.; Taylor, W.F. Mean amplitude of glycemic excursions, a measure of diabetic instability. Diabetes 1970, 19, 644–655. [Google Scholar] [CrossRef]
- Baghurst, P.A. Calculating the mean amplitude of glycemic excursion from continuous glucose monitoring data: An automated algorithm. Diabetes Technol. Ther. 2011, 13, 296–302. [Google Scholar] [CrossRef]
- Fritzsche, G.; Kohnert, K.D.; Heinke, P.; Vogt, L.; Salzsieder, E. The use of a computer program to calculate the mean amplitude of glycemic excursions. Diabetes Technol. Ther. 2011, 13, 319–325. [Google Scholar] [CrossRef]
- Yu, X.; Lin, L.; Shen, J.; Chen, Z.; Jian, J.; Li, B.; Xin, S.X. Calculating the mean amplitude of glycemic excursions from continuous glucose data using an open-code programmable algorithm based on the integer nonlinear method. Comput. Math. Methods Med. 2018, 2018, 6286893. [Google Scholar] [CrossRef]
- Bergenstal, R.M.; Beck, R.W.; Close, K.L.; Grunberger, G.; Sacks, D.B.; Kowalski, A.; Brown, A.S.; Heinemann, L.; Aleppo, G.; Ryan, D.B.; et al. Glucose management indicator (GMI): A new term for estimating A1C from continuous glucose monitoring. Diabetes Care 2018, 41, 2275–2280. [Google Scholar] [CrossRef] [Green Version]
- Bent, B.; Henriquez, M.; Dunn, J.P. Cgmquantify: Python and R Software Packages for Comprehensive Analysis of Interstitial Glucose and Glycemic Variability from Continuous Glucose Monitor Data. IEEE Open J. Eng. Med. Biol. 2021, 2, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Saisho, Y.; Tanaka, C.; Tanaka, K.; Roberts, R.; Abe, T.; Tanaka, M.; Meguro, S.; Irie, J.; Kawai, T.; Itoh, H. Relationships among different glycemic variability indices obtained by continuous glucose monitoring. Prim. Care Diabetes 2015, 9, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, R.A.; Shi, H.; Yuan, L.H.; Brehm, W.; Pop-Busui, R.; Nelson, P.W. Translating Glucose Variability Metrics into the Clinic via C ontinuous G lucose M onitoring: AG raphical U ser I nterface for D iabetes E valuation (CGM-GUIDE©). Diabetes Technol. Ther. 2011, 13, 1241–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attaye, I.; van der Vossen, E.W.; Mendes Bastos, D.N.; Nieuwdorp, M.; Levin, E. Introducing the Continuous Glucose Data Analysis (CGDA) R Package: An Intuitive Package to Analyze Continuous Glucose Monitoring Data. J. Diabetes Sci. Technol. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Moscardó, V.; Giménez, M.; Oliver, N.; Hill, N.R. Updated software for automated assessment of glucose variability and quality of glycemic control in diabetes. Diabetes Technol. Ther. 2020, 22, 701–708. [Google Scholar] [CrossRef]
- Vigers, T.; Chan, C.L.; Snell-Bergeon, J.; Bjornstad, P.; Zeitler, P.S.; Forlenza, G.; Pyle, L. cgmanalysis: An R package for descriptive analysis of continuous glucose monitor data. PLoS ONE 2019, 14, e0216851. [Google Scholar] [CrossRef]
- Czerwoniuk, D.; Fendler, W.; Walenciak, L.; Mlynarski, W. GlyCulator: A glycemic variability calculation tool for continuous glucose monitoring data. J. Diabetes Sci. Technol. 2011, 5, 447–451. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, I.B.; Brownlee, M. Should minimal blood glucose variability become the gold standard of glycemic control? J. Diabetes Its Complicat. 2005, 19, 178–181. [Google Scholar] [CrossRef]
- Cobelli, C.; Facchinetti, A. Yet another glucose variability index: Time for a paradigm change? Diabetes Technol. Ther. 2018, 20, 1–3. [Google Scholar] [CrossRef]
- Kilpatrick, E.S.; Rigby, A.S.; Atkin, S.L. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care 2006, 29, 1486–1490. [Google Scholar] [CrossRef] [Green Version]
- Frontoni, S.; Di Bartolo, P.; Avogaro, A.; Bosi, E.; Paolisso, G.; Ceriello, A. Glucose variability: An emerging target for the treatment of diabetes mellitus. Diabetes Res. Clin. Pract. 2013, 102, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Sechterberger, M.K.; Luijf, Y.M.; DeVries, J.H. Poor agreement of computerized calculators for mean amplitude of glycemic excursions. Diabetes Technol. Ther. 2014, 16, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, N.J.; Nguyen, N.; Chun, E.; Punjabi, N.M.; Gaynanova, I. Open-Source Algorithm to Calculate Mean Amplitude of Glycemic Excursions Using Short and Long Moving Averages. J. Diabetes Sci. Technol. 2022, 16, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Marling, C.R.; Shubrook, J.H.; Vernier, S.J.; Wiley, M.T.; Schwartz, F.L. Characterizing blood glucose variability using new metrics with continuous glucose monitoring data. J. Diabetes Sci. Technol. 2011, 5, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Buse, J.B.; Freeman, J.L.; Edelman, S.V.; Jovanovic, L.; McGill, J.B. Serum 1, 5-anhydroglucitol (GlycoMark™): A short-term glycemic marker. Diabetes Technol. Ther. 2003, 5, 355–363. [Google Scholar] [CrossRef]
- Dovc, K.; Cargnelutti, K.; Sturm, A.; Selb, J.; Bratina, N.; Battelino, T. Continuous glucose monitoring use and glucose variability in pre-school children with type 1 diabetes. Diabetes Res. Clin. Pract. 2019, 147, 76–80. [Google Scholar] [CrossRef] [Green Version]
- Service, F.J. Glucose variability. Diabetes 2013, 62, 1398–1404. [Google Scholar] [CrossRef] [Green Version]
- Siegelaar, S.E.; Holleman, F.; Hoekstra, J.B.; DeVries, J.H. Glucose variability; Does it matter? Endocr. Rev. 2010, 31, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Nightscoutfoundation/Dataxfer: POC to Develop a Web-Based Data Transfer Tool from Nightscout DBS to Other Platforms. Available online: https://github.com/NightscoutFoundation/dataxfer (accessed on 25 April 2022).
- Lewis, D. OpenHumansDataTools. 2018. Available online: https://github.com/danamlewis/OpenHumansDataTools/blob/master/bin/unzip-split-csvify-OpenHumans-data.sh (accessed on 25 April 2022).
- Assessment, G. 6. Glycemic Targets: Standards of Medical Care in Diabetes—2022. Diabetes Care 2022, 45, S83. [Google Scholar]
- Lewis, D.M. Errors of commission or omission: The net risk safety analysis conversation we should be having around automated insulin delivery systems. Diabet. Med. 2021, 39, e14687. [Google Scholar] [CrossRef]
- Toeller, M.; Buyken, A.; Heitkamp, G.; Cathelineau, G.; Ferriss, B. Nutrient intakes as predictors of body weight in European people with type 1 diabetes. Int. J. Obes. 2001, 25, 1815–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szadkowska, A.; Madej, A.; Ziólkowska, K.; Szymanska, M.; Jeziorny, K.; Mianowska, B.; Pietrzak, I. Gender and Age–Dependent effect of type 1 diabetes on obesity and altered body composition in young adults. Ann. Agric. Environ. Med. 2015, 22, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asarani, N.; Reynolds, A.; Elbalshy, M.; Burnside, M.; de Bock, M.; Lewis, D.; Wheeler, B. Efficacy, safety, and user experience of DIY or open-source artificial pancreas systems: A systematic review. Acta Diabetol. 2021, 58, 539–547. [Google Scholar] [CrossRef]
- Palmer, W.; Greeley, S.A.W.; Letourneau-Freiberg, L.R.; Naylor, R.N. Using a do-it-yourself artificial pancreas: Perspectives from patients and diabetes providers. J. Diabetes Sci. Technol. 2020, 14, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Kershenbaum, A.; Kershenbaum, A.; Tarabeia, J.; Stein, N.; Lavi, I.; Rennert, G. Unraveling seasonality in population averages: An examination of seasonal variation in glucose levels in diabetes patients using a large population-based data set. Chronobiol. Int. 2011, 28, 352–360. [Google Scholar] [CrossRef]
- Levy, C.; O’Malley, G.; Raghinaru, D.; Kudva, Y.C.; Laffel, L.M.; Pinsker, J.E.; Lum, J.; Brown, S. Insulin Delivery and Glucose Variability throughout the Menstrual Cycle on Closed Loop Control for Women with Type 1 Diabetes. Diabetes Technol. Ther. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Herranz, L.; Saez-de Ibarra, L.; Hillman, N.; Gaspar, R.; Pallardo, L.F. Glycemic changes during menstrual cycles in women with type 1 diabetes. Med. Clin. 2016, 146, 287–291. [Google Scholar] [CrossRef]
- Mewes, D.; Wäldchen, M.; Knoll, C.; Raile, K.; Braune, K. Variability of Glycemic Outcomes and Insulin Requirements Throughout the Menstrual Cycle: A Qualitative Study on Women With Type 1 Diabetes Using an Open-Source Automated Insulin Delivery System. J. Diabetes Sci. Technol. 2022, in press. [Google Scholar] [CrossRef]
- Burnside, M.; Lewis, D.; Crocket, H.; Wilson, R.; Williman, J.; Jefferies, C.; Paul, R.; Wheeler, B.J.; de Bock, M. Create (community derived automated insulin delivery) trial. randomised parallel arm open label clinical trial comparing automated insulin delivery using a mobile controller (anydana-loop) with an open-source algorithm with sensor augmented pump therapy in type 1 diabetes. J. Diabetes Metab. Disord. 2020, 19, 1615–1629. [Google Scholar] [CrossRef]
- Lewis, D.M.; Leibrand, S.; Street, T.J.; Phatak, S.S. Detecting insulin sensitivity changes for individuals with type 1 diabetes. Diabetes 2018, 67, 79-LB. [Google Scholar] [CrossRef]
| Metric | Acronym | Definition |
|---|---|---|
| Average daily risk range | ADRR | Assessment of overall total daily glucose variations within risk range [30]. Risk scores are defined relative to a target. |
| Continuous overall net glycemic action | CONGA | A GV metric similar to standard deviation (SD) that assesses glucose fluctuations for a predetermined interval [31]. |
| Mean amplitude of glycemic excursion | MAGE | Mean of blood glucose values that exceed one SD from the 24 h mean blood glucose value [32]. Multiple implementations of automatically calculating MAGE are available in the literature [33,34,35]. |
| Glycemic management indicator | GMI | Indicates the expected mean hemoglobin A1C using mean glucose of individuals with diabetes [36]. |
| High blood glucose index | HBGI | A quantifying metric indicating the risk of hyperglycemia calculated using self-monitoring of blood glucose (SMBG) samples [30]. |
| Low blood glucose index | LBGI | A quantifying metric indicating the risk of hypoglycemia calculated using SMBG samples [30]. |
| Coefficient of variation | CV | A statistical metric to compute the diversity of glucose data. Commonly used sub-metrics for glucose data include the interday and intraday CV in CGM data [37]. |
| Glycemic variability metric | J_index | A quality assessment metric of glucose management using a combination of information from the mean and SD [38]. |
| Time in range | TIR | A quantifiable metric to calculate the percentage of time spent within normal glucose levels, i.e., a target range defined between 70 mg/DL to 180 mg/dL. |
| Time outside range | TOR | A quantifiable metric to calculate the percentage of time spent outside normal glucose levels, i.e., either less than 70 mg/DL or greater than 180 mg/dL. |
| Demographic Features | Number of Available Reports | Missing Reports |
|---|---|---|
| Total Number of Individuals | 122 | 0 |
| Diagnosed Date | 122 | 0 |
| Date of Pump Use | 103 | 19 |
| Date of CGM Use | 105 | 17 |
| Date of Closed Loop Initiation | 102 | 20 |
| Open-Source AID Type | 107 | 15 |
| Date of Birth | 117 | 5 |
| Country | 121 | 1 |
| Weight | 118 | 4 |
| Height | 119 | 3 |
| Total Daily Insulin Units | 114 | 8 |
| Daily Basal Insulin Units | 119 | 3 |
| Total Daily Carbs | 105 | 17 |
| Last HbA1C | 116 | 6 |
| Last A1C Date | 116 | 6 |
| Gender | 78 | 44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, A.; Lewis, D.M. Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems. Nutrients 2022, 14, 1906. https://doi.org/10.3390/nu14091906
Shahid A, Lewis DM. Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems. Nutrients. 2022; 14(9):1906. https://doi.org/10.3390/nu14091906
Chicago/Turabian StyleShahid, Arsalan, and Dana M. Lewis. 2022. "Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems" Nutrients 14, no. 9: 1906. https://doi.org/10.3390/nu14091906
APA StyleShahid, A., & Lewis, D. M. (2022). Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems. Nutrients, 14(9), 1906. https://doi.org/10.3390/nu14091906








