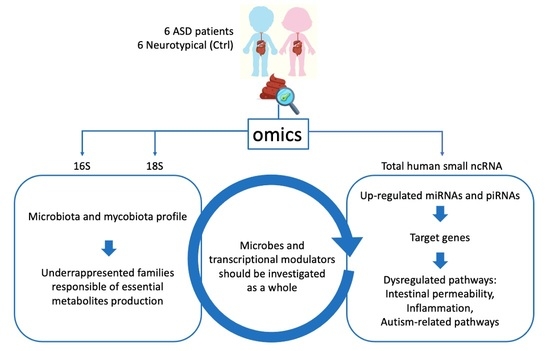
Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Ethical Committees
2.3. Sample Collection
2.4. DNA and SmallRNA Extraction from Stool
2.5. 16S and 18S Sequencing
2.6. SmallRNA Sequencing
2.7. Metataxonomic Bioinformatics Analysis
2.8. Small, Non-Coding RNA Data Analysis
2.9. Identification of sncRNA Targets and Relative Pathways
3. Results
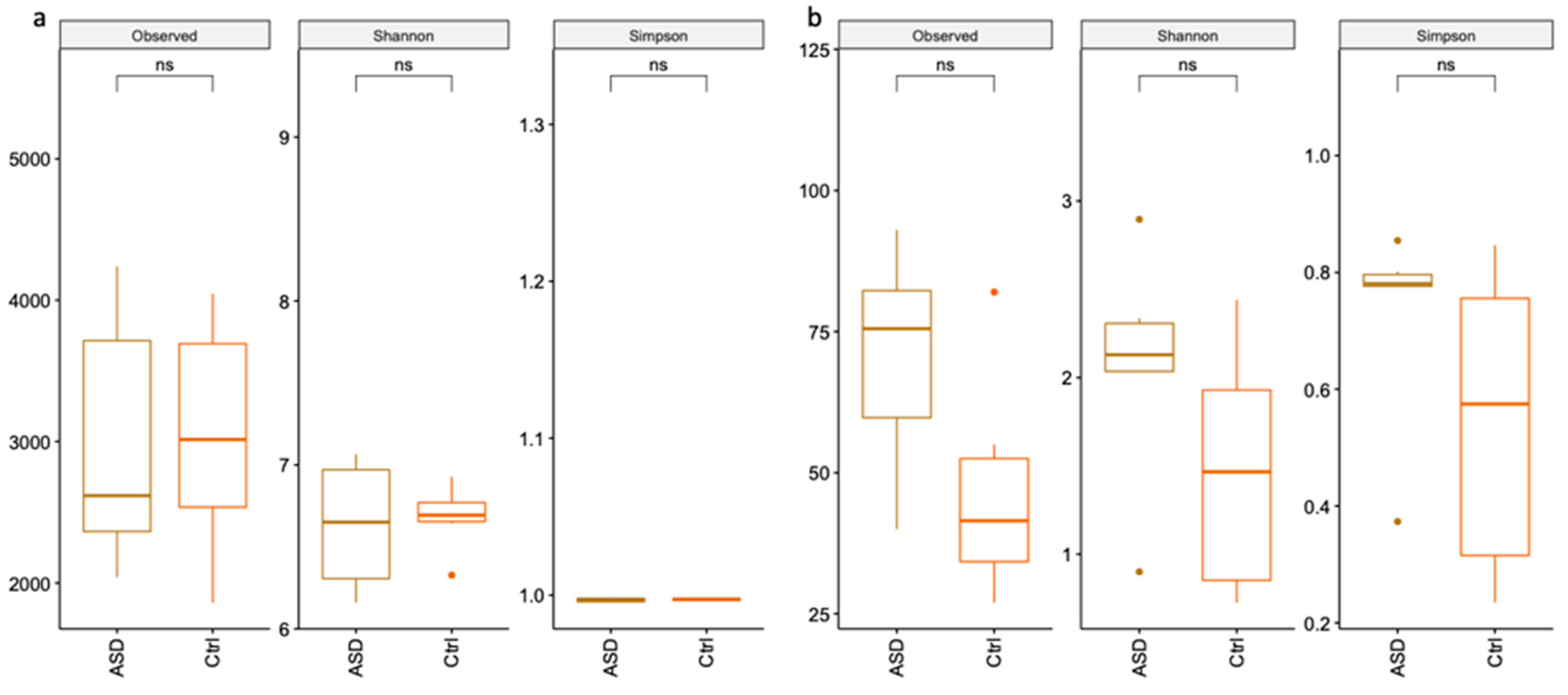
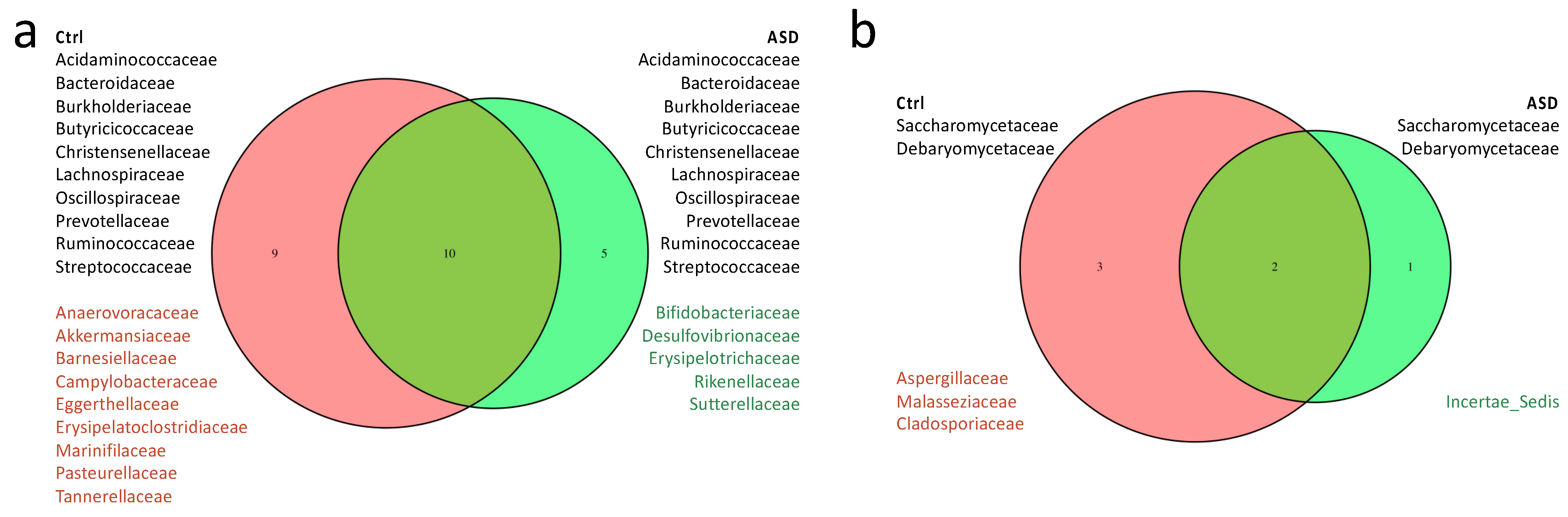
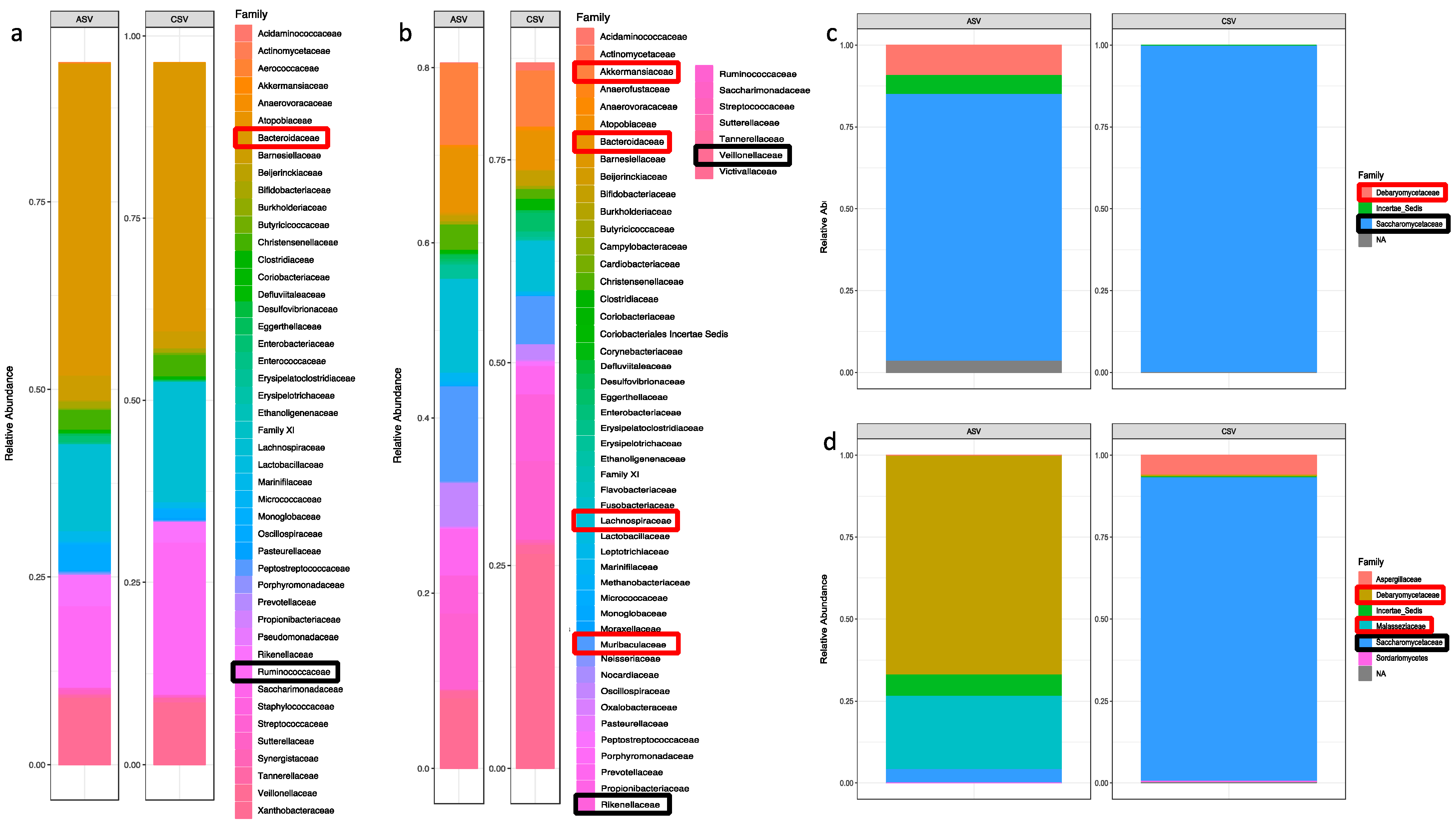
3.1. Microbiota Analysis
3.1.1. Bacteria Profiling: Metataxonomic Analysis
3.1.2. Fungi Profiling: Metataxonomic Analysis
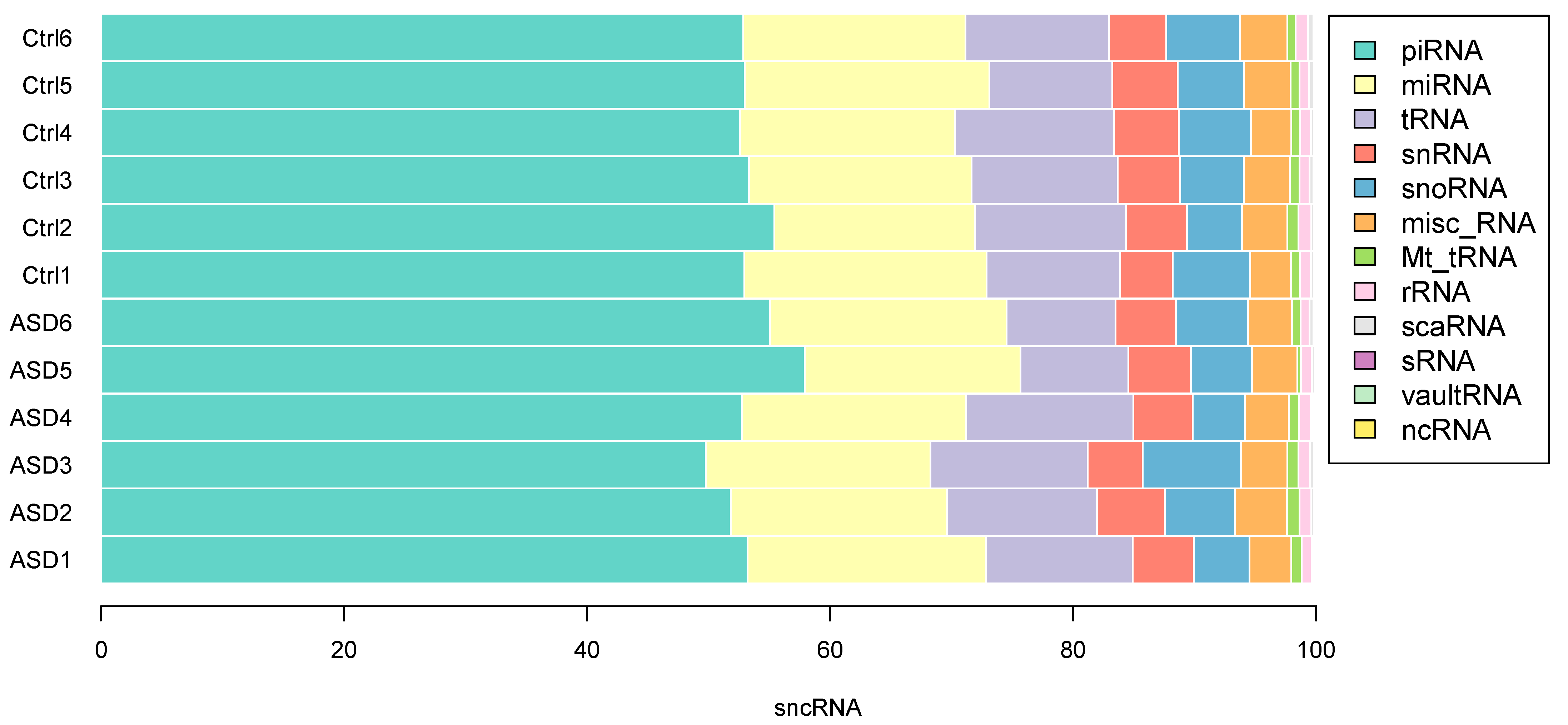
3.2. sncRNA Profiling
3.3. Case Study: Analysis of Siblings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bougeard, C.; Picarel-Blanchot, F.; Schmid, R.; Campbell, R.; Buitelaar, J. Prevalence of Autism Spectrum Disorder and Co-morbidities in Children and Adolescents: A Systematic Literature Review. Front. Psychiatry 2021, 12, 744709. [Google Scholar] [CrossRef] [PubMed]
- Mezzelani, A.; Landini, M.; Facchiano, F.; Raggi, M.E.; Villa, L.; Molteni, M.; de Santis, B.; Brera, C.; Caroli, A.M.; Milanesi, L.; et al. Environment, dysbiosis, immunity and sex-specific susceptibility: A translational hypothesis for regressive autism pathogenesis. Nutr. Neurosci. 2015, 18, 145–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troisi, J.; Autio, R.; Beopoulos, T.; Bravaccio, C.; Carraturo, F.; Corrivetti, G.; Cunningham, S.; Devane, S.; Fallin, D.; Fetissov, S.; et al. Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach. Brain Sci. 2020, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef]
- Chaste, P.; Leboyer, M. Autism risk factors: Genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar] [CrossRef]
- De Angelis, M.; Francavilla, R.; Piccolo, M.; de Giacomo, A.; Gobbetti, M. Autism spectrum disorders and intestinal microbiota. Gut Microbes 2015, 6, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kriss, M.; Hazleton, K.Z.; Nusbacher, N.M.; Martin, C.G.; Lozupone, C.A. Low diversity gut microbiota dysbiosis: Drivers, functional implications and recovery. Curr. Opin. Microbiol. 2018, 44, 34–40. [Google Scholar] [CrossRef]
- Li, M.; Chen, W.-D.; Wang, Y.-D. The roles of the gut microbiota-miRNA interaction in the host pathophysiology. Mol. Med. 2020, 26, 101. [Google Scholar] [CrossRef] [PubMed]
- Heiss, C.N.; Olofsson, L.E. The role of the gut microbiota in development, function and disorders of the central nervous system and the enteric nervous system. J. Neuroendocrinol. 2019, 31, e12684. [Google Scholar] [CrossRef] [PubMed]
- Sabit, H.; Tombuloglu, H.; Rehman, S.; Almandil, N.B.; Cevik, E.; Abdel-Ghany, S.; Rashwan, S.; Abasiyanik, M.F.; Yee Waye, M.M. Gut microbiota metabolites in autistic children: An epigenetic perspective. Heliyon 2021, 7, e06105. [Google Scholar] [CrossRef] [PubMed]
- Petra, A.I.; Panagiotidou, S.; Hatziagelaki, E.; Stewart, J.M.; Conti, P.; Theoharides, T.C. Gut-Microbiota-Brain Axis and Its Effect on Neuropsychiatric Disorders with Suspected Immune Dysregulation. Clin. Ther. 2015, 37, 984–995. [Google Scholar] [CrossRef] [Green Version]
- Kelly, J.R.; Minuto, C.; Cryan, J.F.; Clarke, G.; Dinan, T.G. Cross Talk: The Microbiota and Neurodevelopmental Disorders. Front. Neurosci. 2017, 11, 490. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, E.Y. Gastrointestinal issues in autism spectrum disorder. Harv. Rev. Psychiatry 2014, 22, 104–111. [Google Scholar] [CrossRef]
- Adams, J.B.; Audhya, T.; McDonough-Means, S.; Rubin, R.A.; Quig, D.; Geis, E.; Gehn, E.; Loresto, M.; Mitchell, J.; Atwood, S.; et al. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutr. Metab. 2011, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Shi, K.; Liu, X.; Dai, Y.; Liu, Y.; Zhang, L.; Du, X.; Zhu, T.; Yu, J.; Fang, S.; et al. Gut Microbial Profile Is Associated with the Severity of Social Impairment and IQ Performance in Children With Autism Spectrum Disorder. Front. Psychiatry 2021, 12, 789864. [Google Scholar] [CrossRef]
- Ho, L.K.H.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut microbiota changes in children with autism spectrum disorder: A systematic review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The Gut Microbiota and Autism Spectrum Disorders. Front. Cell. Neurosci. 2017, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.D.; Bernstein, C.N.; Tremlett, H.; van Domselaar, G.; Knox, N.C. A Fungal World: Could the Gut Mycobiome Be Involved in Neurological Disease? Front. Microbiol. 2018, 9, 3249. [Google Scholar] [CrossRef]
- Tang, J.; Iliev, I.D.; Brown, J.; Underhill, D.M.; Funari, V.A. Mycobiome: Approaches to analysis of intestinal fungi. J. Immunol. Methods 2015, 421, 112–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Matijašić, M.; Meštrović, T.; Paljetak, H.Č.; Perić, M.; Barešić, A.; Verbanac, D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 2020, 21, 2668. [Google Scholar] [CrossRef] [Green Version]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect. Immun. 2021, 89, e00648-20. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; de Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantarcioglu, A.S.; Kiraz, N.; Aydin, A. Microbiota-Gut-Brain Axis: Yeast Species Isolated from Stool Samples of Children with Suspected or Diagnosed Autism Spectrum Disorders and In Vitro Susceptibility Against Nystatin and Fluconazole. Mycopathologia 2016, 181, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Strati, F.; Cavalieri, D.; Albanese, D.; de Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Burns, M.B.; Subramanian, S.; Blekhman, R. Interaction between Host MicroRNAs and the Gut Microbiota in Colorectal Cancer. mSystems 2018, 3, e00205-17. [Google Scholar] [CrossRef] [Green Version]
- Larsson, E.; Tremaroli, V.; Lee, Y.S.; Koren, O.; Nookaew, I.; Fricker, A.; Nielsen, J.; Ley, R.E.; Bäckhed, F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 2012, 61, 1124–1131. [Google Scholar] [CrossRef]
- Williams, M.R.; Stedtfeld, R.D.; Tiedje, J.M.; Hashsham, S.A. MicroRNAs-Based Inter-Domain Communication between the Host and Members of the Gut Microbiome. Front. Microbiol. 2017, 8, 1896. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Shao, S.; Liu, N.; Liu, Q.; Jacquemyn, H.; Xing, X. Extracellular Enzyme Activities and Carbon/Nitrogen Utilization in Mycorrhizal Fungi Isolated from Epiphytic and Terrestrial Orchids. Front. Microbiol. 2021, 12, 787820. [Google Scholar] [CrossRef]
- Ji, Y.; Li, X.; Zhu, Y.; Li, N.; Zhang, N.; Niu, M. Faecal microRNA as a biomarker of the activity and prognosis of inflammatory bowel diseases. Biochem. Biophys. Res. Commun. 2018, 503, 2443–2450. [Google Scholar] [CrossRef]
- Nakata, K.; Sugi, Y.; Narabayashi, H.; Kobayakawa, T.; Nakanishi, Y.; Tsuda, M.; Hosono, A.; Kaminogawa, S.; Hanazawa, S.; Takahashi, K. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J. Biol. Chem. 2017, 292, 15426–15433. [Google Scholar] [CrossRef] [Green Version]
- Ragusa, M.; Santagati, M.; Mirabella, F.; Lauretta, G.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; Domini, C.N.; Gulisano, M.; Barone, R.; et al. Potential Associations Among Alteration of Salivary miRNAs, Saliva Microbiome Structure, and Cognitive Impairments in Autistic Children. Int. J. Mol. Sci. 2020, 21, 6203. [Google Scholar] [CrossRef] [PubMed]
- Wakisaka, K.T.; Imai, Y. The dawn of pirna research in various neuronal disorders. Front. Biosci. 2019, 24, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- First, M.B. Diagnostic and statistical manual of mental disorders, and clinical utility. J. Nerv. Ment. Dis. 2013, 201, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- McLaren, M.R.; Callahan, B.J. Silva 138.1 Prokaryotic SSU Taxonomic Training Data Formatted for DADA2 [Data Set]. Zenodo 2021. [Google Scholar] [CrossRef]
- Morien, E.; Parfrey, L.W. SILVA v128 and v132 dada2 Formatted 18s “Train Sets” [Data Set]. Zenodo 2018. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2018, 27, 66–79. [Google Scholar] [CrossRef]
- Bonnici, V.; De Caro, G.; Constantino, G.; Liuni, S.; D’Elia, D.; Bombieri, N.; Licciulli, F.; Giugno, R. Arena-Idb: A platform to build human non-coding RNA interaction networks. BMC Bioinform. 2018, 19, 350. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Consiglio, A.; Mencar, C.; Grillo, G.; Marzano, F.; Caratozzolo, M.F.; Liuni, S. A fuzzy method for RNA-Seq differential expression analysis in presence of multireads. BMC Bioinform. 2016, 17, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, E.; Jiang, T.; Kaloshian, I.; Girke, T. SEED: Efficient clustering of next-generation sequences. Bioinformatics 2011, 27, 2502–2509. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Smyth, G.K. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 2007, 23, 2881–2887. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008, 9, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.-P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- Iglesias-Vázquez, L.; van Ginkel Riba, G.; Arija, V.; Canals, J. Composition of Gut Microbiota in Children with Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 792. [Google Scholar] [CrossRef] [Green Version]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; Comegna, M.; Buommino, E.; et al. Gut Microbiota Features in Young Children with Autism Spectrum Disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Ma, W.; Zhang, J.; He, Y.; Wang, J. Analysis of gut microbiota profiles and microbe-disease associations in children with autism spectrum disorders in China. Sci. Rep. 2018, 8, 13981. [Google Scholar] [CrossRef] [Green Version]
- Tarallo, S.; Ferrero, G.; Gallo, G.; Francavilla, A.; Clerico, G.; Realis Luc, A.; Manghi, P.; Thomas, A.M.; Vineis, P.; Segata, N.; et al. Altered Fecal Small RNA Profiles in Colorectal Cancer Reflect Gut Microbiome Composition in Stool Samples. mSystems 2019, 4, e00289-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segatto, M.; Tonini, C.; Pfrieger, F.W.; Trezza, V.; Pallottini, V. Loss of Mevalonate/Cholesterol Homeostasis in the Brain: A Focus on Autism Spectrum Disorder and Rett Syndrome. Int. J. Mol. Sci. 2019, 20, 3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoch, K.; Meng, L.; Szelinger, S.; Bearden, D.R.; Stray-Pedersen, A.; Busk, O.L.; Stong, N.; Liston, E.; Cohn, R.D.; Scaglia, F.; et al. A Recurrent De Novo Variant in NACC1 Causes a Syndrome Characterized by Infantile Epilepsy, Cataracts, and Profound Developmental Delay. Am. J. Hum. Genet. 2017, 100, 343–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artemios, P.; Areti, S.; Katerina, P.; Helen, F.; Eirini, T.; Charalambos, P. Autism Spectrum Disorder and Psychiatric Comorbidity in a Patient with Myhre Syndrome. J. Autism Dev. Disord. 2019, 49, 3031–3035. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Q.; Gong, N.N.; Kurolap, A.; Feldman, H.B.; Boy, N.; Brugger, M.; Grand, K.; McWalter, K.; Guillen Sacoto, M.J.; et al. Pathogenic variants in SMARCA5, a chromatin remodeler, cause a range of syndromic neurodevelopmental features. Sci. Adv. 2021, 7, eabf2066. [Google Scholar] [CrossRef]
- Tawamie, H.; Martianov, I.; Wohlfahrt, N.; Buchert, R.; Mengus, G.; Uebe, S.; Janiri, L.; Hirsch, F.W.; Schumacher, J.; Ferrazzi, F.; et al. Hypomorphic Pathogenic Variants in TAF13 Are Associated with Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2017, 100, 555–561. [Google Scholar] [CrossRef] [Green Version]
- Eising, E.; Carrion-Castillo, A.; Vino, A.; Strand, E.A.; Jakielski, K.J.; Scerri, T.S.; Hildebrand, M.S.; Webster, R.; Ma, A.; Mazoyer, B.; et al. A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry 2019, 24, 1065–1078. [Google Scholar] [CrossRef] [Green Version]
- Granadillo, J.L.; P A Stegmann, A.; Guo, H.; Xia, K.; Angle, B.; Bontempo, K.; Ranells, J.D.; Newkirk, P.; Costin, C.; Viront, J.; et al. Pathogenic variants in TNRC6B cause a genetic disorder characterised by developmental delay/intellectual disability and a spectrum of neurobehavioural phenotypes including autism and ADHD. J. Med. Genet. 2020, 57, 717–724. [Google Scholar] [CrossRef]
- Huang, Z.-X.; Chen, Y.; Guo, H.-R.; Chen, G.-F. Systematic Review and Bioinformatic Analysis of microRNA Expression in Autism Spectrum Disorder Identifies Pathways Associated with Cancer, Metabolism, Cell Signaling, and Cell Adhesion. Front. Psychiatry 2021, 12, 630876. [Google Scholar] [CrossRef]
- Puri, V.; McQuillin, A.; Choudhury, K.; Datta, S.; Pimm, J.; Thirumalai, S.; Krasucki, R.; Lawrence, J.; Quested, D.; Bass, N.; et al. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol. Psychiatry 2007, 61, 873–879. [Google Scholar] [CrossRef]
- Puri, V.; McQuillin, A.; Datta, S.; Choudhury, K.; Pimm, J.; Thirumalai, S.; Krasucki, R.; Lawrence, J.; Quested, D.; Bass, N.; et al. Confirmation of the genetic association between the U2AF homology motif (UHM) kinase 1 (UHMK1) gene and schizophrenia on chromosome 1q23.3. Eur. J. Hum. Genet. 2008, 16, 1275–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Li, J.; Wu, F.; Zheng, H.; Peng, Q.; Zhou, H. Altered composition and function of intestinal microbiota in autism spectrum disorders: A systematic review. Transl. Psychiatry 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Yi, X.; Zhang, X.; Wang, H.; Liu, H.; Mou, W.-W. Imbalance in the Gut Microbiota of Children with Autism Spectrum Disorders. Front. Cell. Infect. Microbiol. 2021, 11, 572752. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; Labaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [Green Version]
- Dan, Z.; Mao, X.; Liu, Q.; Guo, M.; Zhuang, Y.; Liu, Z.; Chen, K.; Chen, J.; Xu, R.; Tang, J.; et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 2020, 11, 1246–1267. [Google Scholar] [CrossRef]
- Hua, X.; Zhu, J.; Yang, T.; Guo, M.; Li, Q.; Chen, J.; Li, T. The Gut Microbiota and Associated Metabolites Are Altered in Sleep Disorder of Children with Autism Spectrum Disorders. Front. Psychiatry 2020, 11, 855. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef]
- Tavella, T.; Rampelli, S.; Guidarelli, G.; Bazzocchi, A.; Gasperini, C.; Pujos-Guillot, E.; Comte, B.; Barone, M.; Biagi, E.; Candela, M.; et al. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes 2021, 13, 1–19. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Zhang, Y.; Wang, X.; Yang, R.; Zhu, X.; Zhang, Y.; Chen, C.; Yuan, H.; Yang, Z.; Sun, L. Gut bacteria Akkermansia is associated with reduced risk of obesity: Evidence from the American Gut Project. Nutr. Metab. 2020, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Lin, J.; Isnard, S.; Fombuena, B.; Peng, X.; Marette, A.; Routy, B.; Messaoudene, M.; Chen, Y.; Routy, J.-P. The Bacterium Akkermansia muciniphila: A Sentinel for Gut Permeability and Its Relevance to HIV-Related Inflammation. Front. Immunol. 2020, 11, 645. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl. Environ. Microbiol. 2011, 77, 6718–6721. [Google Scholar] [CrossRef] [Green Version]
- Rubic, T.; Lametschwandtner, G.; Jost, S.; Hinteregger, S.; Kund, J.; Carballido-Perrig, N.; Schwärzler, C.; Junt, T.; Voshol, H.; Meingassner, J.G.; et al. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat. Immunol. 2008, 9, 1261–1269. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef] [Green Version]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; de Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 2017, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, N.; Yang, J.-J.; Zhao, D.-M.; Chen, B.; Zhang, G.-Q.; Chen, S.; Cao, R.-F.; Yu, H.; Zhao, C.-Y.; et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol. Res. 2020, 157, 104784. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [Green Version]
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358. [Google Scholar] [CrossRef]
- Alonso, R.; Pisa, D.; Fernández-Fernández, A.M.; Carrasco, L. Infection of Fungi and Bacteria in Brain Tissue from Elderly Persons and Patients with Alzheimer’s Disease. Front. Aging Neurosci. 2018, 10, 159. [Google Scholar] [CrossRef] [Green Version]
- Spatz, M.; Richard, M.L. Overview of the Potential Role of Malassezia in Gut Health and Disease. Front. Cell. Infect. Microbiol. 2020, 10, 201. [Google Scholar] [CrossRef]
- Hughes, H.K.; Ashwood, P. Anti-Candida albicans IgG Antibodies in Children with Autism Spectrum Disorders. Front. Psychiatry 2018, 9, 627. [Google Scholar] [CrossRef]
- De Magistris, L.; Familiari, V.; Pascotto, A.; Sapone, A.; Frolli, A.; Iardino, P.; Carteni, M.; de Rosa, M.; Francavilla, R.; Riegler, G.; et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 418–424. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Huang, H.; Chen, X.; Chen, S.; Yu, L.; Wang, C.; Liu, Y.; Zhang, K.; Wu, L.; Han, Z.-C.; et al. The delivery of hsa-miR-11401 by extracellular vesicles can relieve doxorubicin-induced mesenchymal stem cell apoptosis. Stem Cell Res. Ther. 2021, 12, 77. [Google Scholar] [CrossRef]
- Yang, Q.; Wei, B.; Peng, C.; Wang, L.; Li, C. Identification of serum exosomal miR-98-5p, miR-183-5p, miR-323-3p and miR-19b-3p as potential biomarkers for glioblastoma patients and investigation of their mechanisms. Curr. Res. Transl. Med. 2022, 70, 103315. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Z.; Liu, G.; Jin, C.; Zhang, Q.; Man, S.; Wang, Z. miR-657 Promotes Macrophage Polarization toward M1 by Targeting FAM46C in Gestational Diabetes Mellitus. Mediat. Inflamm. 2019, 2019, 4851214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, B.; Deng, F.; Yan, S.; Huang, K.; Wu, X.; Tao, X.; Wang, S.; Tao, F. Gestational diabetes mellitus, autistic traits and ADHD symptoms in toddlers: Placental inflammatory and oxidative stress cytokines do not play an intermediary role. Psychoneuroendocrinology 2021, 134, 105435. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.R.; Kimchi, E.T.; Manjunath, Y.; Gajagowni, S.; Stuckel, A.J.; Kaifi, J.T. RNA cargos in extracellular vesicles derived from blood serum in pancreas associated conditions. Sci. Rep. 2020, 10, 2800. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Chiu, P.K.-F.; Wong, C.Y.-P.; Cheng, C.K.-L.; Teoh, J.Y.-C.; Ng, C.-F. Identification of piRNA Targets in Urinary Extracellular Vesicles for the Diagnosis of Prostate Cancer. Diagnostics 2021, 11, 1828. [Google Scholar] [CrossRef]
- Abu-Amero, K.K.; Hellani, A.M.; Salih, M.A.; Seidahmed, M.Z.; Elmalik, T.S.; Zidan, G.; Bosley, T.M. A de novo marker chromosome derived from 9p in a patient with 9p partial duplication syndrome and autism features: Genotype-phenotype correlation. BMC Med. Genet. 2010, 11, 135. [Google Scholar] [CrossRef] [Green Version]
- Capkova, Z.; Capkova, P.; Srovnal, J.; Adamova, K.; Prochazka, M.; Hajduch, M. Duplication of 9p24.3 in three unrelated patients and their phenotypes, considering affected genes, and similar recurrent variants. Mol. Genet. Genom. Med. 2021, 9, e1592. [Google Scholar] [CrossRef]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef] [Green Version]
- Klip, A.; McGraw, T.E.; James, D.E. Thirty sweet years of GLUT4. J. Biol. Chem. 2019, 294, 11369–11381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gitlin, A.D.; Heger, K.; Schubert, A.F.; Reja, R.; Yan, D.; Pham, V.C.; Suto, E.; Zhang, J.; Kwon, Y.C.; Freund, E.C.; et al. Integration of innate immune signalling by caspase-8 cleavage of N4BP1. Nature 2020, 587, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Garbett, K.; Ebert, P.J.; Mitchell, A.; Lintas, C.; Manzi, B.; Mirnics, K.; Persico, A.M. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol. Dis. 2008, 30, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Gregg, J.P.; Lit, L.; Baron, C.A.; Hertz-Picciotto, I.; Walker, W.; Davis, R.A.; Croen, L.A.; Ozonoff, S.; Hansen, R.; Pessah, I.N.; et al. Gene expression changes in children with autism. Genomics 2008, 91, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Ginsberg, M.R.; Rubin, R.A.; Falcone, T.; Ting, A.H.; Natowicz, M.R. Brain transcriptional and epigenetic associations with autism. PLoS ONE 2012, 7, e44736. [Google Scholar] [CrossRef] [Green Version]
- Cioana, M.; Michalski, B.; Fahnestock, M. Insulin-Like Growth Factor and Insulin-Like Growth Factor Receptor Expression in Human Idiopathic Autism Fusiform Gyrus Tissue. Autism Res. 2020, 13, 897–907. [Google Scholar] [CrossRef]
- Kojima, K.; Musch, M.W.; Ren, H.; Boone, D.L.; Hendrickson, B.A.; Ma, A.; Chang, E.B. Enteric flora and lymphocyte-derived cytokines determine expression of heat shock proteins in mouse colonic epithelial cells. Gastroenterology 2003, 124, 1395–1407. [Google Scholar] [CrossRef]
- Yan, J.; Charles, J.F. Gut Microbiota and IGF-1. Calcif. Tissue Int. 2018, 102, 406–414. [Google Scholar] [CrossRef]
- Barone, M.V.; Troncone, R.; Auricchio, S. Gliadin peptides as triggers of the proliferative and stress/innate immune response of the celiac small intestinal mucosa. Int. J. Mol. Sci. 2014, 15, 20518–20537. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Borek, D.; Padrick, S.B.; Gomez, T.S.; Metlagel, Z.; Ismail, A.M.; Umetani, J.; Billadeau, D.D.; Otwinowski, Z.; Rosen, M.K. Structure and control of the actin regulatory WAVE complex. Nature 2010, 468, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.C.A.; de Oliveira Sousa, V.; Romão, L. Emerging roles for TGF-beta1 in nervous system development. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2005, 23, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Pozzo-Miller, L. Dysfunction of the corticostriatal pathway in autism spectrum disorders. J. Neurosci. Res. 2020, 98, 2130–2147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ben Zablah, Y.; Zhang, H.; Jia, Z. Rho Signaling in Synaptic Plasticity, Memory, and Brain Disorders. Front. Cell Dev. Biol. 2021, 9, 729076. [Google Scholar] [CrossRef]
- Vahdatpour, C.; Dyer, A.H.; Tropea, D. Insulin-Like Growth Factor 1 and Related Compounds in the Treatment of Childhood-Onset Neurodevelopmental Disorders. Front. Neurosci. 2016, 10, 450. [Google Scholar] [CrossRef] [Green Version]
- Cartagena-Rivera, A.X.; van Itallie, C.M.; Anderson, J.M.; Chadwick, R.S. Apical surface supracellular mechanical properties in polarized epithelium using noninvasive acoustic force spectroscopy. Nat. Commun. 2017, 8, 1030. [Google Scholar] [CrossRef] [Green Version]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Kittana, M.; Ahmadani, A.; Al Marzooq, F.; Attlee, A. Dietary Fat Effect on the Gut Microbiome, and Its Role in the Modulation of Gastrointestinal Disorders in Children with Autism Spectrum Disorder. Nutrients 2021, 13, 3818. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Manlove, L.S.; Farrar, M.A. Interleukin-2 and STAT5 in regulatory T cell development and function. JAK-STAT 2013, 2, e23154. [Google Scholar] [CrossRef] [Green Version]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17 (Suppl. S2), 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Nanni, N.; Bersanelli, M.; Cupaioli, F.A.; Milanesi, L.; Mezzelani, A.; Mosca, E. Network-Based Integrative Analysis of Genomics, Epigenomics and Transcriptomics in Autism Spectrum Disorders. Int. J. Mol. Sci. 2019, 20, 3363. [Google Scholar] [CrossRef] [PubMed] [Green Version]

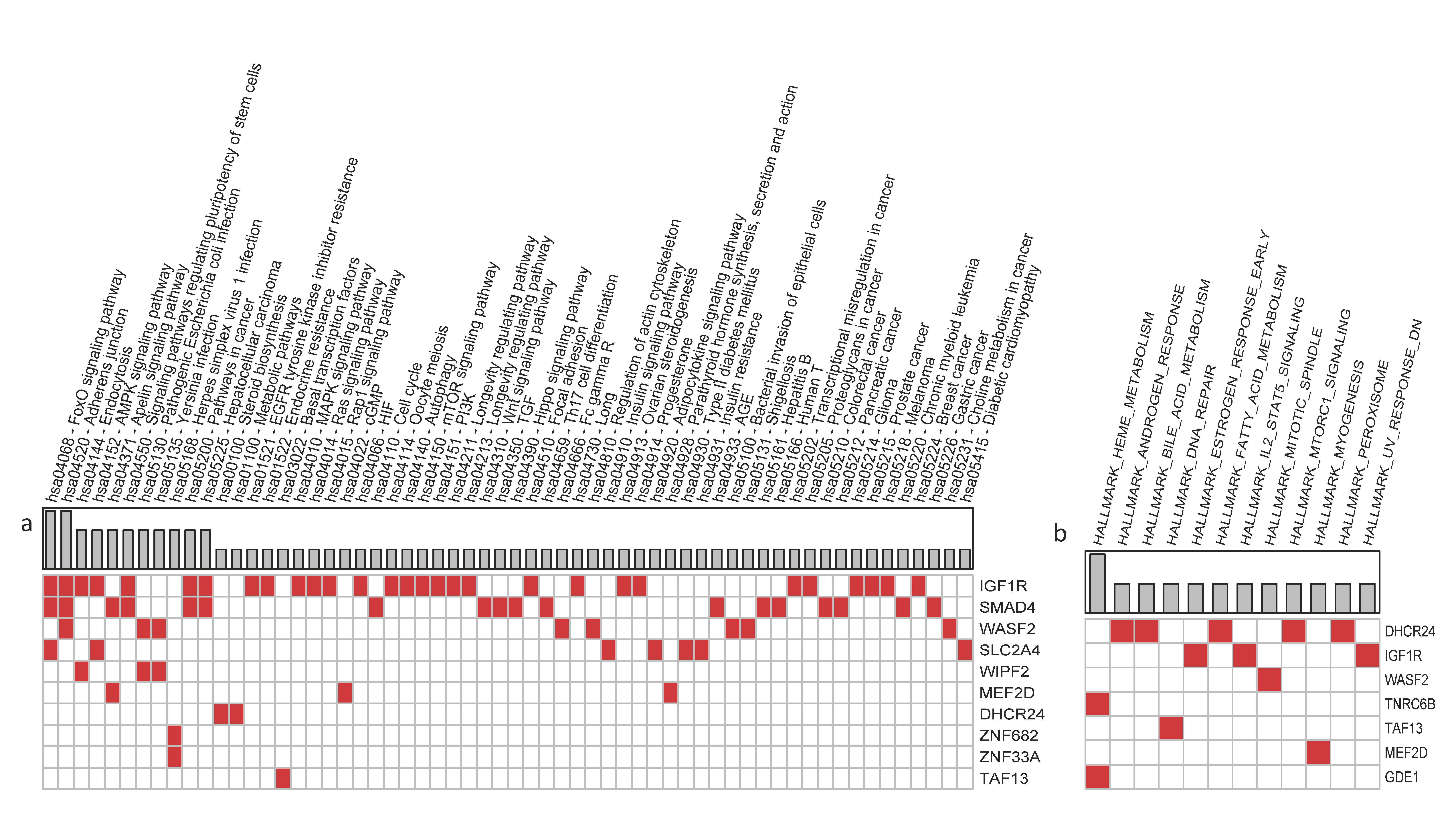
| Small ncRNAs | Target Gene | Tissue Expression Cluster (RNA) | Single-Cell Type Specificity (Enhanced in) | Tissue Specificity (RNA) | GI RNA Expression (Score) | GI Protein Expression (Score) | Biological Process | Molecular Function | Autism-Related Disorders | SFARI (Score Categories) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | hsa-miR-182-3p hsa-miR-99a-5p hsa-miR-4758-5p | CBWD1 | Intestine—Vesicular transport | Nonspecific | Low | Low | Medium-low | NA | ATP binding | ||
| hsa-miR-3911 hsa-miR-99a-5p hsa-miR-595 | DHCR24 | Non-specific—Unknown function | Hepatocytes, Alveolar cells type 2, Theca cells, Alveolar cells type 1 | Adrenal gland, liver | Low | NA | Cholesterol biosynthesis, Cholesterol metabolism, Lipid biosynthesis, Lipid metabolism, Steroid biosynthesis, Steroid metabolism, Sterol biosynthesis, Sterol metabolism | Oxidoreductase | Desmosterolosis (OMIM 602398) [62] | ||
| hsa-miR-4674 hsa-miR-4494 hsa-miR-6841-3p hsa-miR-99a-5p hsa-miR-4487 hsa-miR-3613-3p | GDE1 | Non-specific—Mitochondria | Syncytiotrophoblasts | Low | High | NA | Lipid metabolism | Hydrolase | |||
| hsa-miR-4742-3p hsa-miR-5689 hsa-miR-766-3p | HSBP1 | Non-specific—Mitochondria | Respiratory epithelial cells | Low | High | NA | Negative regulator of the heat shock response | Identical protein binding; transcription corepressor activity | |||
| hsa-miR-182-5p hsa-miR-96-5p hsa-miR-99a-5p hsa-miR-3613-3p hsa-miR-8071 | IGF1R | Ciliated cells—Cilium assembly | Oligodendrocytes, microglial cells, excitatory neurons, oligodendrocyte precursor cells, inhibitory neurons | Low | Medium | High | Host-virus interaction | Kinase, receptor, transferase, tyrosine-protein kinase | |||
| hsa-miR-4712-3p hsa-miR-1324 hsa-miR-99a-5p | MEF2D | Non-specific—Translation | Cone photoreceptor cells, sertoli cell; cluster in intestinal epithelial cells | Skeletal muscle | Low | High | Apoptosis, differentiation, neurogenesis, transcription, transcription regulation | Activator, developmental protein, DNA-binding | |||
| hsa-miR-4712-3p hsa-miR-6865-5p hsa-miR-3911 hsa-miR-595 hsa-miR-2110 hsa-miR-144-3p hsa-miR-3615 | NACC1 | Non-specific—Unknown function | Non-specific | Low | Medium | Medium | Transcription, transcription regulation | Repressor | Disease mutation, epilepsy, mental retardation [63] | 1S | |
| hsa-miR-4712-3p hsa-miR-5689 hsa-miR-96-3p | OLA1 | Non-specific—Mitochondria | Non-specific; cluster in smooth muscle cells | Low | Medium | High | ATP metabolic processes | Hydrolase | |||
| hsa-miR-4712-3p hsa-miR-4742-5p hsa-miR-99a-5p hsa-miR-144-3p hsa-miR-182-5p hsa-miR-96-5p hsa-miR-766-3p | RPL7L1 | Non-specific—Mitochondria | Non-specific | Low | Medium | NA | Blastocyst formation; maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | Ribonucleoprotein, ribosomal protein | |||
| hsa-miR-182-5p hsa-miR-144-3p hsa-miR-933 hsa-miR-154-5p | SMAD4 | Non-specific—Translation | Granulosa cells | Low | Medium | High | Transcription, transcription regulation | DNA-binding | Myhre Syndrome [64]: Juvenile polyposis syndrome | 2 | |
| hsa-miR-4487 hsa-miR-766-3p hsa-miR-144-3p hsa-miR-99a-5p | SMARCA5 | Immune cells—Transcription, translation | Alveolar cells type 1 | Low | Medium | High | Host-virus interaction | Chromatin regulator, helicase, hydrolase | Neurodevelopmental syndrome [65] | ||
| hsa-miR-3613-3p hsa-miR-766-3p hsa-miR-5689 hsa-miR-96-3p hsa-miR-144-3p | TAF13 | Non-specific—Translation | Suprabasal keratinocytes; cluster in macrophages | Low | Medium | Medium | Transcription, transcription regulation | DNA binding | Mental retardation, autosomal recessive 60 (OMIM 617432), Autosomal-Recessive Intellectual Disability [66] | ||
| hsa-miR-4742-5p hsa-miR-99a-5p hsa-miR-2113 hsa-miR-766-5p | TNRC6B | Bone marrow, brain—smell perception, nucleosome | Non-specific | Low | Low | High | RNA-mediated gene silencing, translation regulation | RNA-binding | Complex neurodevelopmental disorder involving spoken language, intellectual disability, neurobehavioural phenotype (ASD), and epilepsy [67,68,69] | 2 | |
| hsa-miR-4742-3p hsa-miR-1324 hsa-miR-5689 hsa-miR-6865-3p | UHMK1 | Non-specific—Unknown function | Non-specific | Low | Medium-high | NA | Neuron projection development | Kinase, RNA-binding, serine/threonine-protein kinase, transferase | Schizophrenia [70,71] | ||
| hsa-miR-3613-3p hsa-miR-766-3p hsa-miR-3615 | WDR12 | Non-specific—Mitochondria | Non-specific; cluster in Smooth muscle cells | Low | Medium-high | Medium-high | Ribosome biogenesis, rRNA processing | Ribonucleoprotein complex binding | |||
| hsa-miR-3613-3p hsa-miR-99a-3p hsa-miR-6939-5p hsa-miR-4758-3p hsa-miR-766-5p | WIPF2 | Bone marrow—Differentiation | Non-specific; cluster in intestinal epithelial cells | Low | High | High | Actin filament-based movement | Actin-binding | |||
| hsa-miR-766-3p hsa-miR-3615 hsa-miR-4712-5p hsa-miR-595 | ZNF682 | Skin—Unknown function | Oligodendrocytes | Low | Medium-low | NA | Transcription, transcription regulation | DNA-binding | |||
| hsa-miR-96-3p hsa-miR-5689 hsa-miR-2110 hsa-miR-6760-5p | ZNF703 | Striated muscle—Muscle contraction | Syncytiotrophoblasts | Skeletal muscle | Medium-low | Medium | Transcription, transcription regulation | Repressor | |||
| piRNA | hsa-piR-16407 hsa-piR-18524 | CFLAR | Lung—Lung homeostasis | Langerhans cells, urothelial cells; cluster in macrophages | Low | Low | High | Apoptosis, host-virus interaction | Cysteine-type endopeptidase activity involved in apoptotic signalling pathway | ||
| hsa-piR-21363 | GOLGA6L2 | Testis—Meiosis | Early spermatids | Testis | NA | NA | NA | NA | |||
| hsa-piR-21363 | SLC2A4 | Striated muscle—Muscle contraction | Cardiomyocytes | Heart muscle, skeletal muscle | Low | NA | Transcription, transcription regulation | DNA-binding | |||
| mirRNA/ piRNA | hsa-miR-708-3p hsa-miR-766-3p hsa-piR-9505 | N4BP1 | Skin—Epithelial junctions | Alveolar cells type 1, glandular and luminal cells, cluster in endometrium | Low | Medium | High | Immunity, innate immunity | Hydrolase, nuclease, RNA-binding | ||
| hsa-miR-766-5p hsa-piR-21363 | SLC2A4 | Striated muscle—Muscle contraction | Cardiomyocytes | Heart muscle, skeletal muscle | Low | NA | Transcription, transcription regulation | DNA-binding | |||
| hsa-miR-5689 hsa-piR-2001 | SLC12A6 | Immune cells—Transcription, Translation | Cone photoreceptor cells, rod photoreceptor cells; cluster in B-cells | Low | Low | Medium | Ion transport, potassium transport, symport, transport | potassium:chloride symporter activity | Andermann syndrome (OMIM #218000) | ||
| hsa-miR-144-3p hsa-piR-13910 | TTN | Striated muscle—Muscle contraction | Cardiomyocytes | Skeletal muscle, tongue | Very low | NA | Cardiac muscle tissue morphogenesis, skeletal muscle thin filament assembly | Calmodulin-binding, Kinase, serine/threonine-protein kinase, transferase | 3S | ||
| hsa-miR-3911 hsa-miR-4487 hsa-piR-433 | ZNF33A | Non-specific—Transport via ER | Non-specific | Low | Medium | Low | Transcription, transcription regulation | DNA-binding | |||
| Common to ASD and Ctrl | Only in | |||||
|---|---|---|---|---|---|---|
| Total | Significantly Up-Regulated | Significantly Down-Regulated | ASD | Ctrl | ||
| miRNA | Couple #1 | 207 (60) | 32 | 28 | 226 (15) | 317 (18) |
| Couple #2 | 152 (51) | 11 | 40 | 245 (19) | 184 (23) | |
| piRNA | Couple #1 | 380 (164) | 112 | 52 | 787 (109) | 1010 (56) |
| Couple #2 | 509 (222) | 66 | 156 | 757 (33) | 617 (85) | |
| Couple #1 | Couple #2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene Name | ASD | Ctrl | log2FC | Fisher_p | ASD | Ctrl | log2FC | Fisher_p |
| hsa-miR-10b-5p | 27.1 | 79.7 | −1.52 | 2.9 × 10−7 | 0.0 | 29.7 | −4.94 | 2.0 × 10−9 |
| hsa-mir-192 | 33.2 | 94.1 | −1.48 | 5.7 × 10−8 | 0.3 | 8.0 | −2.77 | 7.8 × 10−3 |
| hsa-miR-22-3p | 11.3 | 28.7 | −1.26 | 6.4 × 10−3 | 0.0 | 7.2 | −3.04 | 1.6 × 10−2 |
| hsa-miR-192-5p | 90.8 | 185.4 | −1.02 | 1.6 × 10−8 | 0.0 | 20.8 | −4.45 | 9.5 × 10−7 |
| hsa-miR-6760-5p | 7.9 | 0.9 | 2.22 | 3.9 × 10−2 | 11.4 | 0.0 | 3.63 | 9.8 × 10−4 |
| hsa-miR-6766 | 23.6 | 1.8 | 3.14 | 1.0 × 10−5 | 27.0 | 0.0 | 4.81 | 1.5 × 10−8 |
| hsa-miR-6839 | 7.9 | 0.0 | 3.15 | 7.8 × 10−3 | 113.3 | 0.0 | 6.84 | 0.0 × 100 |
| hsa-miR-3976 | 14.8 | 0.0 | 3.99 | 6.1 × 10−5 | 36.1 | 14.4 | 1.27 | 2.6 × 10−3 |
| hsa-piR-28021 | 0.0 | 429.1 | −8.75 | 0.0 × 100 | 0.3 | 57.7 | −5.47 | 0.0 × 100 |
| hsa-piR-8876 | 0.0 | 46.6 | −5.57 | 0.0 × 100 | 0.0 | 9.6 | −3.41 | 2.0 × 10−3 |
| hsa-piR-12132 | 0.0 | 43.9 | −5.49 | 0.0 × 100 | 14.3 | 804.0 | −5.72 | 0.0 × 100 |
| hsa-piR-32989 | 0.0 | 9.9 | −3.44 | 2.0 × 10−3 | 2.3 | 37.7 | −3.56 | 1.0 × 10−9 |
| hsa-piR-5819 | 0.0 | 7.2 | −3.03 | 1.6 × 10−2 | 0.7 | 8.8 | −2.57 | 2.1 × 10−2 |
| hsa-piR-14261 | 612.0 | 3592.7 | −2.55 | 0.0 × 100 | 9.1 | 20.8 | −1.11 | 4.3 × 10−2 |
| hsa-piR-33186 | 4681.1 | 23,105.1 | −2.30 | 0.0 × 100 | 0.0 | 5260.6 | −12.36 | 0.0 × 100 |
| hsa-piR-33033 | 6091.9 | 20,508.4 | −1.75 | 0.0 × 100 | 2.9 | 1094.1 | −8.12 | 0.0 × 100 |
| hsa-piR-5751 | 291.6 | 907.4 | −1.63 | 0.0 × 100 | 0.0 | 32.1 | −5.05 | 0.0 × 100 |
| hsa-piR-8213 | 11.3 | 28.7 | −1.26 | 6.4 × 10−3 | 1.0 | 8.0 | −2.19 | 3.9 × 10−2 |
| hsa-piR-32837 | 413.8 | 944.1 | −1.19 | 0.0 × 100 | 1.6 | 74.5 | −4.85 | 0.0 × 100 |
| hsa-piR-32914 | 413.8 | 944.1 | −1.19 | 0.0 × 100 | 1.6 | 74.5 | −4.85 | 0.0 × 100 |
| hsa-piR-28066 | 1331.4 | 2873.5 | −1.11 | 0.0 × 100 | 1.0 | 40.9 | −4.41 | 0.0 × 100 |
| hsa-piR-31090 | 39.3 | 18.8 | 1.02 | 1.2 × 10−2 | 38.4 | 16.8 | 1.14 | 6.4 × 10−3 |
| hsa-piR-32953 | 534.3 | 253.5 | 1.07 | 0.0 × 100 | 588.9 | 8.0 | 6.03 | 0.0 × 100 |
| hsa-piR-26659 | 267.1 | 121.8 | 1.13 | 0.0 × 100 | 608.1 | 283.0 | 1.10 | 0.0 × 100 |
| hsa-piR-21363 | 433.9 | 137.0 | 1.66 | 0.0 × 100 | 2058.3 | 484.9 | 2.08 | 0.0 × 100 |
| hsa-piR-31508 | 18.3 | 4.5 | 1.82 | 4.3 × 10−3 | 35.2 | 2.4 | 3.41 | 1.0 × 10−8 |
| hsa-piR-16407 | 14.0 | 1.8 | 2.42 | 4.2 × 10−3 | 230.8 | 10.4 | 4.34 | 0.0 × 100 |
| hsa-piR-2750 | 280.2 | 45.7 | 2.59 | 0.0 × 100 | 121.4 | 16.8 | 2.78 | 0.0 × 100 |
| hsa-piR-30491 | 68.1 | 8.1 | 2.93 | 0.0 × 100 | 157.9 | 55.3 | 1.50 | 0.0 × 100 |
| hsa-piR-22093 | 7.0 | 0.0 | 3.00 | 1.6 × 10−2 | 24.7 | 3.2 | 2.61 | 2.7 × 10−5 |
| hsa-piR-18568 | 7.0 | 0.0 | 3.00 | 1.6 × 10−2 | 13.0 | 0.8 | 2.96 | 1.8 × 10−3 |
| hsa-piR-21890 | 7.9 | 0.0 | 3.15 | 7.8 × 10−3 | 27.0 | 4.0 | 2.48 | 3.4 × 10−5 |
| hsa-piR-13475 | 18.3 | 0.9 | 3.35 | 7.6 × 10−5 | 39.4 | 12.0 | 1.63 | 2.0 × 10−4 |
| hsa-piR-8932 | 66.3 | 5.4 | 3.40 | 0.0 × 100 | 86.6 | 40.1 | 1.09 | 3.7 × 10−5 |
| hsa-piR-26586 | 9.6 | 0.0 | 3.41 | 2.0 × 10−3 | 10.1 | 0.0 | 3.47 | 2.0 × 10−3 |
| hsa-piR-12718 | 9.6 | 0.0 | 3.41 | 2.0 × 10−3 | 232.1 | 46.5 | 2.30 | 0.0 × 100 |
| hsa-piR-33000 | 9.6 | 0.0 | 3.41 | 2.0 × 10−3 | 560.5 | 68.1 | 3.02 | 0.0 × 100 |
| hsa-piR-30677 | 11.3 | 0.0 | 3.63 | 9.8 × 10−4 | 75.2 | 6.4 | 3.36 | 0.0 × 100 |
| hsa-piR-33037 | 14.8 | 0.0 | 3.99 | 6.1 × 10−5 | 16.6 | 4.0 | 1.81 | 7.2 × 10−3 |
| hsa-piR-24148 | 18.3 | 0.0 | 4.27 | 7.6 × 10−6 | 44.3 | 0.0 | 5.50 | 0.0 × 100 |
| hsa-piR-3308 | 18.3 | 0.0 | 4.27 | 7.6 × 10−6 | 21.2 | 4.0 | 2.15 | 9.1 × 10−4 |
| hsa-piR-2934 | 73.3 | 2.7 | 4.33 | 0.0 × 100 | 197.9 | 56.1 | 1.80 | 0.0 × 100 |
| hsa-piR-33019 | 20.1 | 0.0 | 4.40 | 1.9 × 10−6 | 392.3 | 0.0 | 8.62 | 0.0 × 100 |
| hsa-piR-3864 | 60.2 | 1.8 | 4.46 | 0.0 × 100 | 99.3 | 4.0 | 4.32 | 0.0 × 100 |
| hsa-piR-25822 | 22.7 | 0.0 | 4.57 | 2.4 × 10−7 | 108.7 | 28.1 | 1.92 | 0.0 × 100 |
| hsa-piR-11291 | 46.3 | 0.9 | 4.64 | 0.0 × 100 | 123.0 | 45.7 | 1.41 | 3.0 × 10−9 |
| hsa-piR-4194 | 48.9 | 0.9 | 4.72 | 0.0 × 100 | 59.9 | 24.8 | 1.24 | 1.9 × 10−4 |
| hsa-piR-9502 | 26.2 | 0.0 | 4.77 | 3.0 × 10−8 | 22.5 | 0.0 | 4.55 | 4.8 × 10−7 |
| hsa-piR-8096 | 27.9 | 0.0 | 4.86 | 7.0 × 10−9 | 83.0 | 11.2 | 2.78 | 0.0 × 100 |
| hsa-piR-30323 | 634.7 | 19.7 | 4.94 | 0.0 × 100 | 595.1 | 8.0 | 6.05 | 0.0 × 100 |
| hsa-piR-32334 | 34.0 | 0.0 | 5.13 | 0.0 × 100 | 15.6 | 0.0 | 4.06 | 3.1 × 10−5 |
| hsa-piR-892 | 151.9 | 0.0 | 7.26 | 0.0 × 100 | 18.6 | 3.2 | 2.22 | 8.5 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiappori, F.; Cupaioli, F.A.; Consiglio, A.; Di Nanni, N.; Mosca, E.; Licciulli, V.F.; Mezzelani, A. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk. Nutrients 2022, 14, 1340. https://doi.org/10.3390/nu14071340
Chiappori F, Cupaioli FA, Consiglio A, Di Nanni N, Mosca E, Licciulli VF, Mezzelani A. Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk. Nutrients. 2022; 14(7):1340. https://doi.org/10.3390/nu14071340
Chicago/Turabian StyleChiappori, Federica, Francesca Anna Cupaioli, Arianna Consiglio, Noemi Di Nanni, Ettore Mosca, Vito Flavio Licciulli, and Alessandra Mezzelani. 2022. "Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk" Nutrients 14, no. 7: 1340. https://doi.org/10.3390/nu14071340
APA StyleChiappori, F., Cupaioli, F. A., Consiglio, A., Di Nanni, N., Mosca, E., Licciulli, V. F., & Mezzelani, A. (2022). Analysis of Faecal Microbiota and Small ncRNAs in Autism: Detection of miRNAs and piRNAs with Possible Implications in Host–Gut Microbiota Cross-Talk. Nutrients, 14(7), 1340. https://doi.org/10.3390/nu14071340