3.1. Effects of Hazelnut Thermal/Pressure Treatment on Patient Response
First, we sought to assess whether the modified hazelnut extracts were associated with a reduced patient response in vivo. To this end, 22 patients with diagnosed hazelnut allergy (16 females namely 72.7%; mean age, 28.1 ± 9.4 years; range: 17–48 years) underwent skin prick testing [
11]. Specifically, 19 (86.4%) of 22 patients reported urticaria/angioedema, three reported oral allergy syndrome (13.6%), and one reported both manifestations (4.5%), suggesting that most patients were likely sensitized to hazelnut proteins such as Cor a 8, Cor a 9, Cor a 11 that are known to be responsible for systemic symptoms (
Table 1).
The SPT reactivity (defined as the mean wheal area of two independent measurements) reported with the hazelnut extract A1 (unmodified hazelnut seed extracts-Corylus avellana, var. Italiana) was 24.6 ± 12.3 mm2, comparable to that produced by the three commercial extracts (ALK, 26.5 ± 13.2 mm2; Stallergenes, 22.8 ± 12.3 mm2; Lofarma, 18.7 ± 9.0 mm2). This suggested that the native A1 extract was effective and suitable for use for comparison with the modified extracts. Negative controls (either saline or urea solutions) did not elicit any skin reaction (no wheal, erythema, or pruritus).
Only six patients (27.3%) showed reactivity to the A2 extract. This reactivity appeared lower than that of the native extract (the mean wheal area of the six reactive patients was 15.2 ± 4.1 mm
2;
p for A1 vs. A2 = 0.005) (
Figure 2;
Table 2).
Only two of the six patients reactive to A2 showed reactivity to the A3 extract. The mean wheal area of the two reactive patients was 13.5 mm
2 (patient #15) and 4.5 mm
2 (patient #20), respectively (
Figure 2;
Table 2).
Finally, only one patient (patient #15) showed SPT reactivity (wheal area, 13.5 mm2) to the A4 extract.
Collectively, these results suggest that all of the three different treatments are effective in reducing hazelnut allergenicity in a meaningful proportion of patients. Treatments 3 and 4 appeared to be the most effective. Notably, one patient out of 22 did not show a significant reduction in SPT reactivity even with the A4 extract. Further analyses remain necessary in order to define the specific epitopes this patient was reactive to.
3.2. Effect of Thermal/Pressure Treatment on Hazelnut Protein Solubility
Food processing is known to alter the final structure and function of proteins, modifying such crucial properties as final solubility. Denaturation, hydrolysis of peptide bonds, restructuring of disulphide bonds, and interaction with other components (i.e., carbohydrates and lipids) can frequently occur during treatment and are among the causes leading to reduced protein solubility [
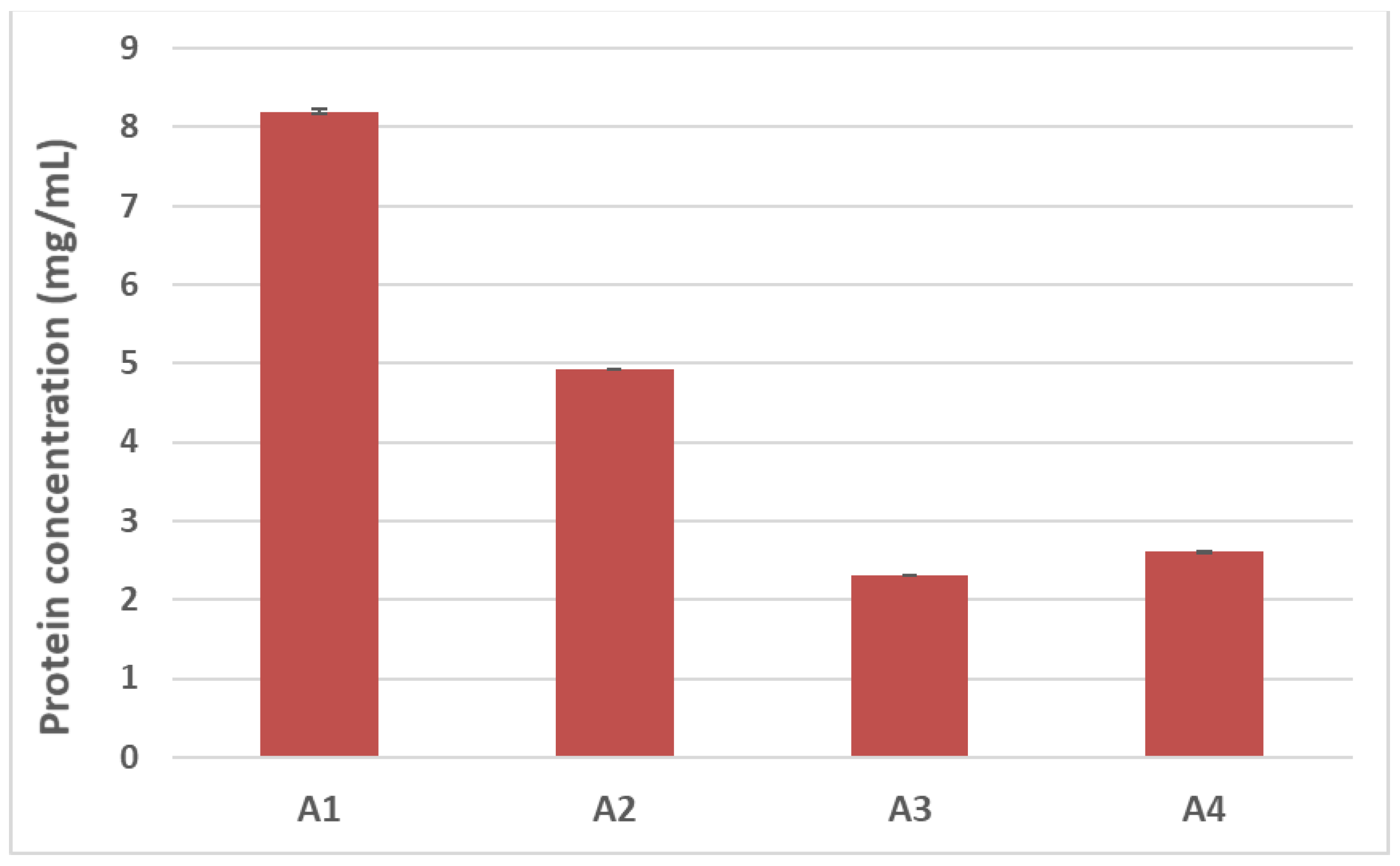
12]. In light of this, we first sought to evaluate how autoclaving/pressure processing could alter the final solubility of hazelnut proteins by estimating the total protein content of samples undergoing the different treatments. The Bradford assay was used for this purpose (
Figure 3). A progressive reduction in protein recovery was shown in the treated hazelnut samples compared to the untreated counterpart. The total protein content of the autoclaved samples (A2) appeared to be 40% lower than the control sample (A1), with a more dramatic reduction observed for the pre-hydrated/autoclaved sample (A3) and the pre-hydrated/autoclaved/dried samples (A4), which showed a 70% decrease in protein recovery compared to the untreated hazelnut sample (
Figure 3).
The results presented here are in accordance with previous studies reporting the effects of autoclaving on other nuts species such as almonds or peanuts [
6,
8,
13,
14]. In general, it has been demonstrated that autoclaving-based treatments produce a decrease in protein recovery and that this trend is more marked when treatment is accomplished under harsher conditions or when preceded by a hydration step. This phenomenon may be attributed to the numerous biochemical and structural modifications occurring on the protein moiety taking place during combined heat and pressure treatment. When combined with aggregation phenomena due to the interaction between proteins or protein-food matrix (intra- or intermolecular covalent and non-covalent interactions), this could promote protein precipitation and a consequent decrease in protein solubility and protein content in the final extract [
15]. As for hazelnuts, in 2012 Lopez et al. demonstrated that autoclaving could alter the secondary and tertiary structure of tree nut proteins, inducing a glycosylation reaction [
16]. In the current study, the reduced protein content observed in the treated hazelnut extracts confirms that autoclave-based treatments can modify the protein structure of hazelnut proteins, leading to a reduction in their solubility. This effect appears to be enhanced by preceding autoclaving with water incubation. In addition to reduced solubility, fragmentation of allergen proteins is likely to occur during autoclaving due to the application of pressure and temperature. It is already known that according to some Oral Food Challenge (OFC) studies [
17,
18] new thresholds might be proposed for allergens undergoing heating/baking treatments in food matrices, as these can induce modification of protein structure and decrease allergenicity. In line with this, it is reasonable that the reduced protein extraction or protein degradation after physical treatments might have an influence on overall SPT reactivity.
3.3. SDS-PAGE Profiling of Processed Hazelnuts and Protein Identifications
The raw (A1) and different processed hazelnut extracts (A2, A3 and A4) were electrophoretically profiled in order to evaluate possible changes in hazelnut protein content and structure induced by the different treatments. Specifically, 10 µg of untreated (A1) and treated hazelnut proteins (A2, A3 and A4, respectively) were analyzed by SDS-PAGE and protein profiles were observed for each sample analyzed (
Figure 4). In the untreated sample (
Figure 4A, lane A1), several bands in the region of 30–50 kDa and 10–22 kDa appear. As known from the literature, in the absence of reducing agents Cor a 9 (11S legumins) is organized in a hexameric structure made up of six subunits interacting non-covalently and arranged in an open ring conformation with 360 kDa [
19].
Each subunit is composed by an acidic polypeptide (30–40 kDa) linked to a basic polypeptide (around 20 kDa) by a disulphide bond [
20]. Under reducing conditions, acid and basic subunits are released; these are clearly visible in the lane of the untreated sample (
Figure 4A, lane A1). Other bands are visible over the 37 kDa region and below 20 kDa; these are likely to be attributed to Cor a 11, Cor a 8 and Cor a 14, the MWs of which are reported to be approximately 48 Da, 9 kDa and 15–16 kDa, respectively [
20]. After autoclaving (
Figure 4A, lane A2), a general decrease in band intensity was recorded, with a concomitant disappearance of the protein bands at 50 kDa and below 20 kDa that were putatively attributed to the Cor a 11, Cor a 8 and Cor a 14 allergens. As for Cor a 9, a marked reduction in signals corresponding to acid and basic subunits was observed, likely attributable to reduced content of the allergen following thermal/pressure treatment. On the contrary, the protein profiles of hazelnut samples incubated with water before autoclaving (
Figure 4A, lanes A3 and A4) appeared as a smear of peptides with a low MW (10–20 kDa), probably produced by fragmentation occurring during the treatments. It is worth noting that the drying process after autoclaving (A4 treatment) did not produce any significant difference in protein profile with respect to the A3 sample.
SDS-PAGE profiles are in accordance with protein assay results (
Figure 3); a progressive reduction in protein content can be seen among the treated hazelnut samples, with a more marked decrease observed for the pre-hydrated/autoclaved and pre-hydrated/autoclaved/dried samples.
In order to obtain insight into the protein content of the specific bands identified by SDS-PAGE, the most relevant bands which were expressed differently after different treatments (
Figure 4B, lanes A2, A3 and A4) were excised from the gel (the excised bands are numbered from 1 to 6), submitted to tryptic digestion, and analyzed with untargeted High Resolution Mass Spectrometry. MS spectra were then processed using Proteome Discoverer software for protein identification. Specifically, the Uniprot database referred to
Corylus avellana (last accessed on 26 November 2021) was interrogated; the results are shown in
Table 3.
As expected, bands 1 and 2 of the autoclaved sample (
Figure 4B, lane A2) were attributed to Cor a 9 allergen, suggesting the resistance of this allergen to autoclave processing (although the reduced intensity of the relative bands leads to the supposition that some partial degradation/structural alteration of the proteins likely occurred during the treatment). Cor a 9 allergen was found in bands 3 and 4 as well (
Figure 4B, lane A2). The smeared bands visible in the protein profiles of the pre-hydrated/autoclaved samples, whether followed or not by drying (approximately 10–20 kDa, bands 5 and 6, respectively) (
Figure 4B, lanes A3 and A4) were instead produced by a mix of peptides belonging to Cor a 9, Cor a 11 and Cor a 14 allergens (
Table 3).
The impact of autoclaving on the protein stability of tree nuts and peanuts has already been reported in the literature [
6,
8,
16,
21,
22], with some papers exploring the effects of water incubation before thermal/pressure treatment [
6,
8]. Similar to the work described here, these authors observed that samples pretreated with water before autoclaving showed a more fragmented SDS-PAGE protein profile and were degraded in comparison with their autoclaved counterparts. To explain these phenomena, they suggested that water absorbed by seeds during incubation may facilitate the propagation of heat in the inner part of the seed, promoting protein disaggregation and a consequent decrease in band intensity. In addition, our results are in line with those described by Lopez et al. in 2012, in a study of the effects of different autoclaving and high-pressure treatment conditions on the final allergenicity of hazelnut flour. The authors highlighted the disappearance of the main hazelnut allergen protein bands in the SDS-PAGE profile of hazelnuts autoclaved at 131 °C for 15 or 20 min, ascribing these results to molecular alterations or post-translational modifications (PTMs, e.g., glycosylation) that occur during autoclaving. Indeed, by generating a series of homology-based 3D bioinformatics models, they found that the structure of the main hazelnut allergens (Cor a 8, Cor a 9) were altered after autoclaving, and found with a new glycosylation site in the Cor a 11 allergen [
16].
3.4. Immunoblotting Experiments
Immunoblotting experiments were performed to assess the immunogenicity and the allergenicity of hazelnut seeds subjected to the different autoclaving treatments (A2, A3, A4). Sera from 14 patients showing different both systemic and OAS clinical symptoms were used for the immunoblotting experiments (
Figure 5).
As a preliminary investigatory step, the sera of the 14 allergic patients were pooled together and used as the primary antibody, with the final aim of obtaining an overview of the effects of the autoclaving-based treatments here investigated on the final allergenicity of the hazelnut.
The membranes were then incubated with a goat anti-rabbit IgG Ab (
Figure 5A) or an anti-human IgE Ab (ε-chain specific) (
Figure 5B) as the secondary antibodies. Finally, to increase the assay sensitivity two additional subgroups with seven pooled sera per group (
Figure 5C,D) were incubated with a goat anti-rabbit IgG Ab (vide infra).
Figure 5A shows one main reactive band with MW of approximately 20 kDa in untreated hazelnut (lane A1), along with weak intensity bands at 50 kDa and in the range of 10 and 15 kDa. Two additional blurred signals between 21–22 kDa are displayed in the A1 sample as well. According to the literature, protein banding at 20 kDa can be putatively attributed to the basic subunit Cor a 9 hazelnut allergens, as can bands in the range of 21–22 kDa. On the contrary, the reactivity displayed at 50 kDa could be ascribable to the Cor a 11 allergen, which has an original MW of 48–50 kDa, while the weak signal in the range of 10–15 kDa could be attributed to Cor a 8 or Cor a 14 allergens, which have MWs falling in this range. After autoclaving treatment (
Figure 5A, lane A2), the weak reactivity of the signals at 50 kDa and below 15 kDa became negligible, while the intense spot at 20 kDa and bands in the range of 21–22 kDa persisted. On the basis of proteomic investigation accomplished with SDS-PAGE protein profiles, these bands can be attributed to Cor a 9 and its isoform allergen Cor a 9.0101 (
Table 3), confirming that these allergens survived the autoclaving processing and preserving their allergenic potential. On the contrary, no reactive signal was displayed for the band at 50 kDa, putatively ascribed to Cor a 11, or for the 10–15 kDa hazelnut allergens (likely Cor a 8 and Cor a 14), confirming the susceptibility of these proteins to autoclave processing (
Figure 5A). As for the pre-hydrated/autoclaved (
Figure 5A, lane A3) and pre-hydrated/autoclaved/dried (
Figure 5A, lane A4) samples, no clearly identifiable reactive bands were shown, thus confirming the key role of the water imbibition step in the structural and conformational alteration/degradation phenomena induced and enhanced by the thermal/pressure treatment applied on these proteins. Cor a 9 was demonstrated to be a very well-structured protein, enriched with a beta-sheet core and containing long unstructured loops. These loop regions were found to be structurally unstable, and were predicted to retain linear epitopes located at the external faces of the protein and thus exposed to solvent [
16]. After submitting hazelnut samples to autoclaving (121 °C or 138 °C for 15 or 30 min), Lopez and co-workers obtained a reduction of Cor a 9 allergenicity, with no bands corresponding to this protein visible in the SDS-PAGE analysis. In light of this, the authors hypothesized that the allergenicity of Cor a 9 could be predominantly ascribed to structural conformation and not to linear epitopes [
16]. Our results seem to support this hypothesis. Indeed, the enhanced action of previous water incubation on autoclaving (A3, A4) seems effective in impairing the structure of Cor a 9, with no reactive epitopes surviving after processing, which is different from the results for the solely autoclaved sample (
Figure 5A, lane A2). On visual inspection of the SDS-PAGE (pictured in
Figure 4B), despite the visible smear bands below 20 kDa produced by a mixture of Cor a 9 and Cor a 14 allergens peptides in the A3 sample and by Cor a 9, Cor a 14 and Cor a 11 allergens peptides in the A4 sample (as highlighted by proteomic investigation in
Table 3), no reactive signals were displayed in the putatively corresponding lanes of the immunoblotting profiles. As already indicated by the electrophoretic analysis, no significant changes in immunoblotting profile or in the consequent final allergenicity of pre-hydrated/autoclaved hazelnut were found by drying the sample (
Figure 5, lane A3).
Similar results were obtained by incubating the A1-A4 samples with anti-human IgE Ab (vide infra) (ε-chain specific) secondary antibody, confirming that a specific IgE can bind protein bands detected in untreated and autoclaved samples, and thus likely trigger an allergic reaction in vivo. On the contrary, no IgE reactivity was displayed for the A3-A4 samples, indicating their likely lack of allergic potential.
Considering that sensitization to specific hazelnut proteins could vary among allergic individuals, additional immunoblotting experiments were performed by dividing the 14 patient sera into two groups of seven patient sera. The aim was to unveil possible different patterns of sensitization in different patient subgroups. In particular,
Figure 5C,D shows immunoblotting experiments conducted using two different serum pools, each of which was made up of seven patients’ sera. These immunoblots showed different sensitivities compared to those shown in
Figure 5A,B, which used a pool made up of all 14 patients’ sera, depending on the differential antibody concentration of each serum sample in the subgroups.
As a matter of the fact, the reactivity profile varied among the different pools.
Figure 5C shows more intense bands at approximately 50 kDa (Cor a 11), 37 kDa (Cor a 9 acid subunit), 20–22 kDa (Cor a 9 basic subunit/Cor a 1.04) and 10 kDa (Cor a 8/Cor a 14) when compared to
Figure 5A,B,D. This was expected, as pools represent patients’ average response and can therefore vary along with the differential pool array. Further analyses of individual patient sera are necessary in order to accurately identify the individual reactivity profile and how it could be modified by specific treatments. This study is a preliminary analysis primarily focused on the general effectiveness of the physical treatment used to reduce allergenicity. A pool of sera containing different IgE antibodies specificities was considered useful in answering this research question; however, the findings provided here will be expanded upon in subsequent studies in which immunoblot analyses will be performed on individual patients in parallel with specific IgE profiles based on component-resolved diagnosis. Using this approach together with Mass Spectrometry analysis of protein fragments should permit better understanding of which specific allergenic proteins are more liable to degradation, and consequently which specific IgE reactivities will be lost. This will enable prediction of individual patients’ absence of reactivity to modified hazelnut (i.e., no reaction after exposure to modified hazelnut) by analyzing individual IgE profiles against each antigenic protein obtained by component-resolved diagnosis.
The effect of the autoclaving on the final allergenicity of hazelnuts was already investigated in 2012 by Lopez et al. The authors submitted hazelnut defatted flour to autoclave processing under different conditions (121 °C 15 min, 121 °C 30 min, 138 °C 15 min, 138 °C 30 min) and investigated the IgE-reactivity of fifteen allergic patients via Western blot experiments. In addition, they studied the changes to the conformational structure of hazelnut allergens induced by autoclaving by generating a series of homology-based 3D bioinformatics models for the allergens Cor a 1, Cor a 8, Cor a 9, and Cora 11. As result, the authors observed that under harsher conditions (138 °C 15 min, 138 °C 30 min) autoclaving induced a severe reduction in hazelnut allergenicity in the patients studied. Indeed, the specific IgE binding of certain immunoreactive hazelnut protein bands such as Cor a 1, Cor a 8, Cor a 9 and (vide infra) Cor a 11 all decreased. Moreover, the structural analysis (3D modelling) of these allergens highlighted that, vide infra, relevant glycosylation occurred in the protein allergen Cora 11 after autoclaving, suggesting that the combination of temperature and pressure could promote the interaction of protein and matrix, likely altering the final allergenicity of the protein [
16].
Very recently, Cuadraro et al. investigated the effects of autoclaving on the final allergenicity of the Cor a 9, Cor a 14 and Cor a 8 hazelnut allergens by testing two different autoclaving temperatures, 121 °C and 138 °C, for 30 min. Whole hazelnut seeds were processed and, similar to the results described by Lopez et al., they observed in the different immunoblot profiles a marked reduction of Cor a 9, Cor a 14, and Cor a 8 reactivity after autoclaving hazelnut material at 138 °C for 30 min [
23].
In the present investigation, we observed that by autoclaving hazelnut seeds at 134 °C for 10 min the intensity of Cor a 9 band appeared to be reduced, with the disappearance of the 50 kDa and 10 kDa bands putatively ascribed to the Cor a 11 and Cor a 8 allergens (as shown in the SDS-PAGE picture in
Figure 3, lane A2). These results are comparable to those obtained by the aforementioned authors when applying similar autoclaving conditions, that is, a temperature of 121 °C for 30 min [
16,
23].
Furthermore, the immunoblotting profiles presented here confirm the IgG and IgE reactivity of Cor a 9 basic subunits after the autoclaving of hazelnut material only (
Figure 5A–D). On the contrary, by incubating hazelnut seeds with water before autoclaving the full degradation/fragmentation of the proteins was observed, with the disappearance of the main allergenic bands in both the corresponding SDS-PAGE (
Figure 3, lane A3 and A4) and the immunoblotting profile (
Figure 5A–D). These results are comparable to those observed by Lopez et al. and Cuadraro et al. when analyzing hazelnut autoclaved at 138 °C for 30 min via SDS-PAGE and Western blotting [
16,
23]. In the light of this, it is reasonable to suppose that water incubation strengthens the alterative phenomena induced by autoclaving on proteins, obtaining the same effects as autoclaving of hazelnut under such harsh conditions as 138 °C for 30 min.