Association between the rs3812316 Single Nucleotide Variant of the MLXIPL Gene and Alpha-Linolenic Acid Intake with Triglycerides in Mexican Mestizo Women
Abstract
1. Introduction
2. Materials and Methods
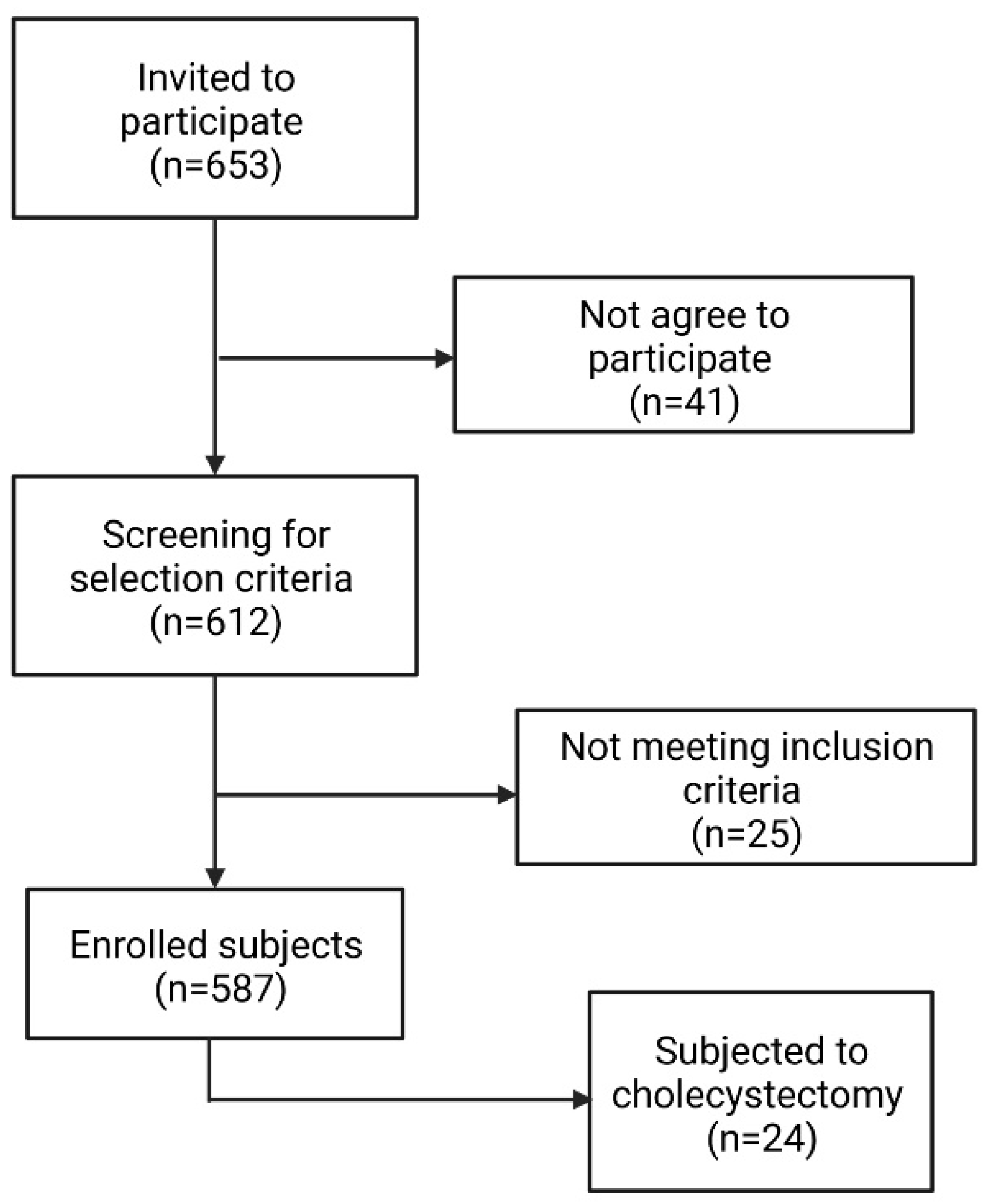
2.1. Subjects
2.2. Ethical Considerations
2.3. Anthropometric Measurements
2.4. Nutritional Assessment
- Adequate total fat intake (Ad fat): 20–35% of kcal;
- Excessive total fat intake (Ex fat): >35% of kcal;
- Adequate total saturated fatty acid intake (Ad SFA): <7% of kcal;
- Excessive total saturated fatty acids intake (Ex SFA): >7% of kcal;
- Adequate total monounsaturated fatty acids intake (Ad MUFA): 15–25% of kcal;
- Insufficient total monounsaturated fatty acids intake (Ins MUFA): <15% of kcal;
- Adequate total polyunsaturated fatty acids intake (Ad PUFA): 3–10% of kcal;
- Insufficient total polyunsaturated fatty acids intake (Ins PUFA): <3% of kcal;
- Adequate total alpha-linolenic acid intake (Ad ALA): 0.6–1.2% of kcal;
- Insufficient total alpha-linolenic acid intake (Ins ALA): <0.6% of kcal.
2.5. Biochemical Measurements and Definitions
2.6. DNA Extraction and Genotyping
2.7. RNA Extraction of Tissue Samples
2.8. qPCR Real-Time
2.9. In Silico Analysis of rs3812316 SNV of MLXIPL Gene
2.10. Statistical Analyses
3. Results
3.1. Characteristics of the Study Population
3.2. Frequencies of SNVs rs17145750 and rs3812316 of MLXIPL Gene
3.3. Anthropometric, Biochemical, and Nutritional Variables by rs17145750 and rs3812316 SNVs
3.4. Association of rs17145750 and rs3812316 SNV’s of MLXIPL Gene with Dyslipidemias
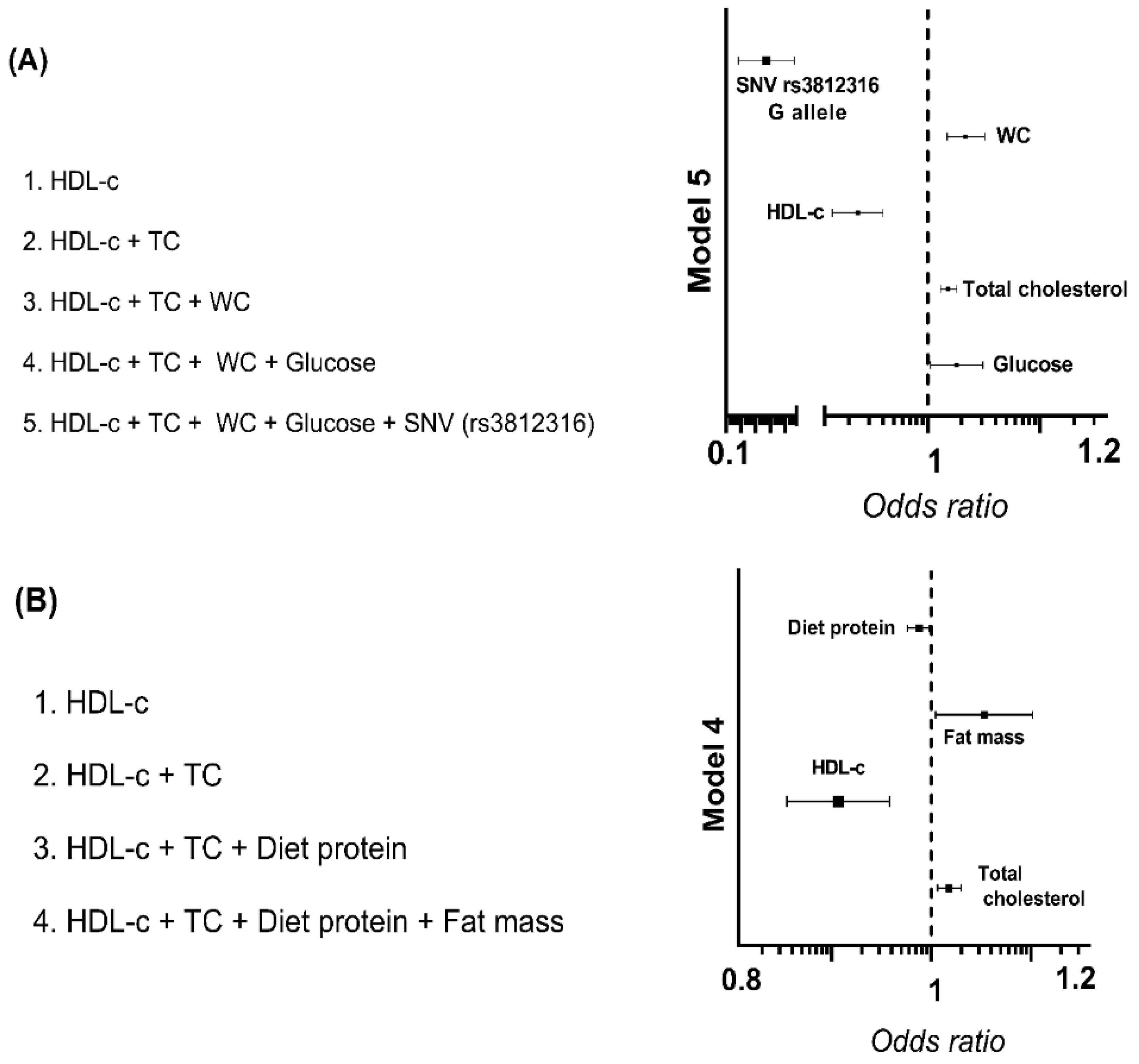
3.5. Association of rs3812316 SNV of MLXIPL Gene with Triglyceride Levels
3.6. SNVs and Nutrient Association
3.7. Expression of ChREBP α and β Isoforms by Genotype
3.8. In Silico Analysis of the SNV rs3812316
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diamond, J. The double puzzle of diabetes. Nature 2003, 423, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.R.; Novotny, J.A.; Rumpler, W.V. Effects of the interaction of sex and food intake on the relation between energy expenditure and body composition. Am. J. Clin. Nutr. 2004, 79, 385–389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Villalobos, A.; Aguilar-Salinas, C.A.; Martínez, M.R.; Rojas-Martínez, R. Perfiles poblacionales asociados con la asistencia a servicios preventivos para tamizaje de diabetes e hipertensión. Ensanut 2018–2019. Salud Pública Méx. 2020, 62, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Gomez, B.; Almeda-Valdés, P.; Tussié-Luna, M.T.; Aguilar-Salinas, C.A. Dyslipidemia in Mexico, a Call for Action. Rev. Investig. Clin. 2018, 70, 211–216. [Google Scholar] [CrossRef]
- García-Chávez, C.G.; Rivera, J.A.; Monterrubio-Flores, E.; Rodríguez-Ramírez, S. Dietary patterns are associated with obesity in Mexican schoolchildren. Eur. J. Clin. Nutr. 2020, 74, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Barquera, S.; Hernández-Barrera, L.; Tolentino, M.L.; Espinosa, J.; Ng, S.W.; Rivera, J.A.; Popkin, B.M. Energy Intake from Beverages Is Increasing among Mexican Adolescents and Adults. J. Nutr. 2008, 138, 2454–2461. [Google Scholar] [CrossRef]
- Sayón-Orea, C.; Bes-Rastrollo, M.; Carlos, S.; Beunza, J.J.; Basterra-Gortari, F.J.; Martínez-González, M.A. Association between Sleeping Hours and Siesta and the Risk of Obesity: The SUN Mediterranean Cohort. Obes. Facts 2013, 6, 337–347. [Google Scholar] [CrossRef]
- Kooner, J.S.; Chambers, J.C.; Aguilar-Salinas, C.A.; Hinds, D.; Hyde, C.L.; Warnes, G.R.; Pérez, F.J.G.; Frazer, K.; Elliott, P.; Scott, J.; et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008, 40, 149–151. [Google Scholar] [CrossRef]
- Uyeda, K.; Yamashita, H.; Kawaguchi, T. Carbohydrate responsive element-binding protein (ChREBP): A key regulator of glucose metabolism and fat storage. Biochem. Pharmacol. 2002, 63, 2075–2080. [Google Scholar] [CrossRef]
- Herman, M.A.; Peroni, O.D.; Villoria, J.; Schön, M.R.; Abumrad, N.A.; Blüher, M.; Klein, S.; Kahn, B.B. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 2012, 484, 333–338. [Google Scholar] [CrossRef]
- Jois, T.; Howard, V.; Youngs, K.; Cowley, M.A.; Sleeman, M.W. Dietary Macronutrient Composition Directs ChREBP Isoform Expression and Glucose Metabolism in Mice. PLoS ONE 2016, 11, e0168797. [Google Scholar] [CrossRef]
- Guo, S.; Zheng, F.; Qiu, X.; Yang, N. ChREBP gene polymorphisms are associated with coronary artery disease in Han population of Hubei province. Clin. Chim. Acta 2011, 412, 1854–1860. [Google Scholar] [CrossRef] [PubMed]
- Polgár, N.; Járomi, L.; Csöngei, V.; Maász, A.; Sipeky, C.; Sáfrány, E.; Szabó, M.; Melegh, B. Triglyceride level modifying functional variants of GALTN2 and MLXIPL in patients with ischaemic stroke. Eur. J. Neurol. 2010, 17, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Vrablik, M.; Ceska, R.; Adamkova, V.; Peasey, A.; Pikhart, H.; Kubinova, R.; Marmot, M.; Bobak, M.; Hubacek, J.A. MLXIPL variant in individuals with low and high triglyceridemia in white population in Central Europe. Qual. Life Res. 2008, 124, 553–555. [Google Scholar] [CrossRef] [PubMed]
- Sumegi, K.; Jaromi, L.; Magyari, L.; Kovesdi, E.; Duga, B.; Szalai, R.; Maasz, A.; Matyas, P.; Janicsek, I.; Melegh, B. Functional Variants of Lipid Level Modifier MLXIPL, GCKR, GALNT2, CILP2, ANGPTL3 and TRIB1 Genes in Healthy Roma and Hungarian Populations. Pathol. Oncol. Res. 2015, 21, 743–749. [Google Scholar] [CrossRef]
- Aceves, D.; Ruiz, B.; Nuno, P.; Roman, S.; Zepeda, E.; Panduro, A. Heterogeneity of Apolipoprotein E Polymorphism in Different Mexican Populations. Hum. Biol. 2006, 78, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Villalobos, H.; Muñoz-Valle, J.; González-Martín, A.; Gorostiza, A.; Magaña, M.; Páez-Riberos, L. Genetic admixture, relatedness, and structure patterns among Mexican populations revealed by the Y-chromosome. Am. J. Phys. Anthropol. 2007, 135, 448–461. [Google Scholar] [CrossRef]
- Pérez-Lizaur, A.B.; Kaufer-Horwitz, M.; Plazas, M. Environmental and personal correlates of fruit and vegetable consumption in low income, urban Mexican children. J. Hum. Nutr. Diet. 2007, 21, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Vannice, G.; Rasmussen, H. Position of the Academy of Nutrition and Dietetics: Dietary Fatty Acids for Healthy Adults. J. Acad. Nutr. Diet. 2014, 114, 136–153. [Google Scholar] [CrossRef]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults [M1]. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [Google Scholar] [CrossRef] [PubMed]
- Vaca, A.L.J.; Valdes-Flores, M.; Rivera-Vega, M.D.R.; González-Huerta, L.M.; Kofman-Alfaro, S.H.; Cuevas-Covarrubias, S.A. Deletion Pattern of the STS Gene in X-linked Ichthyosis in a Mexican Population. Mol. Med. 2001, 7, 845–849. [Google Scholar] [CrossRef]
- Ramírez-Meza, S.M.; Maldonado-González, M.; Hernández-Nazara, Z.H.; Martínez-López, E.; Ocampo-González, S.; Bobadilla-Morales, L.; Torres-Baranda, J.R.; Ruíz-Madrigal, B. Development of an effective and rapid qPCR for identifying human ChREBPα/β isoforms in hepatic and adipose tissues. Scand. J. Clin. Lab. Investig. 2019, 79, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Mutant Suite Predictor of Effects of Single Point Protein Mutation. Available online: http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi (accessed on 17 February 2022).
- Ortega-Azorín, C.; Sorlí, J.V.; Estruch, R.; Asensio, E.M.; Coltell, O.; González, J.I.; Martínez-González, M.; Ros, E.; Salas-Salvadó, J.; Fitó, M.; et al. Amino Acid Change in the Carbohydrate Response Element Binding Protein Is Associated With Lower Triglycerides and Myocardial Infarction Incidence Depending on Level of Adherence to the Mediterranean Diet in the PREDIMED Trial. Circ. Cardiovasc. Genet. 2014, 7, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Analysis Tool for Random Samples, by YongYong Shi. Available online: http://analysis.bio-x.cn/myAnalysis.php (accessed on 20 July 2022).
- UniProt. Available online: https://www.uniprot.org/uniprotkb/Q9NP71/entry (accessed on 10 June 2021).
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef]
- De Castro-Orós, I.; Cenarro, A.; Tejedor, M.T.; Baila-Rueda, L.; Mateo-Gallego, R.; Lamiquiz-Moneo, I.; Pocoví, M.; Civeira, F. Common Genetic Variants Contribute to Primary Hypertriglyceridemia Without Differences Between Familial Combined Hyperlipidemia and Isolated Hypertriglyceridemia. Circ. Cardiovasc. Genet. 2014, 7, 814–821. [Google Scholar] [CrossRef][Green Version]
- Aung, L.-H.; Yin, R.-X.; Wu, J.-Z.; Wu, N.-F.; Wang, W.; Li, H. Association between the MLX Interacting Protein-Like, BUD13 Homolog and Zinc Finger Protein 259 Gene Polymorphisms and Serum Lipid Levels. Sci. Rep. 2014, 4, 5565. [Google Scholar] [CrossRef]
- National Health and Nutrition Survey (ENSANUT) 2018. Available online: https://www.inegi.org.mx/programas/ensanut/2018/#Documentation (accessed on 8 April 2021). (In Spanish).
- Aguilar-Salinas, C.A.; Olaiz, G.; Valles, V.; Torres, J.M.R.; Pérez, F.J.G.; Rull, J.A.; Rojas, R.; Franco, A.; Sepulveda, J. High prevalence of low HDL cholesterol concentrations and mixed hyperlipidemia in a Mexican nationwide survey. J. Lipid Res. 2001, 42, 1298–1307. [Google Scholar] [CrossRef]
- Been, L.F.; Nath, S.K.; Ralhan, S.K.; Wander, G.S.; Mehra, N.K.; Singh, J.; Mulvihill, J.J.; Sanghera, D.K. Replication of Association Between a Common Variant Near Melanocortin-4 Receptor Gene and Obesity-related Traits in Asian Sikhs. Obesity 2010, 18, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Miller, B.; Uyeda, K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Metab. 2006, 291, E358–E364. [Google Scholar] [CrossRef] [PubMed]
- Parks, E.J. Effect of Dietary Carbohydrate on Triglyceride Metabolism in Humans. J. Nutr. 2001, 131, 2772S–2774S. [Google Scholar] [CrossRef] [PubMed]
- Santesso, N.; Akl, E.A.; Bianchi, M.; Mente, A.; Mustafa, R.; Heels-Ansdell, D.; Schunemann, H.J. Effects of higher- versus lower-protein diets on health outcomes: A systematic review and meta-analysis. Eur. J. Clin. Nutr. 2012, 66, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Treviño-Villarreal, J.H.; Reynolds, J.S.; Bartelt, A.; Langston, P.K.; MacArthur, M.R.; Arduini, A.; Tosti, V.; Veronese, N.; Bertozzi, B.; Brace, L.E.; et al. Dietary protein restriction reduces circulating VLDL triglyceride levels via CREBH-APOA5–dependent and –independent mechanisms. JCI Insight 2018, 3, 99470. [Google Scholar] [CrossRef]
- Dentin, R.; Denechaud, P.-D.; Benhamed, F.; Girard, J.; Postic, C. Hepatic Gene Regulation by Glucose and Polyunsaturated Fatty Acids: A Role for ChREBP. J. Nutr. 2006, 136, 1145–1149. [Google Scholar] [CrossRef]
- Qian, F.; Korat, A.A.; Malik, V.; Hu, F.B. Metabolic Effects of Monounsaturated Fatty Acid–Enriched Diets Compared with Carbohydrate or Polyunsaturated Fatty Acid–Enriched Diets in Patients With Type 2 Diabetes: A Systematic Review and Meta-analysis of Randomized Controlled Trials. Diabetes Care 2016, 39, 1448–1457. [Google Scholar] [CrossRef]
- Strychar, I.; Cohn, J.S.; Renier, G.; Rivard, M.; Aris-Jilwan, N.; Beauregard, H.; Meltzer, S.; Bélanger, A.; Dumas, R.; Ishac, A.; et al. Effects of a diet higher in carbohydrate/lower in fat versus lower in carbohydrate/higher in monounsaturated fat on post meal triglyceride concentrations and other cardiovascular risk factors in type 1 diabetes. Diabetes Care 2009, 32, 1597–1599. [Google Scholar] [CrossRef]
- Pelkman, C.L.; Fishell, V.K.; Maddox, D.H.; Pearson, T.A.; Mauger, D.T.; Kris-Etherton, P.M. Effects of moderate-fat (from monounsaturated fat) and low-fat weight-loss diets on the serum lipid profile in overweight and obese men and women. Am. J. Clin. Nutr. 2004, 79, 204–212. [Google Scholar] [CrossRef]
- Zheng, C.; Khoo, C.; Furtado, J.; Ikewaki, K.; Sacks, F.M. Dietary monounsaturated fat activates metabolic pathways for triglyceride-rich lipoproteins that involve apolipoproteins E and C-III. Am. J. Clin. Nutr. 2008, 88, 272–281. [Google Scholar] [CrossRef]
- Dentin, R.; Benhamed, F.; Pégorier, J.-P.; Foufelle, F.; Viollet, B.; Vaulont, S.; Girard, J.; Postic, C. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Investig. 2005, 115, 2843–2854. [Google Scholar] [CrossRef]
- Iizuka, K. Protein Amount, Quality, and Physical Activity. Nutrients 2021, 13, 3720. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiao, J.-G.; Gao, S.; Ning, L.-J.; Limbu, S.M.; Qiao, F.; Chen, L.-Q.; Zhang, M.-L.; Du, Z.-Y. Dietary oils modify lipid molecules and nutritional value of fillet in Nile tilapia: A deep lipidomics analysis. Food Chem. 2019, 277, 515–523. [Google Scholar] [CrossRef]
- Chilton, F.H.; Dutta, R.; Reynolds, L.M.; Sergeant, S.; Mathias, R.A.; Seeds, M.C. Precision Nutrition and Omega-3 Polyunsaturated Fatty Acids: A Case for Personalized Supplementation Approaches for the Prevention and Management of Human Diseases. Nutrients 2017, 9, 1165. [Google Scholar] [CrossRef] [PubMed]
- Li, M.V.; Chang, B.; Imamura, M.; Poungvarin, N.; Chan, L. Glucose-Dependent Transcriptional Regulation by an Evolutionarily Conserved Glucose-Sensing Module. Diabetes 2006, 55, 1179–1189. [Google Scholar] [CrossRef] [PubMed]



| Variable | Total n = 587 | Women n = 415 | Men n = 172 | p-Value |
|---|---|---|---|---|
| Age (y) Age range | 40.93 ± 11.86 18–65 | 41.69 ± 11.71 18–65 | 39.14 ± 12.04 18–65 | 0.001 |
| Anthropometric variables | ||||
| BMI (kg/m2) | 30.78 ± 6.77 | 31.38 ± 6.88 | 29.36 ± 6.29 | 0.001 |
| Waist circumference (cm) | 96.12 ± 16.38 | 95.23 ± 16.09 | 98.21 ± 16.93 | 0.043 |
| Hip circumference (cm) | 109.44 ± 13.63 | 111.24 ± 14.17 | 105.17 ± 11.22 | <0.001 |
| WHR | 0.86 ± 0.08 | 0.85 ± 0.08 | 0.91 ± 0.08 | <0.001 |
| Fat mass (g) | 31.1 ± 14.57 | 33.49 ± 13.93 | 25.62 ± 14.56 | <0.001 |
| Body fat percentage (%) | 36.55 ± 10.67 | 40.29 ± 8.71 | 27.77 ± 9.65 | <0.001 |
| Biochemical variables | ||||
| Glucose (mg/dL) | 93.96 ± 18.22 | 93.34 ± 16.8 | 95.44 ± 21.21 | 0.196 |
| Insulin (μU/mL) | 12.74 ± 12.81 | 13.6 ± 12.86 | 10.6 ± 12.47 | 0.001 |
| HOMA-IR | 2.99 ± 3.26 | 3.19 ± 3.36 | 2.48 ± 2.94 | 0.001 |
| Triglycerides (mg/dL) | 141.54 ± 72.54 | 138.56 ± 73.34 | 148.62 ± 70.3 | 0.075 |
| TC (mg/dL) | 187.64 ± 41.47 | 187.73 ± 42.32 | 187.42 ± 39.5 | 0.935 |
| VLDL-c (mg/dL) | 28.3 ± 14.5 | 27.71 ± 14.66 | 29.72 ± 14.06 | 0.123 |
| LDL-c (mg/dL) | 115.91 ± 37.67 | 115.65 ± 38.69 | 116.5 ± 35.23 | 0.803 |
| HDL-c (mg/dL) | 43.42 ± 13.6 | 44.36 ± 14 | 41.19 ± 12.35 | 0.010 |
| Non-HDL-c (mg/dL) | 144.22 ± 41.87 | 143.37 ± 42.39 | 146.23 ± 40.68 | 0.448 |
| Albumin (g/dL) | 4.29 ± 0.48 | 4.19 ± 0.43 | 4.52 ± 0.50 | <0.001 |
| Variable | Total n = 587 | Women n = 415 | Men n = 172 | p-Value |
|---|---|---|---|---|
| energy (kcal) | 2025.0 ± 842.2 | 1866.7 ± 715.2 | 2390.4 ± 989.5 | <0.001 |
| Carbohydrates (kcal) | 1000.6 ± 436.5 | 923.5 ± 377.9 | 1178.5 ± 506.4 | <0.001 |
| Carbohydrates (%) | 49.68 ± 9.32 | 49.76 ± 9.29 | 49.51 ± 9.40 | 0.768 |
| Proteins (kcal) | 350.73 ± 146.99 | 322.27 ± 124.29 | 416.58 ± 172.58 | <0.001 |
| Proteins (%) | 17.81 ± 4.56 | 17.74 ± 4.73 | 17.98 ± 4.15 | 0.570 |
| Fat (kcal) | 672.8 ± 361.58 | 619.83 ± 317.46 | 795.37 ± 423.58 | <0.001 |
| Fat (%) | 32.44 ± 8.68 | 32.41 ± 8.74 | 32.50 ± 8.56 | 0.903 |
| Saturated fat (g) | 25.03 ± 14.78 | 22.98 ± 12.96 | 29.76 ± 17.45 | <0.001 |
| Monounsaturated fat (g) | 22.89 ± 13.23 | 21.03 ± 11.40 | 27.18 ± 15.95 | <0.001 |
| Polyunsaturated fat (g) | 11.9 ± 8.62 | 11.4 ± 7.94 | 13.06 ± 9.95 | 0.034 |
| SFA 6:0 (g) | 0.26 ± 0.28 | 0.24 ± 0.25 | 0.31 ± 0.33 | 0.025 |
| SFA 8:0 (g) | 0.22 ± 0.23 | 0.2 ± 0.21 | 0.26 ± 0.27 | 0.018 |
| SFA 10:0 (g) | 0.38 ± 0.36 | 0.35 ± 0.33 | 0.44 ± 0.40 | 0.021 |
| SFA 14:0 (g) | 1.87 ± 1.50 | 1.73 ± 1.38 | 2.19 ± 1.72 | 0.002 |
| SFA 16:0 (g) | 11.91 ± 7.19 | 10.91 ± 6.18 | 14.21 ± 8.70 | <0.001 |
| SFA 18:0 (g) | 5.19 ± 3.40 | 4.75 ± 2.97 | 6.21 ± 4.05 | <0.001 |
| MUFA 14:1 (g) | 0.06 ± 0.08 | 0.06 ± 0.07 | 0.08 ± 0.09 | 0.006 |
| MUFA 16:1 (g) | 1.14 ± 0.88 | 1.04 ± 0.72 | 1.39 ± 1.12 | <0.001 |
| MUFA 18:1 (g) | 20.05 ± 12.04 | 18.37 ± 10.25 | 23.94 ± 14.71 | <0.001 |
| PUFA 18:2 (g) | 9.96 ± 7.83 | 9.53 ± 7.08 | 10.94 ± 9.27 | 0.049 |
| PUFA 18:3 (g) | 0.95 ± 0.87 | 0.9 ± 0.93 | 1.04 ± 0.72 | 0.087 |
| PUFA 20:4 (g) | 0.14 ± 0.12 | 0.13 ± 0.11 | 0.16 ± 0.13 | 0.005 |
| Total n-3 (g) | 1.05 ± 0.97 | 1.01 ± 1.05 | 1.15 ± 0.75 | 0.114 |
| Total n-6 (g) | 10.1 ± 7.86 | 9.66 ± 7.11 | 11.1 ± 9.3 | 0.045 |
| n-3:n-6 ratio | 11.05 ± 7.13 | 11.03 ± 6.36 | 11.1 ± 8.68 | 0.916 |
| Dietary cholesterol (mg) | 293.08 ± 212.82 | 268.72 ± 182.81 | 349.44 ± 261.8 | <0.001 |
| Dietary fiber (g) | 25.74 ± 13.27 | 24.1 ± 12.09 | 29.52 ± 15.03 | <0.001 |
| Soluble fiber (g) | 0.53 ± 0.86 | 0.46 ± 0.79 | 0.68 ± 0.98 | 0.008 |
| Insoluble fiber (g) | 1.18 ± 1.95 | 1.01 ± 1.79 | 1.58 ± 2.22 | 0.004 |
| Crude fiber (g) | 4.73 ± 13.24 | 4.44 ± 12.91 | 5.4 ± 13.98 | 0.426 |
| Total sugar (g) | 76.67 ± 42.02 | 73.34 ± 40.04 | 84.47 ± 45.49 | 0.006 |
| Added sugars (g) | 21.02 ± 19.49 | 21.71 ± 20.84 | 19.49 ± 16.08 | 0.329 |
| Glucose (g) | 8.08 ± 7.25 | 8.02 ± 7.07 | 8.23 ± 7.65 | 0.753 |
| Galactose (g) | 0.42 ± 3.57 | 0.42 ± 3.68 | 0.42 ± 3.29 | 0.998 |
| Fructose (g) | 10.82 ± 10.35 | 10.56 ± 9.76 | 11.43 ± 11.61 | 0.393 |
| Sucrose (g) | 19.32 ± 19.85 | 19.51 ± 21.13 | 18.9 ± 16.58 | 0.740 |
| Lactose (g) | 10.7 ± 12.26 | 9.25 ± 10.45 | 14 ± 15.16 | <0.001 |
| Total n = 587 n (Proportion) | Women n = 415 n (Proportion) | Men n = 172 n (Proportion) | p-Value | |
|---|---|---|---|---|
| SNV rs17145750 C > T | ||||
| Genotype | ||||
| CC | 490 (0.834) | 342 (0.823) | 148 (0.861) | 0.329 |
| CT | 92 (0.157) | 71 (0.170) | 22 (0.127) | 0.215 |
| TT | 5 (0.009) | 2 (0.007) | 2 (0.012) | 0.585 |
| p (HWE) | 0.767 | 0.407 | 0.266 | |
| Allele | ||||
| C | 1072 (0.91) | 755 (0.91) | 318 (0.92) | 0.493 |
| T | 102 (0.09) | 75 (0.09) | 26 (0.08) | 0.493 |
| Dominant model | ||||
| CC | 490 (0.834) | 342 (0.823) | 148 (0.861) | |
| CT + TT | 97 (0.166) | 73 (0.177) | 24 (0.139) | |
| SNV rs3812316 C > G | ||||
| Genotype | ||||
| CC | 508 (0.865) | 357 (0.861) | 151 (0.878) | 0.598 |
| CG | 78 (0.133) | 57 (0.137) | 21 (0.122) | 0.689 |
| GG | 1 (0.002) | 1 (0.002) | 0 (0) | 1.000 |
| p (HWE) | 0.262 | 0.414 | 0.393 | |
| Allele | ||||
| C | 1094 (0.93) | 771 (0.93) | 323 (0.94) | 0.611 |
| G | 80 (0.07) | 59 (0.07) | 21 (0.06) | 0.611 |
| Dominant model | ||||
| CC | 508 (0.865) | 357 (0.860) | 151 (0.878) | |
| CG + GG | 79 (0.135) | 58 (0.140) | 21 (0.122) |
| B | Standard Error | 95% CI | p-Value | |
|---|---|---|---|---|
| WOMEN | ||||
| Model, R2 = 65.2 | ||||
| HDL-cholesterol (mg/dL) | −0.005 | 0.001 | −0.007–−0.004 | <0.001 |
| Total cholesterol (mg/dL) | 0.002 | 0.000 | 0.001–0.002 | <0.001 |
| Waist circumference (cm) | 0.003 | 0.001 | 0.002–0.004 | <0.001 |
| SNV rs3812316 C > G | −0.070 | 0.027 | −0.124–−0.016 | 0.011 |
| HOMA-IR | 0.007 | 0.003 | 0.001–0.013 | 0.015 |
| MEN | ||||
| Model, R2 = 79.4 | ||||
| HDL-cholesterol (mg/dL) | −0.007 | 0.001 | −0.010–−0.004 | <0.001 |
| Percentage of body fat (%) | 0.007 | 0.002 | 0.003–0.010 | <0.001 |
| Albumin (g) | 0.108 | 0.032 | 0.044–0.171 | 0.001 |
| Total dietary fiber (g) | −0.004 | 0.001 | −0.007–−0.001 | 0.005 |
| Total cholesterol (mg/dL) | 0.001 | 0.000 | 0.000–0.002 | 0.032 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maldonado-González, M.; Hernández-Nazara, Z.H.; Torres-Castillo, N.; Martínez-López, E.; de la Cruz-Color, L.; Ruíz-Madrigal, B. Association between the rs3812316 Single Nucleotide Variant of the MLXIPL Gene and Alpha-Linolenic Acid Intake with Triglycerides in Mexican Mestizo Women. Nutrients 2022, 14, 4726. https://doi.org/10.3390/nu14224726
Maldonado-González M, Hernández-Nazara ZH, Torres-Castillo N, Martínez-López E, de la Cruz-Color L, Ruíz-Madrigal B. Association between the rs3812316 Single Nucleotide Variant of the MLXIPL Gene and Alpha-Linolenic Acid Intake with Triglycerides in Mexican Mestizo Women. Nutrients. 2022; 14(22):4726. https://doi.org/10.3390/nu14224726
Chicago/Turabian StyleMaldonado-González, Montserrat, Zamira H. Hernández-Nazara, Nathaly Torres-Castillo, Erika Martínez-López, Lucia de la Cruz-Color, and Bertha Ruíz-Madrigal. 2022. "Association between the rs3812316 Single Nucleotide Variant of the MLXIPL Gene and Alpha-Linolenic Acid Intake with Triglycerides in Mexican Mestizo Women" Nutrients 14, no. 22: 4726. https://doi.org/10.3390/nu14224726
APA StyleMaldonado-González, M., Hernández-Nazara, Z. H., Torres-Castillo, N., Martínez-López, E., de la Cruz-Color, L., & Ruíz-Madrigal, B. (2022). Association between the rs3812316 Single Nucleotide Variant of the MLXIPL Gene and Alpha-Linolenic Acid Intake with Triglycerides in Mexican Mestizo Women. Nutrients, 14(22), 4726. https://doi.org/10.3390/nu14224726





