Ginger Root Extract Improves GI Health in Diabetic Rats by Improving Intestinal Integrity and Mitochondrial Function
Abstract
1. Introduction
2. Materials and Methods
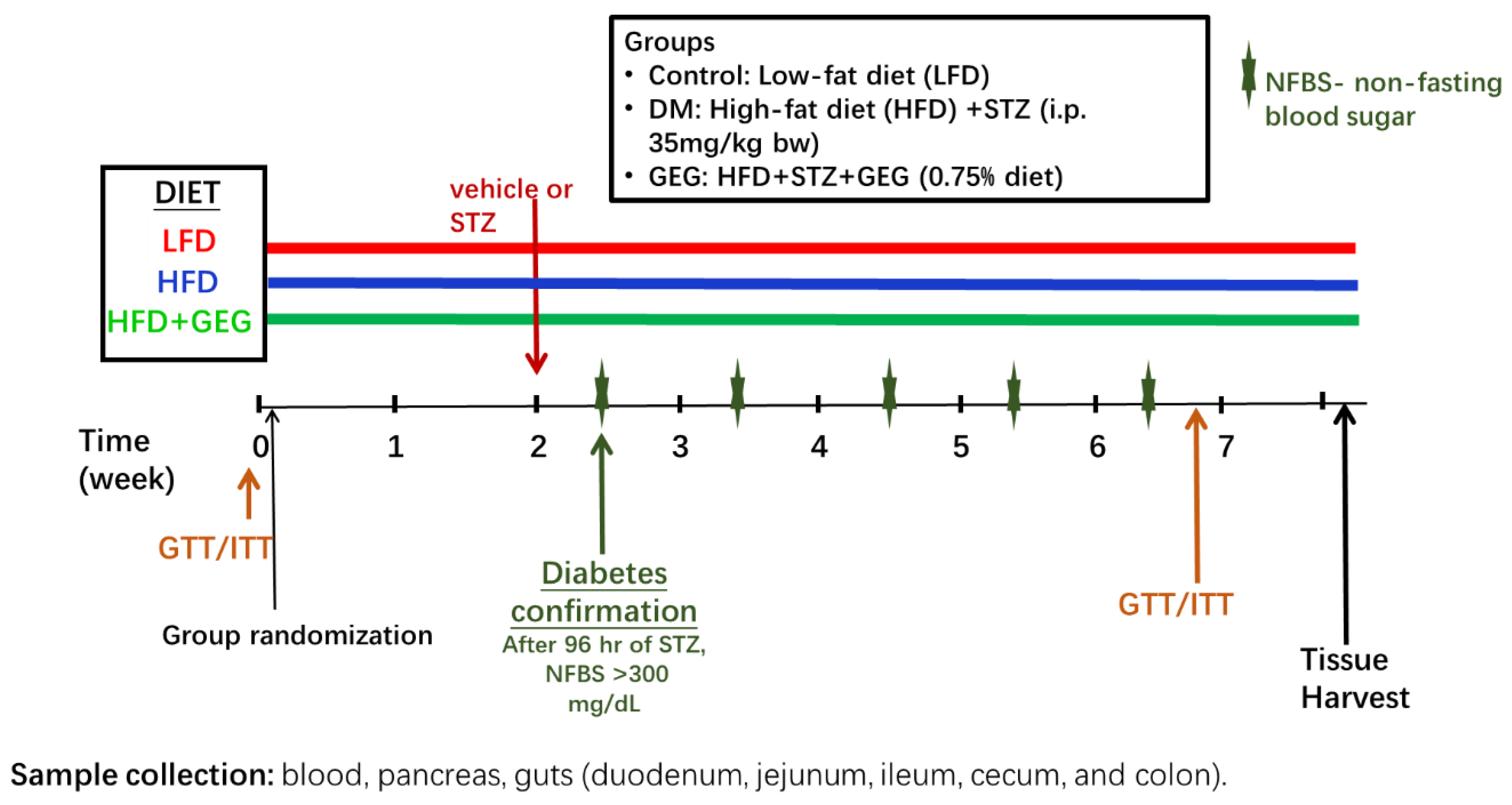
2.1. Animals and Treatments
2.2. In Vivo Glucose and Insulin Tolerance Tests
2.3. Sample Collection
2.4. Insulin Measurement
2.5. Analysis of Pancreatic Tissue
2.6. RNA Isolation and qRT-PCR
2.7. Immunohistochemistry for Claudin-3 and PINK1 in the Colon
2.8. Statistical Analysis
3. Results
3.1. Glucose Homeostasis: Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
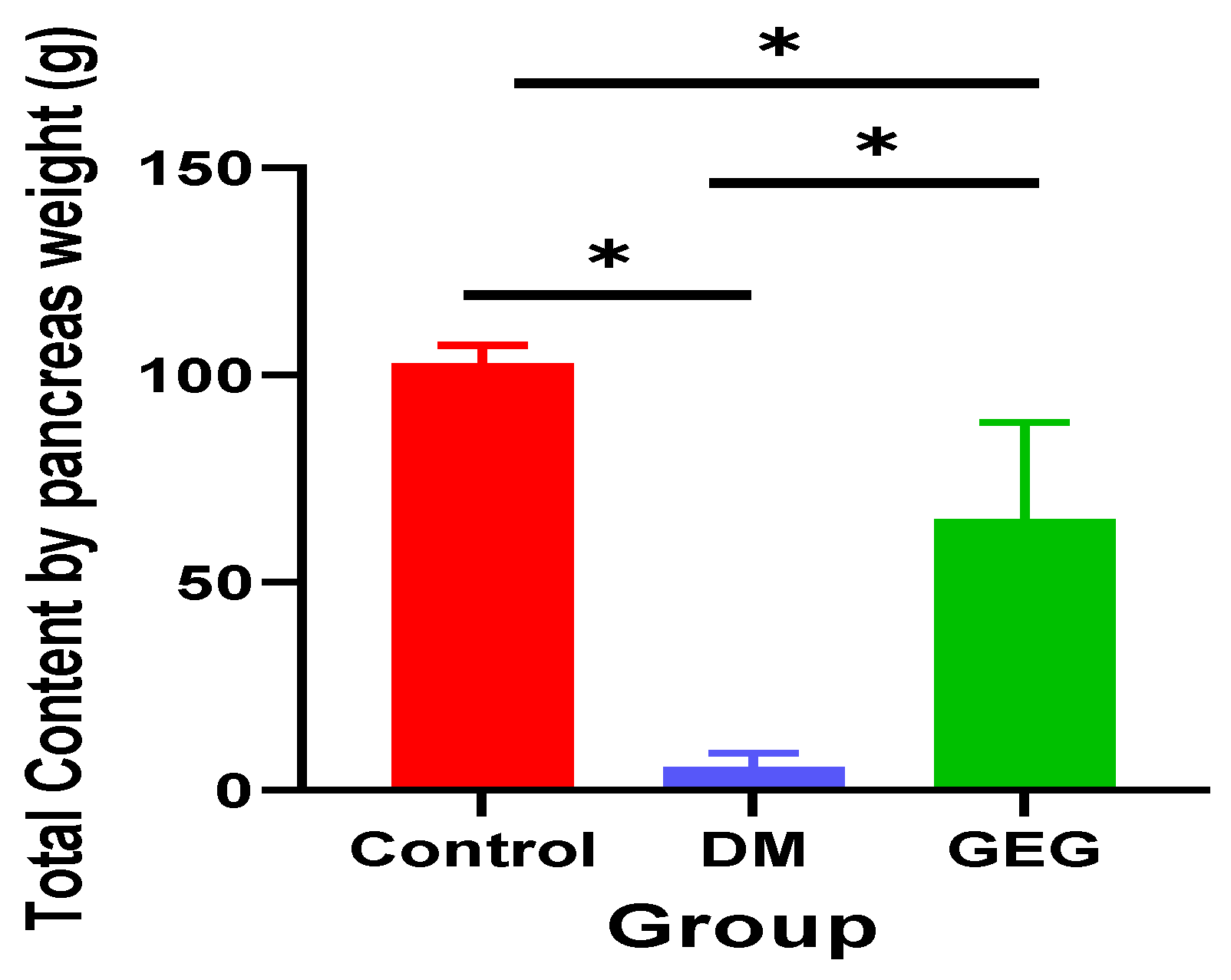
3.2. Insulin and Histological Assessment of Pancreatic Tissue
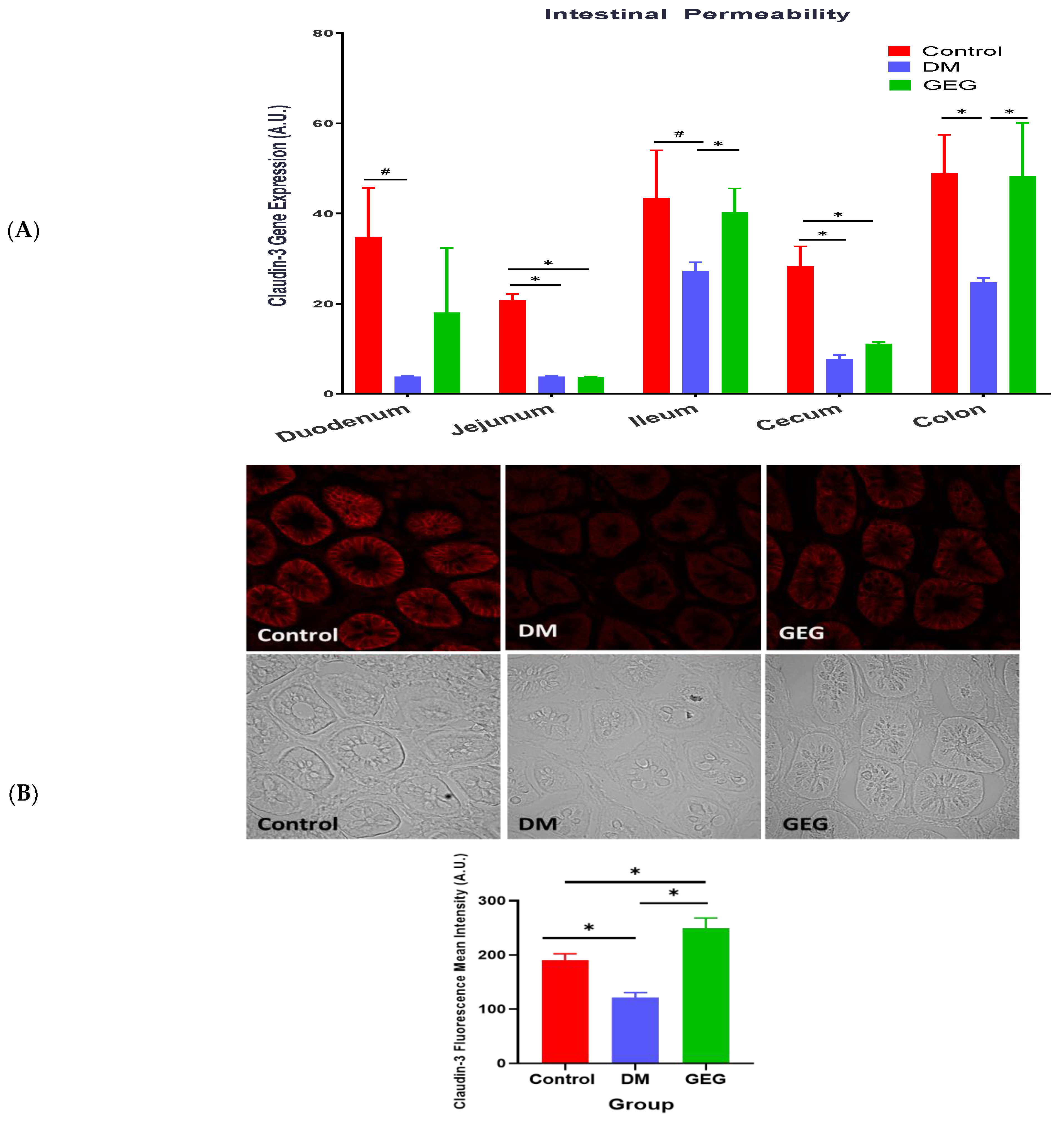
3.3. mRNA Expression of Intestinal Tight Junction Marker Assessment
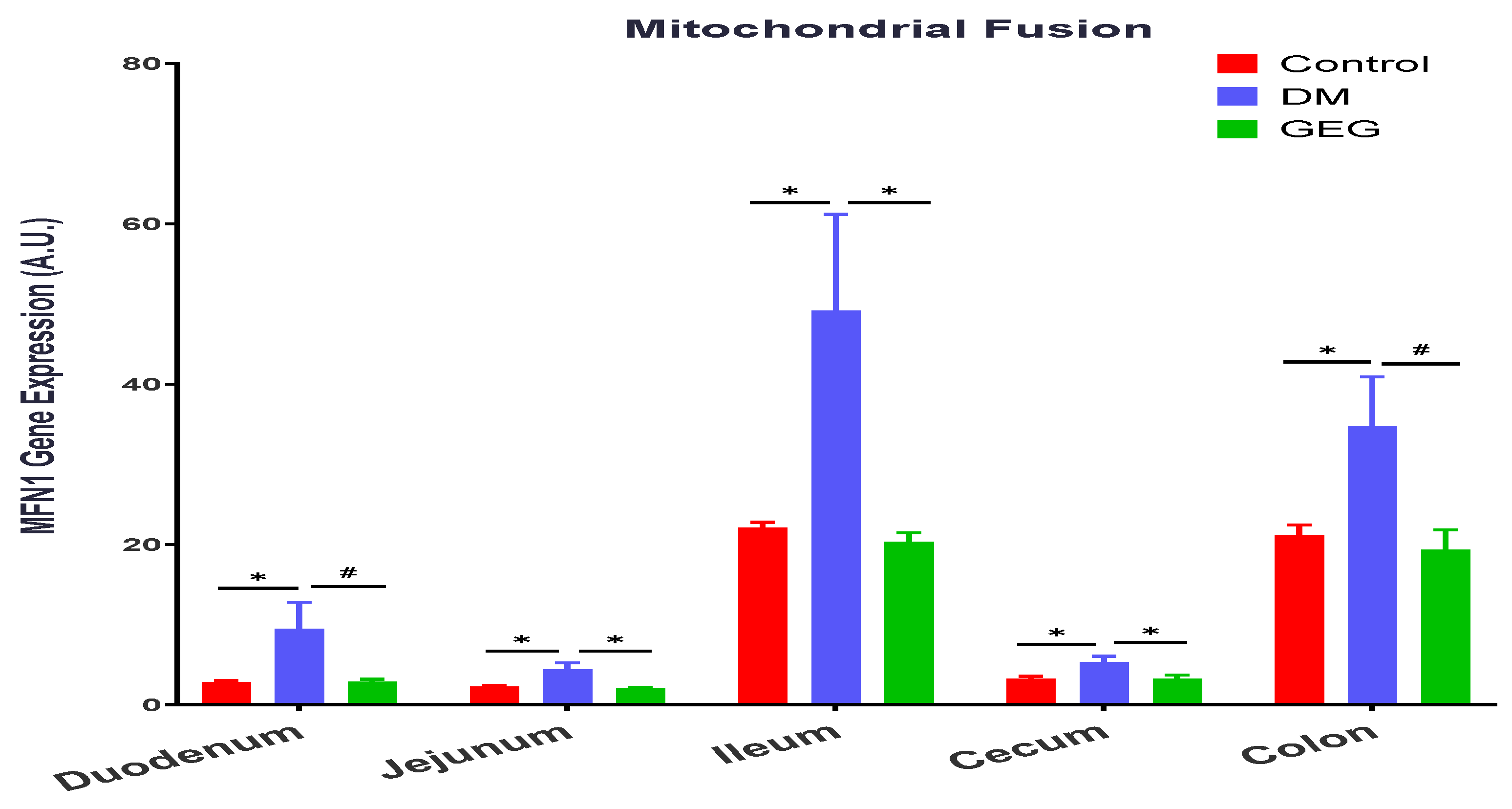
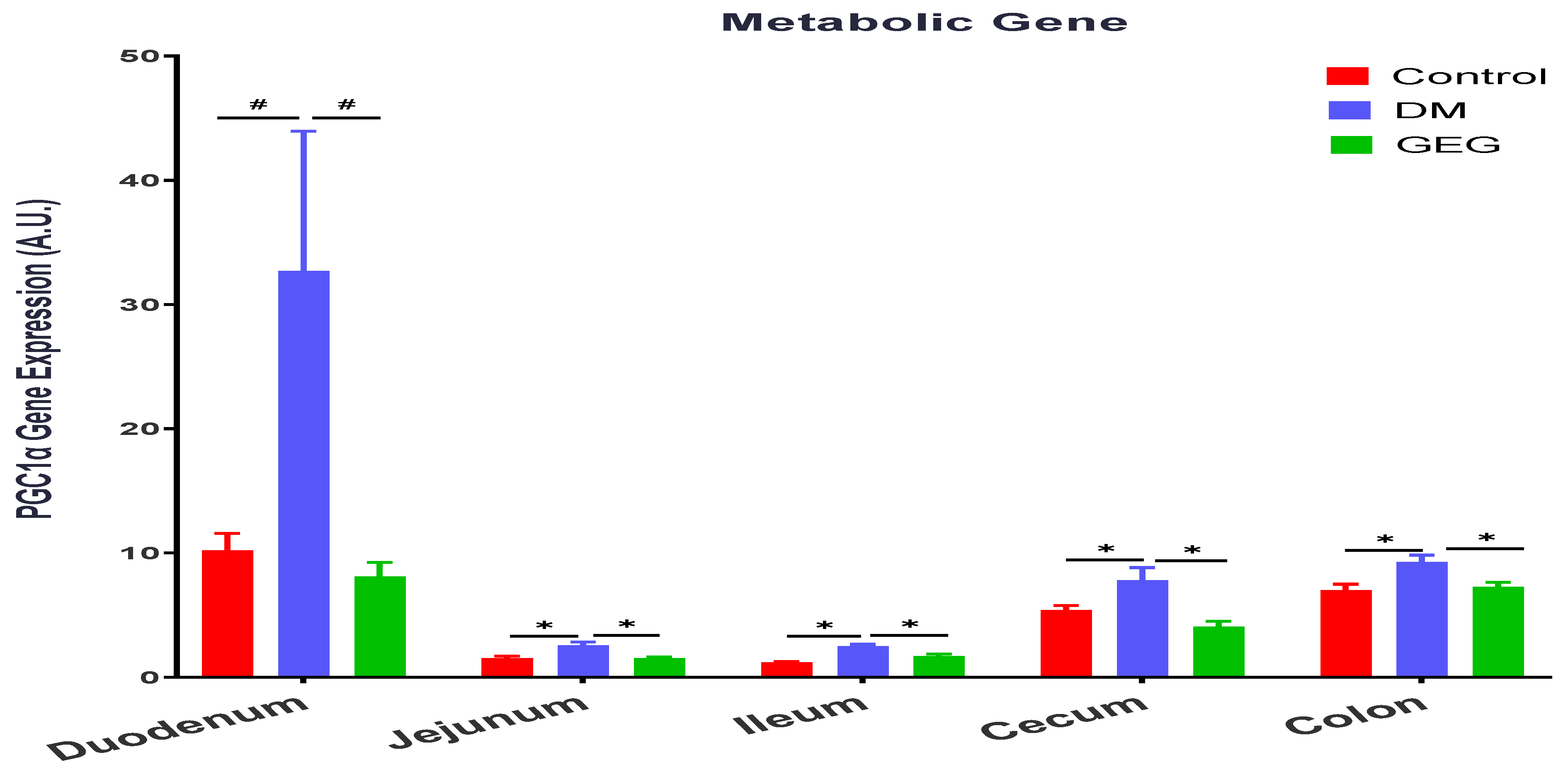
3.4. mRNA Expression of the Intestinal Mitochondrial Fusion, Fission, and Biogenesis Markers
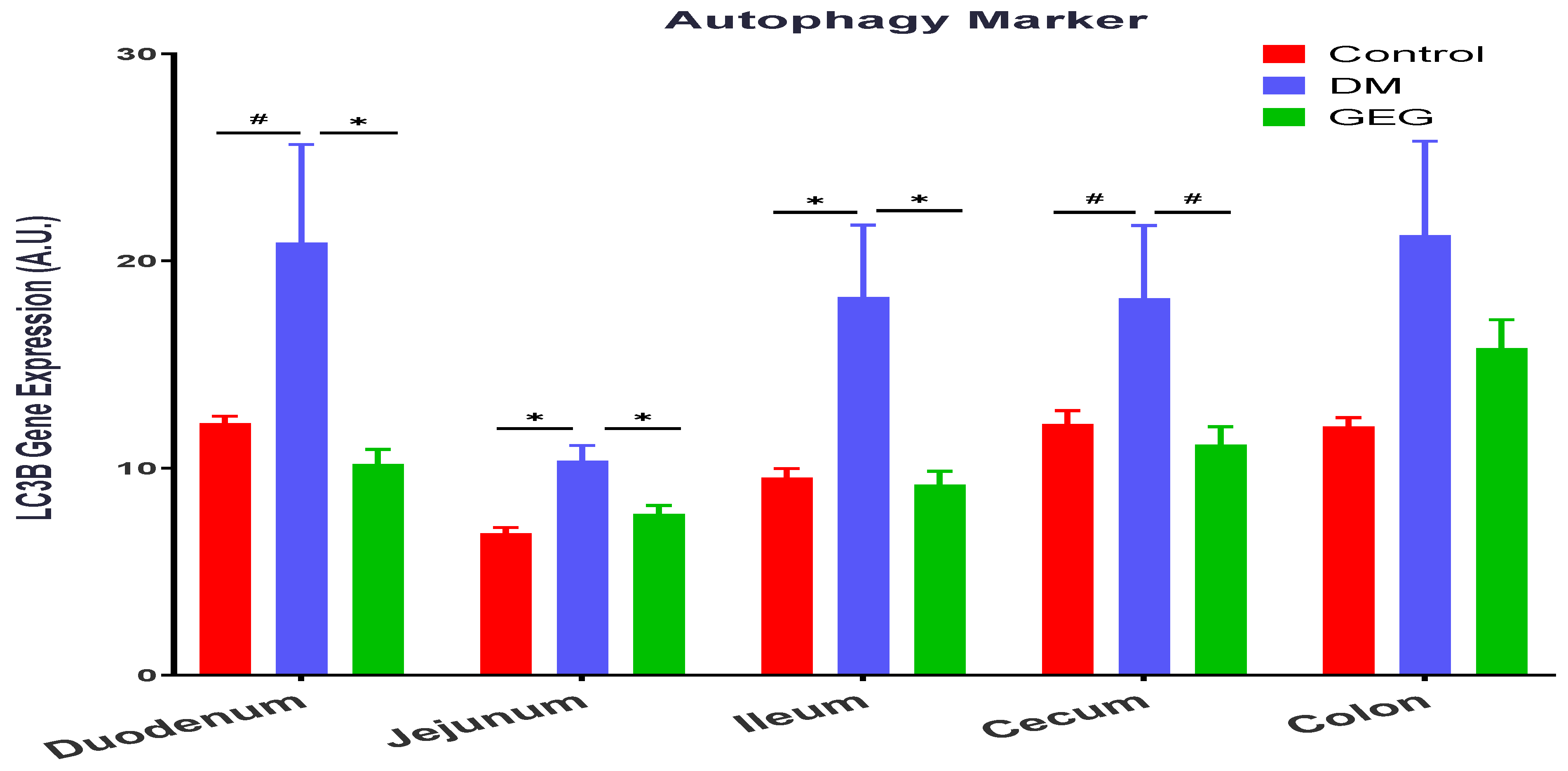
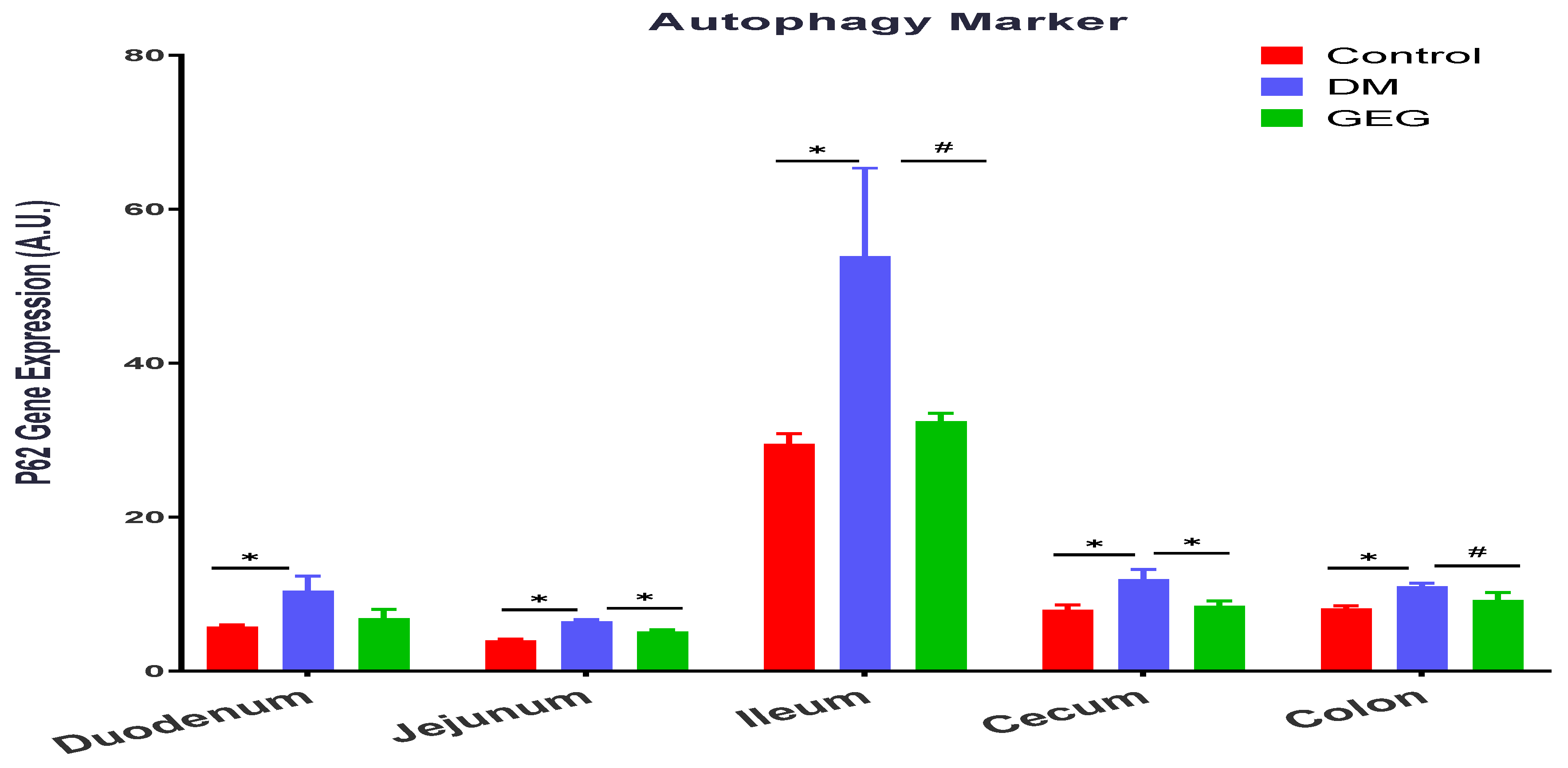
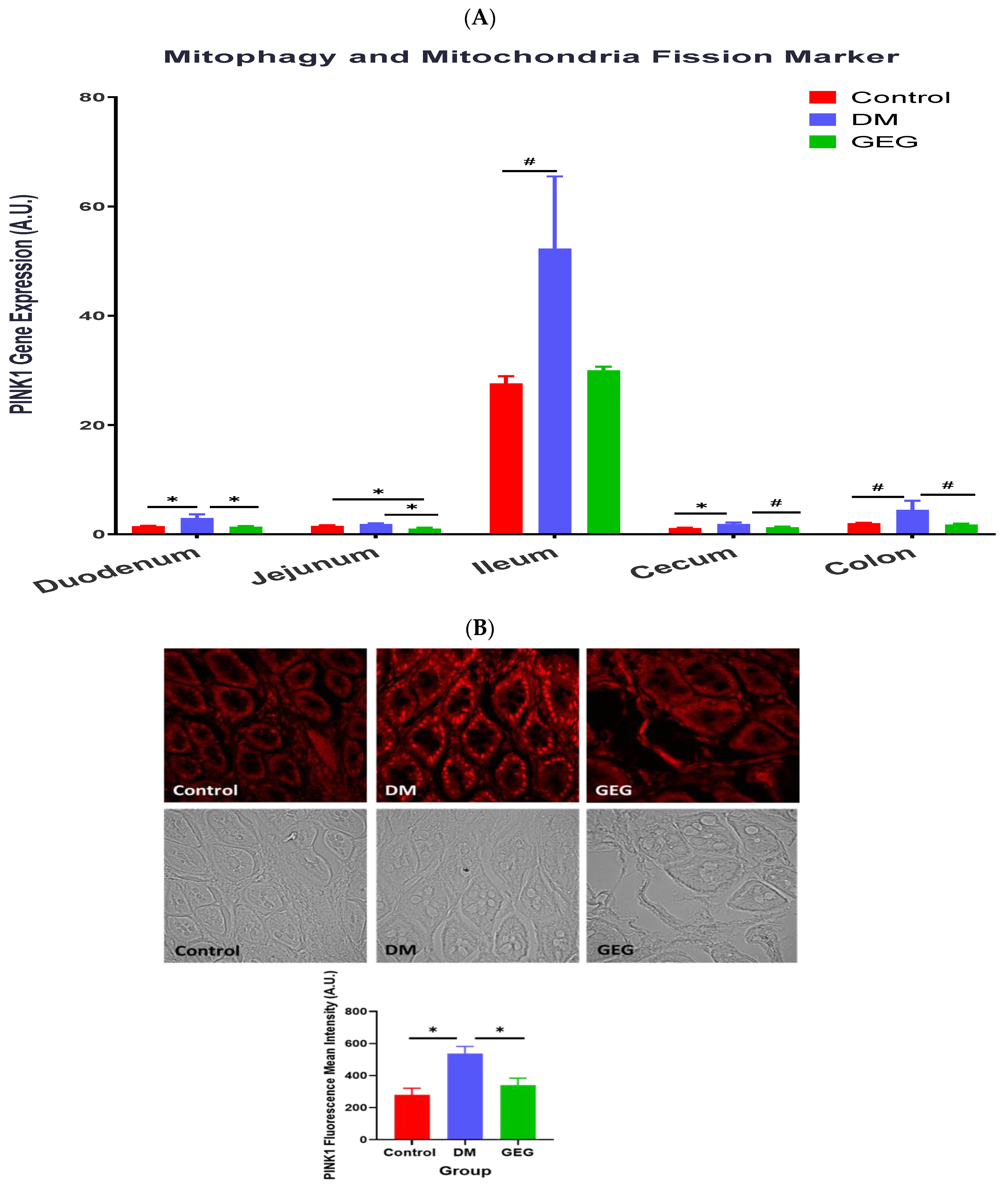
3.5. mRNA Expression of the Intestinal Mitochondrial Mitophagy Markers Assessment
3.6. mRNA Expression of Intestinal Oxidative Stress and Inflammation Markers Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinti, M.V.; Fink, G.K.; Hatha way, Q.A.; Durr, A.J.; Kunovac, A.; Hollander, J.M. Mitochondrial dysfunction in type 2 diabetes mellitus: An organ-based analysis. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E268–E285. [Google Scholar] [CrossRef]
- Rovira-Llopis, S.; Banuls, C.; Diaz-Morales, N.; Hernandez-Mijares, A.; Rocha, M.; Victor, V.M. Mitochondrial dynamics in type 2 diabetes: Pathophysiological implications. Redox Biol. 2017, 11, 637–645. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Grosheva, I.; Zheng, D.; Soffer, E.; Blacher, E.; Braverman, S.; Tengeler, A.C.; Barak, O.; Elazar, M.; et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 2018, 359, 1376–1383. [Google Scholar] [CrossRef]
- Salinas, E.; Reyes-Pavon, D.; Cortes-Perez, N.G.; Torres-Maravilla, E.; Bitzer-Quintero, O.K.; Langella, P.; Bermudez-Humaran, L.G. Bioactive Compounds in Food as a Current Therapeutic Approach to Maintain a Healthy Intestinal Epithelium. Microorganisms 2021, 9, 1634. [Google Scholar] [CrossRef]
- Rocha, M.; Apostolova, N.; Diaz-Rua, R.; Muntane, J.; Victor, V.M. Mitochondria and T2D: Role of Autophagy, ER Stress, and Inflammasome. Trends Endocrinol. Metab. 2020, 31, 725–741. [Google Scholar] [CrossRef]
- Burgos-Moron, E.; Abad-Jimenez, Z.; Maranon, A.M.; Iannantuoni, F.; Escribano-Lopez, I.; Lopez-Domenech, S.; Salom, C.; Jover, A.; Mora, V.; Roldan, I.; et al. Relationship Between Oxidative Stress, ER Stress, and Inflammation in Type 2 Diabetes: The Battle Continues. J. Clin. Med. 2019, 8, 1385. [Google Scholar] [CrossRef]
- Shan, Z.; Fa, W.H.; Tian, C.R.; Yuan, C.S.; Jie, N. Mitophagy and mitochondrial dynamics in type 2 diabetes mellitus treatment. Aging 2022, 14, 2902–2919. [Google Scholar] [CrossRef]
- Shen, C.L.; Wang, R.; Yakhnitsa, V.; Santos, J.M.; Watson, C.; Kiritoshi, T.; Ji, G.; Hamood, A.N.; Neugebauer, V. Gingerol-Enriched Ginger Supplementation Mitigates Neuropathic Pain via Mitigating Intestinal Permeability and Neuroinflammation: Gut-Brain Connection. Front. Pharmacol. 2022, 13, 912609. [Google Scholar] [CrossRef]
- Shen, C.L.; Wang, R.; Ji, G.; Elmassry, M.M.; Zabet-Moghaddam, M.; Vellers, H.; Hamood, A.N.; Gong, X.; Mirzaei, P.; Sang, S.; et al. Dietary supplementation of gingerols- and shogaols-enriched ginger root extract attenuate pain-associated behaviors while modulating gut microbiota and metabolites in rats with spinal nerve ligation. J. Nutr. Biochem. 2022, 100, 108904. [Google Scholar] [CrossRef]
- Vasquez-Reyes, S.; Velazquez-Villegas, L.A.; Vargas-Castillo, A.; Noriega, L.G.; Torres, N.; Tovar, A.R. Dietary bioactive compounds as modulators of mitochondrial function. J. Nutr. Biochem. 2021, 96, 108768. [Google Scholar] [CrossRef]
- Al-Amin, Z.M.; Thomson, M.; Al-Qattan, K.K.; Peltonen-Shalaby, R.; Ali, M. Anti-diabetic and hypolipidaemic properties of ginger (Zingiber officinale) in streptozotocin-induced diabetic rats. Br. J. Nutr. 2006, 96, 660–666. [Google Scholar] [CrossRef]
- Rehman, M.U.; Rashid, S.M.; Rasool, S.; Shakeel, S.; Ahmad, B.; Ahmad, S.B.; Madkhali, H.; Ganaie, M.A.; Majid, S.; Bhat, S.A. Zingerone (4-(4-hydroxy-3-methylphenyl)butan-2-one) ameliorates renal function via controlling oxidative burst and inflammation in experimental diabetic nephropathy. Arch. Physiol. Biochem. 2019, 125, 201–209. [Google Scholar] [CrossRef]
- Saravanan, N.; Patil, M.A.; Kumar, P.U.; Suryanarayana, P.; Reddy, G.B. Dietary ginger improves glucose dysregulation in a long-term high-fat high-fructose fed prediabetic rat model. Indian J. Exp. Biol. 2017, 55, 142–150. [Google Scholar]
- Li, Y.; Tran, V.H.; Kota, B.P.; Nammi, S.; Duke, C.C.; Roufogalis, B.D. Preventative effect of Zingiber officinale on insulin resistance in a high-fat high-carbohydrate diet-fed rat model and its mechanism of action. Basic Clin. Pharmacol. Toxicol. 2014, 115, 209–215. [Google Scholar] [CrossRef]
- Li, X.H.; McGrath, K.C.; Nammi, S.; Heather, A.K.; Roufogalis, B.D. Attenuation of liver pro-inflammatory responses by Zingiber officinale via inhibition of NF-kappa B activation in high-fat diet-fed rats. Basic Clin. Pharmacol. Toxicol. 2012, 110, 238–244. [Google Scholar] [CrossRef]
- Mansour, D.F.; Abdallah, H.M.I.; Ibrahim, B.M.M.; Hegazy, R.R.; Esmail, R.S.E.; Abdel-Salam, L.O. The Carcinogenic Agent Diethylnitrosamine Induces Early Oxidative Stress, Inflammation and Proliferation in Rat Liver, Stomach and Colon: Protective Effect of Ginger Extract. Asian Pac. J. Cancer Prev. 2019, 20, 2551–2561. [Google Scholar] [CrossRef]
- Azizidoost, S.; Nazeri, Z.; Mohammadi, A.; Mohammadzadeh, G.; Cheraghzadeh, M.; Jafari, A.; Kheirollah, A. Effect of Hydroalcoholic Ginger Extract on Brain HMG-CoA Reductase and CYP46A1 Levels in Streptozotocin-induced Diabetic Rats. Avicenna J. Med. Biotechnol. 2019, 11, 234–238. [Google Scholar]
- Marefati, N.; Abdi, T.; Beheshti, F.; Vafaee, F.; Mahmoudabady, M.; Hosseini, M. Zingiber officinale (Ginger) hydroalcoholic extract improved avoidance memory in rat model of streptozotocin-induced diabetes by regulating brain oxidative stress. Horm. Mol. Biol. Clin. Investig. 2021, 43, 15–26. [Google Scholar] [CrossRef]
- Dufour, J.M.; Rajotte, R.V.; Zimmerman, M.; Rezania, A.; Kin, T.; Dixon, D.E.; Korbutt, G.S. Development of an ectopic site for islet transplantation, using biodegradable scaffolds. Tissue Eng. 2005, 11, 1323–1331. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Nascimento, J.C.; Matheus, V.A.; Oliveira, R.B.; Tada, S.F.S.; Collares-Buzato, C.B. High-Fat Diet Induces Disruption of the Tight Junction-Mediated Paracellular Barrier in the Proximal Small Intestine Before the Onset of Type 2 Diabetes and Endotoxemia. Dig. Dis. Sci. 2021, 66, 3359–3374. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, H.M.; Mao, Q.Y.; Cong, X.; Zhang, Y.; Lee, S.W.; Park, K.; Wu, L.L.; Xiang, R.L.; Yu, G.Y. High Glucose Reduces the Paracellular Permeability of the Submandibular Gland Epithelium via the MiR-22-3p/Sp1/Claudin Pathway. Cells 2021, 10, 3230. [Google Scholar] [CrossRef]
- Huang, Y.; Mao, Q.Y.; Shi, X.J.; Cong, X.; Zhang, Y.; Wu, L.L.; Yu, G.Y.; Xiang, R.L. Disruption of tight junctions contributes to hyposalivation of salivary glands in a mouse model of type 2 diabetes. J. Anat. 2020, 237, 556–567. [Google Scholar] [CrossRef]
- Haniadka, R.; Saldanha, E.; Sunita, V.; Palatty, P.L.; Fayad, R.; Baliga, M.S. A review of the gastroprotective effects of ginger (Zingiber officinale Roscoe). Food Funct. 2013, 4, 845–855. [Google Scholar] [CrossRef]
- Nikkhah Bodagh, M.; Maleki, I.; Hekmatdoost, A. Ginger in gastrointestinal disorders: A systematic review of clinical trials. Food Sci. Nutr. 2019, 7, 96–108. [Google Scholar] [CrossRef]
- Hua, S. Advances in Oral Drug Delivery for Regional Targeting in the Gastrointestinal Tract—Influence of Physiological, Pathophysiological and Pharmaceutical Factors. Front Pharmacol. 2020, 11, 524. [Google Scholar] [CrossRef]
- Twig, G.; Elorza, A.; Molina, A.J.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072. [Google Scholar] [CrossRef]
- Wada, J.; Nakatsuka, A. Mitochondrial Dynamics and Mitochondrial Dysfunction in Diabetes. Acta Med. Okayama 2016, 70, 151–158. [Google Scholar] [CrossRef]
- Szendroedi, J.; Phielix, E.; Roden, M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2011, 8, 92–103. [Google Scholar] [CrossRef]
- Williams, M.; Caino, M.C. Mitochondrial Dynamics in Type 2 Diabetes and Cancer. Front. Endocrinol. 2018, 9, 211. [Google Scholar] [CrossRef]
- Kabra, U.D.; Pfuhlmann, K.; Migliorini, A.; Keipert, S.; Lamp, D.; Korsgren, O.; Gegg, M.; Woods, S.C.; Pfluger, P.T.; Lickert, H.; et al. Direct Substrate Delivery Into Mitochondrial Fission-Deficient Pancreatic Islets Rescues Insulin Secretion. Diabetes 2017, 66, 1247–1257. [Google Scholar] [CrossRef]
- Whitaker, R.M.; Corum, D.; Beeson, C.C.; Schnellmann, R.G. Mitochondrial Biogenesis as a Pharmacological Target: A New Approach to Acute and Chronic Diseases. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 229–249. [Google Scholar] [CrossRef]
- Wu, H.; Deng, X.; Shi, Y.; Su, Y.; Wei, J.; Duan, H. PGC-1alpha, glucose metabolism and type 2 diabetes mellitus. J. Endocrinol. 2016, 229, R99–R115. [Google Scholar] [CrossRef]
- Besseiche, A.; Riveline, J.P.; Gautier, J.F.; Breant, B.; Blondeau, B. Metabolic roles of PGC-1alpha and its implications for type 2 diabetes. Diabetes Metab. 2015, 41, 347–357. [Google Scholar] [CrossRef]
- Hancock, C.R.; Han, D.H.; Higashida, K.; Kim, S.H.; Holloszy, J.O. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. Faseb. J. 2011, 25, 785–791. [Google Scholar] [CrossRef]
- Turner, N.; Bruce, C.R.; Beale, S.M.; Hoehn, K.L.; So, T.; Rolph, M.S.; Cooney, G.J. Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: Evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes 2007, 56, 2085–2092. [Google Scholar] [CrossRef]
- Garcia-Roves, P.; Huss, J.M.; Han, D.H.; Hancock, C.R.; Iglesias-Gutierrez, E.; Chen, M.; Holloszy, J.O. Raising plasma fatty acid concentration induces increased biogenesis of mitochondria in skeletal muscle. Proc. Natl. Acad. Sci. USA 2007, 104, 10709–10713. [Google Scholar] [CrossRef]
- Kang, I.; Chu, C.T.; Kaufman, B.A. The mitochondrial transcription factor TFAM in neurodegeneration: Emerging evidence and mechanisms. FEBS Lett. 2018, 592, 793–811. [Google Scholar] [CrossRef]
- Sergi, D.; Naumovski, N.; Heilbronn, L.K.; Abeywardena, M.; O’Callaghan, N.; Lionetti, L.; Luscombe-Marsh, N. Mitochondrial (Dys)function and Insulin Resistance: From Pathophysiological Molecular Mechanisms to the Impact of Diet. Front. Physiol. 2019, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Sireesh, D.; Dhamodharan, U.; Ezhilarasi, K.; Vijay, V.; Ramkumar, K.M. Association of NF-E2 Related Factor 2 (Nrf2) and inflammatory cytokines in recent onset Type 2 Diabetes Mellitus. Sci. Rep. 2018, 8, 5126. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Dong, L.; Hu, X.; Feng, F.; Chen, F. 6-Gingerol, a Functional Polyphenol of Ginger, Promotes Browning through an AMPK-Dependent Pathway in 3T3-L1 Adipocytes. J. Agric. Food Chem. 2019, 67, 14056–14065. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhang, S.; Wu, J.; Sun, X.; Shen, Z.; Dong, J.; Huang, J. Promotion of Mitochondrial Biogenesis via Activation of AMPK-PGC1a Signaling Pathway by Ginger (Zingiber officinale Roscoe) Extract, and Its Major Active Component 6-Gingerol. J. Food. Sci. 2019, 84, 2101–2111. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Xu, C.; Zhang, X.; Long, J.; Rezaeian, A.H.; Liu, C.; Furth, M.E.; Kridel, S.; Pasche, B.; Bian, X.W.; et al. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat. Immunol. 2018, 19, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Pontrelli, P.; Oranger, A.; Barozzino, M.; Conserva, F.; Papale, M.; Gesualdo, L. The pathological role of the ubiquitination pathway in diabetic nephropathy. Minerva. Med. 2018, 109, 53–67. [Google Scholar] [CrossRef]
- Xiang, R.L.; Huang, Y.; Zhang, Y.; Cong, X.; Zhang, Z.J.; Wu, L.L.; Yu, G.Y. Type 2 diabetes-induced hyposalivation of the submandibular gland through PINK1/Parkin-mediated mitophagy. J. Cell Physiol. 2020, 235, 232–244. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Yuan, T.; Yang, T.; Chen, H.; Fu, D.; Hu, Y.; Wang, J.; Yuan, Q.; Yu, H.; Xu, W.; Xie, X. New insights into oxidative stress and inflammation during diabetes mellitus-accelerated atherosclerosis. Redox. Biol. 2019, 20, 247–260. [Google Scholar] [CrossRef]
- Shanmugam, K.R.; Mallikarjuna, K.; Reddy, K.S. Effect of alcohol on blood glucose and antioxidant enzymes in the liver and kidney of diabetic rats. Indian J. Pharmacol. 2011, 43, 330–335. [Google Scholar] [CrossRef]
- Ramudu, S.K.; Korivi, M.; Kesireddy, N.; Lee, L.C.; Cheng, I.S.; Kuo, C.H.; Kesireddy, S.R. Nephro-protective effects of a ginger extract on cytosolic and mitochondrial enzymes against streptozotocin (STZ)-induced diabetic complications in rats. Chin. J. Physiol. 2011, 54, 79–86. [Google Scholar] [CrossRef] [PubMed]






| Gene | Forward | Reverse |
|---|---|---|
| Claudin-3 | 5′-CCC AGC CTA CGG AGT TAC CC-3′ | 5′-TGC CGA TGA ATG CCG AAA CG-3′ |
| MFN1 | 5′-AGC TCG CTG TCA TTG GGG AG-3′ | 5′-TCC CTC CAC ACT CAG GAA GC-3′ |
| FIS1 | 5′-CTG CGG TGC AGG ATG AAA GAC-3′ | 5′-GGC GTA TTC AAA CTG CGT GCT-3′ |
| PGC-1α | 5′-CAG GAG CTG GAT GGC TTG GG-3′ | 5′-GGG CAA AGA GGC TGG TCC T-3′ |
| TFAM | 5′ -GCT TCC AGG GGG CTA AGG ATG-3′ | 5′-TCG CCC AAC TTC AGC CAT TT-3′ |
| P62 | 5′-CTG AGT CGG CTT CTG CTC CA-3′ | 5′-GCG GCT TCT CTT CCC TCC AT-3′ |
| LC3B | 5′-CAT GCC GTC CGA GAA GAC CT-3′ | 5′-CCG GAT GAG CCG GAC ATC TT-3′ |
| PINK1 | 5′ -TCG GCC TGT CAG GAG ATC CA-3′ | 5′-CAT TGC AGC CCT TGC CGA TG-3′ |
| SOD1 | 5′-AGG GCG TCA TTC ACT TCG AG-3′ | 5′-ACA TGC CTC TCT TCA TCC GCT-3′ |
| NF-kB | 5′-CCT CCA CCC CGA CGT ATT GC-3′ | 5′-GCC AAG GCC TGG TTT GAG AT-3′ |
| β-actin | 5′-ACA ACC TTC TTG CAG CTC CTC C-3′ | 5′-TGA CCC ATA CCC ACC ATC ACA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Santos, J.M.; Dufour, J.M.; Stephens, E.R.; Miranda, J.M.; Washburn, R.L.; Hibler, T.; Kaur, G.; Lin, D.; Shen, C.-L. Ginger Root Extract Improves GI Health in Diabetic Rats by Improving Intestinal Integrity and Mitochondrial Function. Nutrients 2022, 14, 4384. https://doi.org/10.3390/nu14204384
Wang R, Santos JM, Dufour JM, Stephens ER, Miranda JM, Washburn RL, Hibler T, Kaur G, Lin D, Shen C-L. Ginger Root Extract Improves GI Health in Diabetic Rats by Improving Intestinal Integrity and Mitochondrial Function. Nutrients. 2022; 14(20):4384. https://doi.org/10.3390/nu14204384
Chicago/Turabian StyleWang, Rui, Julianna Maria Santos, Jannette M. Dufour, Emily R. Stephens, Jonathan M. Miranda, Rachel L. Washburn, Taylor Hibler, Gurvinder Kaur, Dingbo Lin, and Chwan-Li Shen. 2022. "Ginger Root Extract Improves GI Health in Diabetic Rats by Improving Intestinal Integrity and Mitochondrial Function" Nutrients 14, no. 20: 4384. https://doi.org/10.3390/nu14204384
APA StyleWang, R., Santos, J. M., Dufour, J. M., Stephens, E. R., Miranda, J. M., Washburn, R. L., Hibler, T., Kaur, G., Lin, D., & Shen, C.-L. (2022). Ginger Root Extract Improves GI Health in Diabetic Rats by Improving Intestinal Integrity and Mitochondrial Function. Nutrients, 14(20), 4384. https://doi.org/10.3390/nu14204384











