Role of Probiotics in the Management of COVID-19: A Computational Perspective
Abstract
:1. Introduction
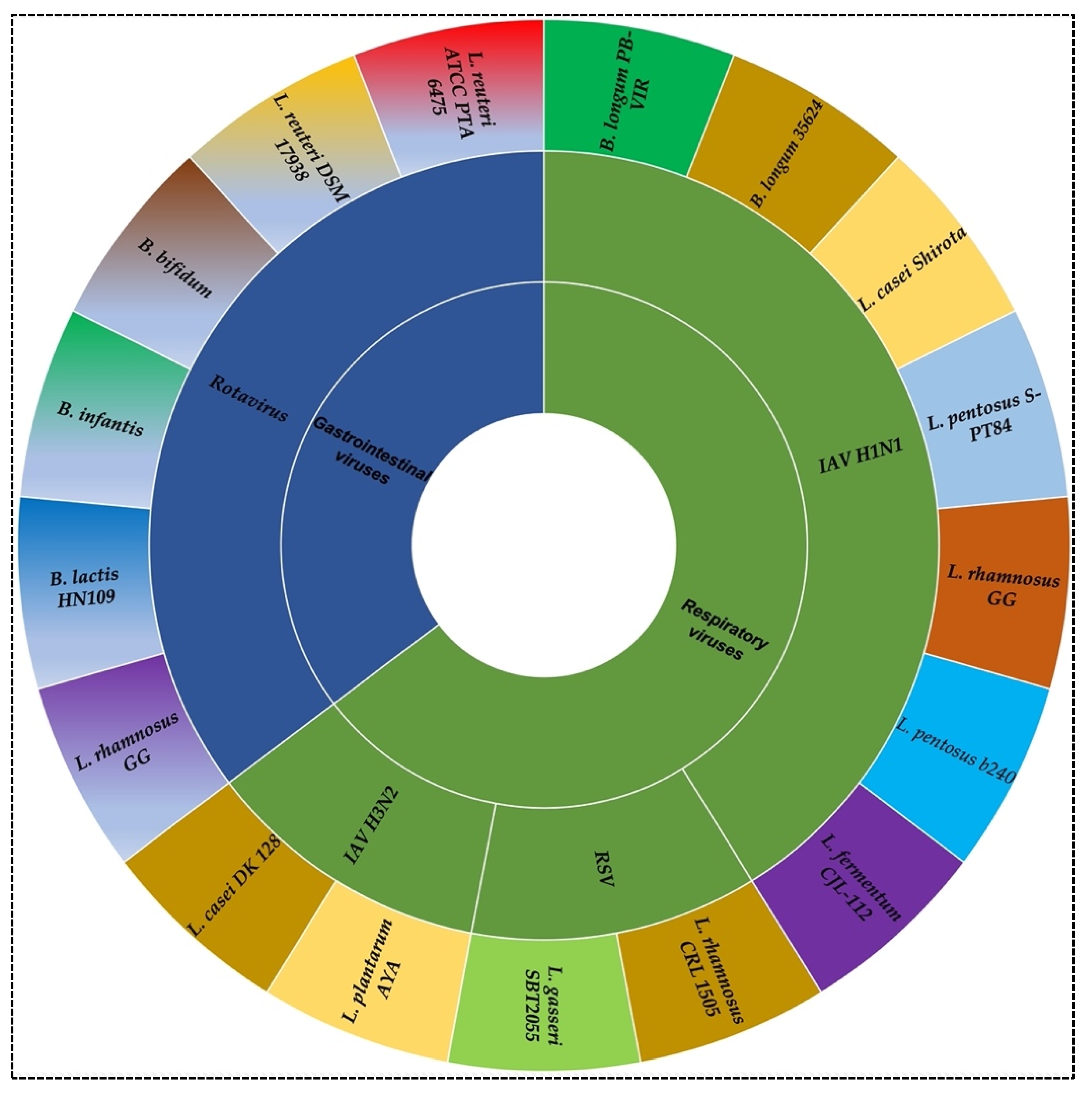
2. Probiotics against Viral Infection
3. Gut-Lung Axis Associated with COVID-19
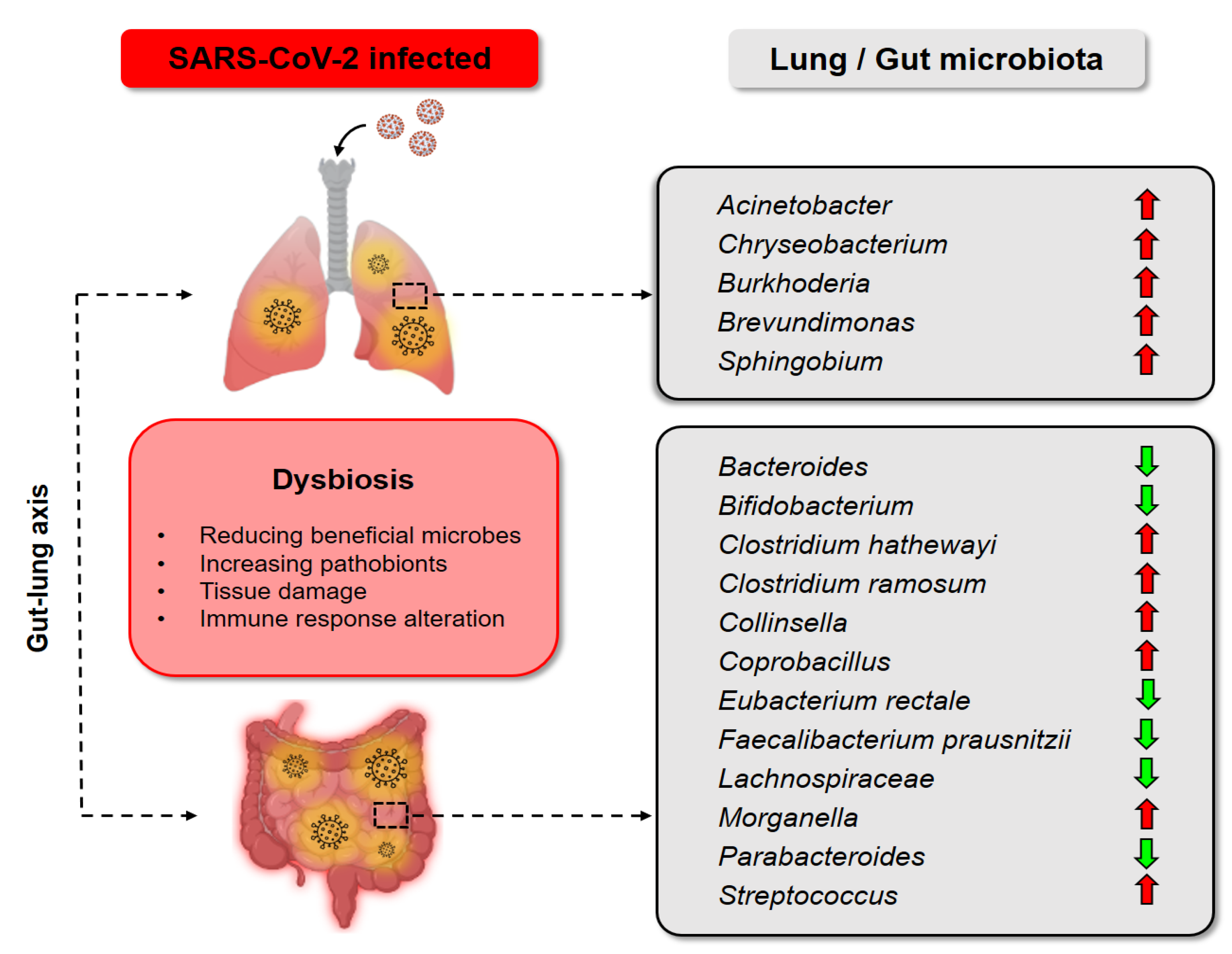
3.1. Rationale of Probiotics as an Adjunctive Treatment for COVID-19
3.2. Clinical Evidence That Supports Probiotics as a Promising Anti-COVID-19 Strategy
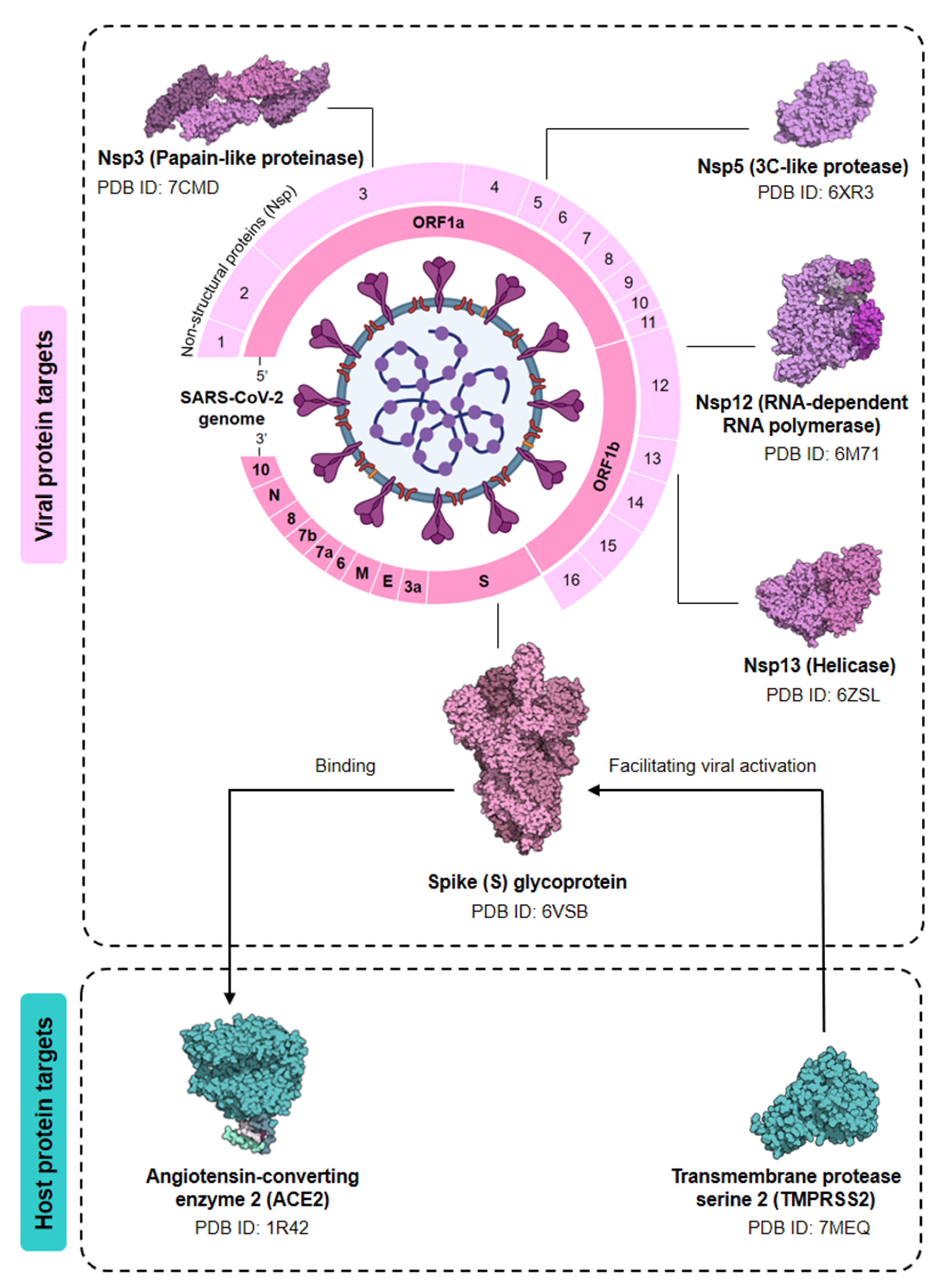
4. Computational Insight: The Future of Probiotics
4.1. Data
4.2. Technology: Microbiome-Driven Approach
4.3. Technology: Ensemble-Driven Docking Approach
| No. | Aim of Research | Computational Methods | Tools [Reference of the Tools] | Reference |
|---|---|---|---|---|
| 1 | Investigate and identify potential hits that could inhibit SARS-CoV-2 by carrying out virtual screening, which included molecular docking, in silico ADMET, and simulation | Screened phytochemicals against five protein targets of COVID-19 (3CLpro, RdRp, ACE2, PLpro, SGp-RBD) Predicted best-docked score phytochemicals in terms of:
| [142] | |
| 2 | Research the role of tea polyphenols on SARS-CoV-2 inhibition |
| [135] | |
| 3 | Study the interaction of luteolin, ribavirin, chloroquine, and remdesivir with the main protease of COVID-19 |
|
| [144] |
| 4 | Investigate the effects of pomegranate peel extract on SARS-CoV-2 spike glycoproteins, furin, ACE2, and transmembrane serine protease 2 |
| [143] | |
| 5 | Investigate the effect of remdesivir, sofosbuvir, ribavirin, galidesivir and tenofovir on RdRp |
| [163] | |
| 6 | Test several anti-polymerase drugs against SARS-CoV-2 RdRp by using computational approaches |
| [166] | |
| 7 | Investigate the effect of grazoprevir (antiviral drug against HCV) on SARS-CoV-2 by using in silico methods |
| [172] | |
| 8 | Investigate the effect of probiotics (Plantaricin JLA-9, Plantaricin W, Plataricin D) on spike protein and the interaction of spike protein with human ACE2 receptor |
| [145] | |
| 9 | Investigate the action of probiotic Lactobacillus plantarum Probio-88 against SARS-CoV-2 replication and immune regulation, with in silico study of metabolite Plantaricin E (PlnE) and Plantaricin F (PlnF) from Lactobacillus plantarum Probio-88 |
| [147] |
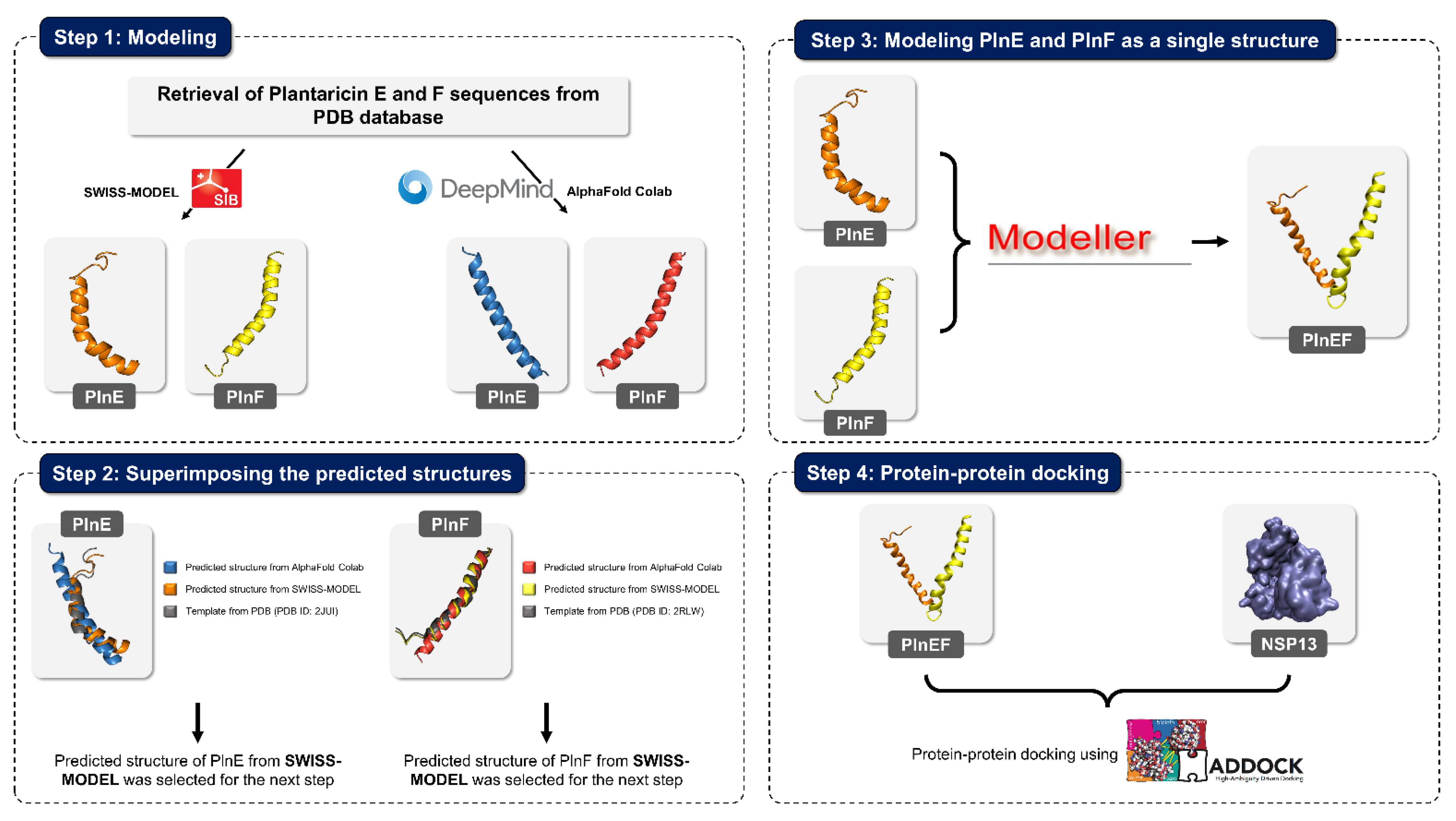
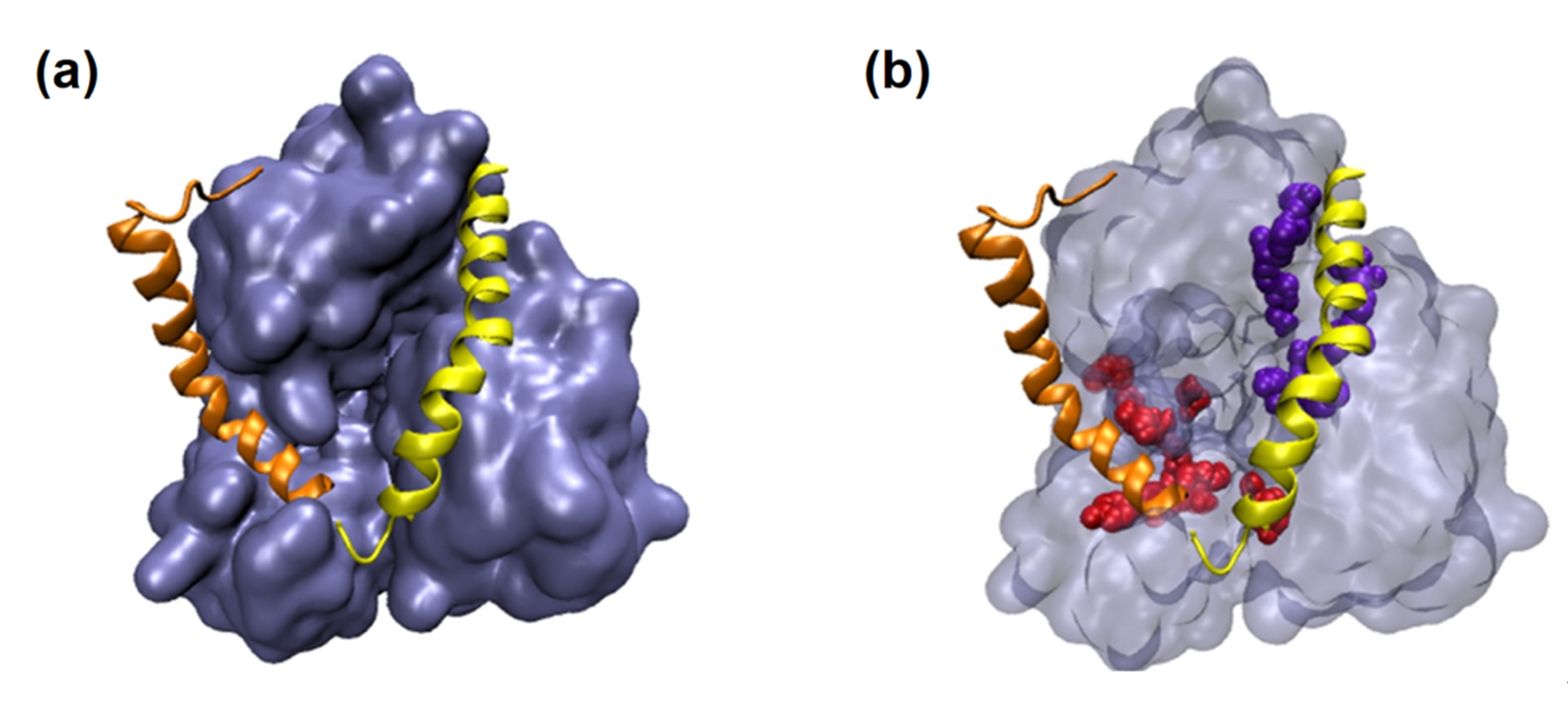
4.4. Case Study: Metabolites of Lactobacillus Plantarum as a Promise to COVID-19
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, S.D. Coronavirus Pandemic: A Serious Threat to Humanity. J. Health Manag. 2020, 22, 1–2. [Google Scholar] [CrossRef]
- Anderson, R.M.; Vegvari, C.; Hollingsworth, T.D.; Pi, L.; Maddren, R.; Ng, C.W.; Baggaley, R.F. The SARS-CoV-2 pandemic: Remaining uncertainties in our understanding of the epidemiology and transmission dynamics of the virus, and challenges to be overcome. Interface Focus 2021, 11, 20210008. [Google Scholar] [CrossRef]
- Meredith, S.; Towey, R. WHO Labels New COVID Strain, Named Omicron, a ‘Variant of Concern,’ Citing Possible Increased Reinfection Risk; CNBC: Englewood Cliffs, NJ, USA, 2021. [Google Scholar]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Gao, Z.; Xu, Y.; Sun, C.; Wang, X.; Guo, Y.; Qiu, S.; Ma, K. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2021, 54, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Deng, L.; Zhang, L.; Cai, Y.; Cheung, C.W.; Xia, Z. Review of the Clinical Characteristics of Coronavirus Disease 2019 (COVID-19). J. Gen. Intern. Med. 2020, 35, 1545–1549. [Google Scholar] [CrossRef] [Green Version]
- Shivakumar, S.; Smibert, O.C.; Trubiano, J.A.; Frauman, A.G.; Liew, D.F. Immunosuppression for COVID-19: Repurposing medicines in a pandemic. Aust. Prescr. 2020, 43, 106–107. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, S.; Chen, Y.; Lu, H.; Shi, D.; Guo, J.; Wu, W.R.; Yang, Y.; Li, Y.; Xu, K.J.; et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 2021, 71, 222–225. [Google Scholar] [CrossRef]
- Xu, K.; Cai, H.; Shen, Y.; Ni, Q.; Chen, Y.; Hu, S.; Li, J.; Wang, H.; Yu, L.; Huang, H.; et al. Management of corona virus disease-19 (COVID-19): The Zhejiang experience. J. Zhejiang Univ. Med. Sci. 2020, 49, 147–157. [Google Scholar]
- Yahfoufi, N.; Mallet, J.F.; Graham, E.; Matar, C. Role of probiotics and prebiotics in immunomodulation. Curr. Opin. Food Sci. 2018, 20, 82–91. [Google Scholar] [CrossRef]
- Plaza-Díaz, J.; Ruiz-Ojeda, F.J.; Vilchez-Padial, L.M.; Gil, A. Evidence of the Anti-Inflammatory Effects of Probiotics and Synbiotics in Intestinal Chronic Diseases. Nutrients 2017, 9, 555. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Al Kassaa, I. Antiviral Probiotics: A New Concept in Medical Sciences. In New Insights on Antiviral Probiotics: From Research to Applications; Springer International Publishing: Cham, The Netherlands, 2017; pp. 1–46. [Google Scholar]
- Mack, D. Probiotics-mixed messages. Can. Fam. Physician Med. Fam. Can. 2005, 51, 1455–1457, 1462. [Google Scholar]
- Bezirtzoglou, E.; Stavropoulou, E. Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe 2011, 17, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Marinova, V.Y.; Rasheva, I.K.; Kizheva, Y.K.; Dermenzhieva, Y.D.; Hristova, P.K. Microbiological quality of probiotic dietary supplements. Biotechnol. Biotechnol. Equip. 2019, 33, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Salminen, S.J.; Gueimonde, M.; Isolauri, E. Probiotics that modify disease risk. J. Nutr. 2005, 135, 1294–1298. [Google Scholar] [CrossRef] [Green Version]
- Mousavi Khaneghah, A.; Abhari, K.; Eş, I.; Soares, M.B.; Oliveira, R.B.A.; Hosseini, H.; Rezaei, M.; Balthazar, C.F.; Silva, R.; Cruz, A.G.; et al. Interactions between probiotics and pathogenic microorganisms in hosts and foods: A review. Trends Food Sci. Technol. 2020, 95, 205–218. [Google Scholar] [CrossRef]
- Wan, L.Y.; Chen, Z.J.; Shah, N.P.; El-Nezami, H. Modulation of Intestinal Epithelial Defense Responses by Probiotic Bacteria. Crit. Rev. Food Sci. Nutr. 2016, 56, 2628–2641. [Google Scholar] [CrossRef]
- Villena, J.; Shimosato, T.; Vizoso-Pinto, M.G.; Kitazawa, H. Editorial: Nutrition, Immunity and Viral Infections. Front. Nutr. 2020, 7, 125. [Google Scholar] [CrossRef]
- Boutayeb, A. The Burden of Communicable and Non-Communicable Diseases in Developing Countries. In Handbook of Disease Burdens and Quality of Life Measures; Springer: Berlin/Heidelberg, Germany, 2010; Volume 100, pp. 532–545. [Google Scholar]
- Rubin, R. COVID-19 Vaccines vs Variants—Determining How Much Immunity Is Enough. JAMA 2021, 325, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Rajput, S.; Paliwal, D.; Naithani, M.; Kothari, A.; Meena, K.; Rana, S. COVID-19 and Gut Microbiota: A Potential Connection. Indian J. Clin. Biochem. IJCB 2021, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, W.J.; Kim, H.W.; Brandt, C.D.; Bise, B.; Kapikian, A.Z.; Chanock, R.M.; Curlin, G.; Parrott, R.H. Rotavirus gastroenteritis in the Washington, DC, area: Incidence of cases resulting in admission to the hospital. Am. J. Dis. Child. 1980, 134, 777–779. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 62 (Suppl. 2), S96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Wu, X.; Nawaz, M.; Li, J.; Yu, P.; Moore, J.E.; Xu, J. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr. Microbiol. 2011, 63, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Preidis, G.A.; Saulnier, D.M.; Blutt, S.E.; Mistretta, T.A.; Riehle, K.P.; Major, A.M.; Venable, S.F.; Barrish, J.P.; Finegold, M.J.; Petrosino, J.F.; et al. Host response to probiotics determined by nutritional status of rotavirus-infected neonatal mice. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Pant, N.; Marcotte, H.; Brüssow, H.; Svensson, L.; Hammarström, L. Effective prophylaxis against rotavirus diarrhea using a combination of Lactobacillus rhamnosus GG and antibodies. BMC Microbiol. 2007, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; Duffy, L.C.; Griffiths, E.; Dryja, D.; Leavens, A.; Rossman, J.; Rich, G.; Riepenhoff-Talty, M.; Locniskar, M. Immune responses in rhesus rotavirus-challenged BALB/c mice treated with bifidobacteria and prebiotic supplements. Pediatric Res. 2002, 51, 750–755. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.A.; Chenoll, E.; Casinos, B.; Bataller, E.; Ramón, D.; Genovés, S.; Montava, R.; Ribes, J.M.; Buesa, J.; Fàbrega, J.; et al. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 2011, 77, 8775–8783. [Google Scholar] [CrossRef] [Green Version]
- Shu, Q.; Qu, F.; Gill, H.S. Probiotic Treatment Using Bifidobacterium lactis HN019 Reduces Weanling Diarrhea Associated with Rotavirus and Escherichia coli Infection in a Piglet Model. J. Pediatric Gastroenterol. Nutr. 2001, 33, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, G.; Wen, K.; Wu, S.; Zhang, Y.; Bui, T.; Yang, X.; Kocher, J.; Sun, J.; Jortner, B.; et al. Lactobacillus rhamnosus GG on rotavirus-induced injury of ileal epithelium in gnotobiotic pigs. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 750–758. [Google Scholar] [CrossRef] [Green Version]
- Ang, L.; Arboleya, S.; Lihua, G.; Chuihui, Y.; Nan, Q.; Suarez, M.; Solís, G.; de los Reyes-Gavilán, C.G.; Gueimonde, M. The establishment of the infant intestinal microbiome is not affected by rotavirus vaccination. Sci. Rep. 2014, 4, 7417. [Google Scholar] [CrossRef] [Green Version]
- García-López, R.; Pérez-Brocal, V.; Diez-Domingo, J.; Moya, A. Gut microbiota in children vaccinated with rotavirus vaccine. Pediatric Infect. Dis. J. 2012, 31, 1300–1302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Azevedo, M.; Wen, K.; Gonzalez, A.; Saif, L.; Li, G.; Yousef, A.; Yuan, L. Probiotic Lactobacillus acidophilus enhances the immunogenicity of an oral rotavirus vaccine in gnotobiotic pigs. Vaccine 2008, 26, 3655–3661. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, A.N.; Chattha, K.S.; Kandasamy, S.; Liu, Z.; Esseili, M.; Shao, L.; Rajashekara, G.; Saif, L.J. Lactobacilli and bifidobacteria promote immune homeostasis by modulating innate immune responses to human rotavirus in neonatal gnotobiotic pigs. PLoS ONE 2013, 8, e76962. [Google Scholar] [CrossRef]
- Lehtoranta, L.; Pitkäranta, A.; Korpela, R. Probiotics in respiratory virus infections. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2014, 33, 1289–1302. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, Y.T.; Ngo, V.L.; Cho, Y.H.; Ko, E.J.; Hong, S.M.; Kim, K.H.; Jang, J.H.; Oh, J.S.; Park, M.K.; et al. Heat-killed Lactobacillus casei confers broad protection against influenza A virus primary infection and develops heterosubtypic immunity against future secondary infection. Sci. Rep. 2017, 7, 17360. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.M.; Smith, A.P. A Critical, Nonlinear Threshold Dictates Bacterial Invasion and Initial Kinetics During Influenza. Sci. Rep. 2016, 6, 38703. [Google Scholar] [CrossRef] [Green Version]
- Tate, M.D.; Schilter, H.C.; Brooks, A.G.; Reading, P.C. Responses of mouse airway epithelial cells and alveolar macrophages to virulent and avirulent strains of influenza A virus. Viral Immunol. 2011, 24, 77–88. [Google Scholar] [CrossRef]
- Harata, G.; He, F.; Hiruta, N.; Kawase, M.; Kubota, A.; Hiramatsu, M.; Yausi, H. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett. Appl. Microbiol. 2010, 50, 597–602. [Google Scholar] [CrossRef]
- Izumo, T.; Maekawa, T.; Ida, M.; Noguchi, A.; Kitagawa, Y.; Shibata, H.; Yasui, H.; Kiso, Y. Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int. Immunopharmacol. 2010, 10, 1101–1106. [Google Scholar] [CrossRef]
- Hori, T.; Kiyoshima, J.; Shida, K.; Yasui, H. Effect of intranasal administration of Lactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin. Diagn. Lab. Immunol. 2001, 8, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Yeo, J.M.; Lee, H.J.; Kim, J.W.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Song, C.S. Lactobacillus fermentum CJL-112 protects mice against influenza virus infection by activating T-helper 1 and eliciting a protective immune response. Int. Immunopharmacol. 2014, 18, 50–54. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Kunitoh-Asari, A.; Hayakawa, K.; Imai, S.; Kasuya, K.; Abe, K.; Adachi, Y.; Fukudome, S.; Takahashi, Y.; Hachimura, S. Oral administration of Lactobacillus plantarum strain AYA enhances IgA secretion and provides survival protection against influenza virus infection in mice. PLoS ONE 2014, 9, e86416. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, N.; Saito, T.; Uematsu, T.; Kishi, K.; Toba, M.; Kohda, N.; Suzuki, T. Oral administration of heat-killed Lactobacillus pentosus strain b240 augments protection against influenza virus infection in mice. Int. Immunopharmacol. 2011, 11, 199–203. [Google Scholar] [CrossRef]
- Groeger, D.; Schiavi, E.; Grant, R.; Kurnik-Łucka, M.; Michalovich, D.; Williamson, R.; Beinke, S.; Kiely, B.; Akdis, C.; Hessel, E.; et al. Intranasal Bifidobacterium longum protects against viral-induced lung inflammation and injury in a murine model of lethal influenza infection. EBioMedicine 2020, 60, 102981. [Google Scholar] [CrossRef]
- Groves, H.T.; Higham, S.L.; Moffatt, M.F.; Cox, M.J.; Tregoning, J.S.; Bomberger, J.M. Respiratory Viral Infection Alters the Gut Microbiota by Inducing Inappetence. mBio 2020, 11, e03236-19. [Google Scholar] [CrossRef] [Green Version]
- Harding, J.N.; Siefker, D.; Vu, L.; You, D.; DeVincenzo, J.; Pierre, J.F.; Cormier, S.A. Altered gut microbiota in infants is associated with respiratory syncytial virus disease severity. BMC Microbiol. 2020, 20, 140. [Google Scholar] [CrossRef]
- Collins, P.L.; Melero, J.A. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res. 2011, 162, 80–99. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; McAllister, D.A.; O’Brien, K.L.; Simoes, E.A.F.; Madhi, S.A.; Gessner, B.D.; Polack, F.P.; Balsells, E.; Acacio, S.; Aguayo, C.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet (Lond. Engl.) 2017, 390, 946–958. [Google Scholar] [CrossRef] [Green Version]
- Eguchi, K.; Fujitani, N.; Nakagawa, H.; Miyazaki, T. Prevention of respiratory syncytial virus infection with probiotic lactic acid bacterium Lactobacillus gasseri SBT2055. Sci. Rep. 2019, 9, 4812. [Google Scholar] [CrossRef]
- Chiba, E.; Tomosada, Y.; Vizoso-Pinto, M.G.; Salva, S.; Takahashi, T.; Tsukida, K.; Kitazawa, H.; Alvarez, S.; Villena, J. Immunobiotic Lactobacillus rhamnosus improves resistance of infant mice against respiratory syncytial virus infection. Int. Immunopharmacol. 2013, 17, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Tomosada, Y.; Chiba, E.; Zelaya, H.; Takahashi, T.; Tsukida, K.; Kitazawa, H.; Alvarez, S.; Villena, J. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013, 14, 40. [Google Scholar] [CrossRef] [Green Version]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Smyk, W.; Janik, M.K.; Portincasa, P.; Milkiewicz, P.; Lammert, F.; Krawczyk, M. COVID-19: Focus on the lungs but do not forget the gastrointestinal tract. Eur. J. Clin. Investig. 2020, 50, e13276. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Fanos, V.; Pintus, M.C.; Pintus, R.; Marcialis, M. Lung microbiota in the acute respiratory disease: From coronavirus to metabolomics. J. Pediatric Neonatal Individ. Med. 2020, 9, e090139. [Google Scholar] [CrossRef]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Raftery, A.L.; Tsantikos, E.; Harris, N.L.; Hibbs, M.L. Links between Inflammatory Bowel Disease and Chronic Obstructive Pulmonary Disease. Front. Immunol. 2020, 11, 2144. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.-Y.; Tso, E.Y.K.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.L.; Chan, P.K.S.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276. [Google Scholar] [CrossRef]
- Yamamoto, S.; Saito, M.; Tamura, A.; Prawisuda, D.; Mizutani, T.; Yotsuyanagi, H. The human microbiome and COVID-19: A systematic review. PLoS ONE 2021, 16, e0253293. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e911. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J.; et al. Clinical Significance of the Correlation between Changes in the Major Intestinal Bacteria Species and COVID-19 Severity. Engineering 2020, 6, 1178–1184. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Gao, Y.; Zhou, J.; Wang, S.; Huang, B.; Wu, J.; Cao, Q.; Chen, Y.; Wang, Z.; et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Infect. 2020, 81, e64–e67. [Google Scholar] [CrossRef]
- Smythies, L.E.; Sellers, M.; Clements, R.H.; Mosteller-Barnum, M.; Meng, G.; Benjamin, W.H.; Orenstein, J.M.; Smith, P.D. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Investig. 2005, 115, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Grayson, M.H.; Camarda, L.E.; Hussain, S.-R.A.; Zemple, S.J.; Hayward, M.; Lam, V.; Hunter, D.A.; Santoro, J.L.; Rohlfing, M.; Cheung, D.S.; et al. Intestinal Microbiota Disruption Reduces Regulatory T Cells and Increases Respiratory Viral Infection Mortality Through Increased IFNγ Production. Front. Immunol. 2018, 9, 1587. [Google Scholar] [CrossRef] [Green Version]
- Zafar, N.; Aslam, M.; Ali, A.; Khatoon, A.; Nazir, A.; Tanveer, Q.; Bilal, M.; Kanwar, R.; Qadeer, A.; Sikandar, M.; et al. Probiotics: Helpful for the prevention of COVID-19? Biomed. Res. Ther. 2020, 7, 4086–4099. [Google Scholar] [CrossRef]
- Raghuvanshi, R.; Grayson, A.G.; Schena, I.; Amanze, O.; Suwintono, K.; Quinn, R.A. Microbial Transformations of Organically Fermented Foods. Metabolites 2019, 9, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kligler, B.; Cohrssen, A. Probiotics. Am. Fam. Physician 2008, 78, 1073–1078. [Google Scholar]
- Snydman, D.R. The safety of probiotics. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2008, 46 (Suppl. 2), S104–S111; discussion S144–S151. [Google Scholar] [CrossRef] [Green Version]
- Gueimonde, M.; Ouwehand, A.C.; Salminen, S. Safety of probiotics. Scand. J. Nutr. 2004, 48, 42–48. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Batista, K.S.; de Albuquerque, J.G.; de Vasconcelos, M.H.A.; Bezerra, M.L.R.; da Silva Barbalho, M.B.; Oliveira, R.P.; Aquino, J.d.S. Probiotics and prebiotics: Potential prevention and therapeutic target for nutritional management of COVID-19? Nutr. Res. Rev. 2021, 1–42. [Google Scholar] [CrossRef] [PubMed]
- Akour, A. Probiotics and COVID-19: Is there any link? Lett. Appl. Microbiol. 2020, 71, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Dhar, D.; Mohanty, A. Gut microbiota and COVID-19- possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cheng, F.; Xu, Q.; Su, Y.; Cai, X.; Zeng, F.; Zhang, Y. The role of probiotics in coronavirus disease-19 infection in Wuhan: A retrospective study of 311 severe patients. Int. Immunopharmacol. 2021, 95, 107531. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Castrellón, P.; Gandara-Martí, T.; Abreu, A.T.; Nieto-Rufino, C.D.; López-Orduña, E.; Jiménez-Escobar, I.; Jiménez-Gutiérrez, C.; López-Velazquez, G.; Espadaler-Mazo, J. Efficacy and safety of novel probiotic formulation in adult Covid19 outpatients: A randomized, placebo-controlled clinical trial. medRxiv 2021. [Google Scholar] [CrossRef]
- Bousquet, J.; Antó, J.; Czarlewski, W.; Haahtela, T.; Fonseca, S.; Iaccarino, G.; Blain, H.; Vidal, A.; Sheikh, A.; Akdis, C.; et al. Cabbage and fermented vegetables: From death rate heterogeneity in countries to candidates for mitigation strategies of severe COVID-19. Allergy 2020, 76, 735–750. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Therap. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Eetemadi, A.; Rai, N.; Pereira, B.M.P.; Kim, M.; Schmitz, H.; Tagkopoulos, I. The Computational Diet: A Review of Computational Methods Across Diet, Microbiome, and Health. Front. Microbiol. 2020, 11, 393. [Google Scholar] [CrossRef] [Green Version]
- Satokari, R. Modulation of Gut Microbiota for Health by Current and Next-Generation Probiotics. Nutrients 2019, 11, 1921. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Chang, H.-W.; Yan, D.; Lee, K.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.; Pfitzner, B.; Neschen, S.; Kahle, M.; Harir, M.; Lucio, M.; Moritz, F.; Tziotis, D.; Witting, M.; Rothballer, M.; et al. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J. 2014, 8, 2380–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Deeke, S.A.; Ning, Z.; Starr, A.E.; Butcher, J.; Li, J.; Mayne, J.; Cheng, K.; Liao, B.; Li, L.; et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 2018, 9, 2873. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota: Beyond metagenomics, metatranscriptomics illuminates microbiome functionality in IBD. Nature reviews. Gastroenterol. Hepatol. 2018, 15, 193–194. [Google Scholar] [CrossRef]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S.S. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Oh, K.; Kim, H.C. Dietary assessment methods in epidemiologic studies. Epidemiol. Health 2014, 36, e2014009. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, L.; Qi, C.; Zhuang, H.; Fu, T.; Zhang, X. gutMDisorder: A comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2019, 48, D554–D560. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jing, Q.; Yan, S.; Liu, X.; Sun, Y.; Zhu, D.; Wang, D.; Hao, C.; Xue, D. Amadis: A Comprehensive Database for Association Between Microbiota and Disease. Front. Physiol. 2021, 12, 697059. [Google Scholar] [CrossRef] [PubMed]
- Hiseni, P.; Rudi, K.; Wilson, R.C.; Hegge, F.T.; Snipen, L. HumGut: A comprehensive human gut prokaryotic genomes collection filtered by metagenome data. Microbiome 2021, 9, 165. [Google Scholar] [CrossRef]
- Chen, Y.-A.; Park, J.; Natsume-Kitatani, Y.; Kawashima, H.; Mohsen, A.; Hosomi, K.; Tanisawa, K.; Ohno, H.; Konishi, K.; Murakami, H.; et al. MANTA, an integrative database and analysis platform that relates microbiome and phenotypic data. PLoS ONE 2020, 15, e0243609. [Google Scholar] [CrossRef]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef] [Green Version]
- Friede, A.; Reid, J.A.; Ory, H.W. CDC WONDER: A comprehensive on-line public health information system of the Centers for Disease Control and Prevention. Am. J. Public Health 1993, 83, 1289–1294. [Google Scholar] [CrossRef] [Green Version]
- Imhann, F.; Van der Velde, K.J.; Barbieri, R.; Alberts, R.; Voskuil, M.D.; Vich Vila, A.; Collij, V.; Spekhorst, L.M.; Van der Sloot, K.W.J.; Peters, V.; et al. The 1000IBD project: Multi-omics data of 1000 inflammatory bowel disease patients; data release 1. BMC Gastroenterol. 2019, 19, 5. [Google Scholar] [CrossRef]
- Freifeld, C.C.; Mandl, K.D.; Reis, B.Y.; Brownstein, J.S. HealthMap: Global Infectious Disease Monitoring through Automated Classification and Visualization of Internet Media Reports. J. Am. Med. Inform. Assoc. 2008, 15, 150–157. [Google Scholar] [CrossRef]
- Kim, S.K.; Guevarra, R.B.; Kim, Y.T.; Kwon, J.; Kim, H.; Cho, J.H.; Kim, H.B.; Lee, J.H. Role of Probiotics in Human Gut Microbiome-Associated Diseases. J. Microbiol. Biotechnol. 2019, 29, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. 1), S38–S44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. 2011, 10, 17. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. Erratum: SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2015, 4, 30. [Google Scholar] [CrossRef] [Green Version]
- Westreich, S.T.; Treiber, M.L.; Mills, D.A.; Korf, I.; Lemay, D.G. SAMSA2: A standalone metatranscriptome analysis pipeline. BMC Bioinform. 2018, 19, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milanese, A.; Mende, D.R.; Paoli, L.; Salazar, G.; Ruscheweyh, H.-J.; Cuenca, M.; Hingamp, P.; Alves, R.; Costea, P.I.; Coelho, L.P.; et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019, 10, 1014. [Google Scholar] [CrossRef]
- Zhang, H.; Ai, J.W.; Yang, W.; Zhou, X.; He, F.; Xie, S.; Zeng, W.; Li, Y.; Yu, Y.; Gou, X.; et al. Metatranscriptomic Characterization of Coronavirus Disease 2019 Identified a Host Transcriptional Classifier Associated With Immune Signaling. Clin. Infect. Dis. 2021, 73, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Xiao, Y.; Kang, L.; Ma, W.; Shi, L.; Zhang, L.; Zhou, Z.; Yang, J.; Zhong, J.; Yang, D.; et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 713–720. [Google Scholar] [CrossRef] [Green Version]
- Mohs, R.C.; Greig, N.H. Drug discovery and development: Role of basic biological research. Alzheimer’s Dement. (N. Y.) 2017, 3, 651–657. [Google Scholar] [CrossRef]
- Dibyajyoti, S.; Bin, E.T.; Swati, P.P. Bioinformatics:The effects on the cost of drug discovery. Galle Med. J. 2013, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- Osakwe, O. The Significance of Discovery Screening and Structure Optimization Studies. In Social Aspects of Drug Discovery, Development and Commercialization; Academic Press: Cambridge, MA, USA, 2016; pp. 109–128. [Google Scholar]
- Clark, D.E. What has computer-aided molecular design ever done for drug discovery? Expert Opin. Drug Discov. 2006, 1, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Distinto, S.; Liedl, K.R.; Markt, P.; Rollinger, J.M.; Schuster, D.; Spitzer, G.M.; Wolber, G. Development of anti-viral agents using molecular modeling and virtual screening techniques. Infect. Disord. Drug Targets 2011, 11, 64–93. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Chandra, S.; Siddiqi, M.I. Recent advances in the development of antiviral agents using computer-aided structure based approaches. Curr. Pharm. Des. 2014, 20, 3488–3499. [Google Scholar] [CrossRef]
- Selvaraj, G.F.; Piramanayagam, S.; Devadasan, V.; Hassan, S.; Krishnasamy, K.; Srinivasan, S. Computational analysis of drug like candidates against Neuraminidase of Human Influenza A virus subtypes. Inform. Med. Unlocked 2020, 18. [Google Scholar] [CrossRef]
- Mallipeddi, P.L.; Kumar, G.; White, S.W.; Webb, T.R. Recent advances in computer-aided drug design as applied to anti-influenza drug discovery. Curr. Top. Med. Chem. 2014, 14, 1875–1889. [Google Scholar] [CrossRef] [PubMed]
- Mottin, M.; Borba, J.; Melo-Filho, C.; Neves, B.; Muratov, E.; Torres, P.; Braga, R.; Perryman, A.; Ekins, S.; Andrade, C. Computational Drug Discovery for the Zika Virus. Braz. J. Pharm. Sci. 2018, 54, e01002. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Maryam, A.; Muneer, I.; Xing, F.; Ashfaq, U.A.; Khan, F.A.; Anwar, F.; Geesi, M.H.; Khalid, R.R.; Rauf, S.A.; et al. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci. Rep. 2019, 9, 1433. [Google Scholar] [CrossRef]
- Alizon, S.; Cazals, F.; Guindon, S.; Lemaitre, C.; Mary-Huard, T.; Niarakis, A.; Salson, M.; Scornavacca, C.; Touzet, H. SARS-CoV-2 Through the Lens of Computational Biology: How Bioinformatics Is Playing a Key Role in the Study of the Virus and Its Origins; CNRS: Paris, France, 2021. [Google Scholar]
- Mhatre, S.; Naik, S.; Patravale, V. A molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2. Comput. Biol. Med. 2021, 129, 104137. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.; Thiessen, P.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2018, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Irwin, J.J.; Shoichet, B.K. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Modeling 2005, 45, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef] [Green Version]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Vardhan, S.; Sahoo, S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020, 124, 103936. [Google Scholar] [CrossRef] [PubMed]
- Suručić, R.; Tubić, B.; Stojiljković, M.P.; Djuric, D.M.; Travar, M.; Grabež, M.; Šavikin, K.; Škrbić, R. Computational study of pomegranate peel extract polyphenols as potential inhibitors of SARS-CoV-2 virus internalization. Mol. Cell. Biochem. 2021, 476, 1179–1193. [Google Scholar] [CrossRef]
- Yu, R.; Chen, L.; Lan, R.; Shen, R.; Li, P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents 2020, 56, 106012. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Altayb, H.N.; Al-Abbasi, F.A.; Al-Malki, A.L.; Kamal, M.A.; Kumar, V. Antiviral effects of probiotic metabolites on COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 4175–4184. [Google Scholar] [CrossRef]
- Balmeh, N.; Mahmoudi, S.; Fard, N.A. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inform. Med. Unlocked 2021, 23, 100515. [Google Scholar] [CrossRef]
- Rather, I.; Choi, S.B.; Kamli, M.; Hakeem, K.; Sabir, J.; Park, Y.-H.; Hor, Y.-Y.; Leitão, J. Potential Adjuvant Therapeutic Effect of Lactobacillus plantarum Probio-88 Postbiotics against SARS-CoV-2. Vaccines 2021, 9, 1067. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Ahmad, A.; Khan, S.A.; Asif, M.; Bhutani, R.; Al-Abbasi, F.A. Synthesis, molecular properties, toxicity and biological evaluation of some new substituted imidazolidine derivatives in search of potent anti-inflammatory agents. Saudi Pharm. J. 2016, 24, 104–114. [Google Scholar] [CrossRef] [Green Version]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonawane, P.; Patel, K.; Vishwakarma, R.; Singh, S.; Khan, B. In Silico mutagenesis and Docking studies of active site residues suggest altered substrate specificity and possible physiological role of Cinnamoyl CoA Reductase 1 (Ll-CCRH1). Bioinformation 2013, 9, 224–232. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web-server for automatic binding site prediction, analysis, and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef] [Green Version]
- Pawar, S.; Rohane, S. Review on Discovery Studio: An important Tool for Molecular Docking. Asian J. Res. Chem. 2021, 14, 1–3. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Williams, C.; Headd, J.; Moriarty, N.; Prisant, M.; Videau, L.; Deis, L.; Verma, V.; Keedy, D.; Hintze, B.; Chen, V.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2017, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [CrossRef]
- Hooft, R.W.; Vriend, G.; Sander, C.; Abola, E.E. Errors in protein structures. Nature 1996, 381, 272. [Google Scholar] [CrossRef] [PubMed]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.-Y.; Sali, A. Protein Structure Modeling with MODELLER. In Structural Proteomics: High-Throughput Methods; Kobe, B., Guss, M., Huber, T., Eds.; Humana Press: Totowa, NJ, USA, 2008; pp. 145–159. [Google Scholar]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2018, 35, 1067–1069. [Google Scholar] [CrossRef]
- Aftab, O.; Ghouri, M.; Masood, M.; Haider, Z.; Khan, Z.; Ahmad, A.; Munawar, N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020, 18, 275. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Schmid, N.; Eichenberger, A.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.; van Gunsteren, W. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. EBJ 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.K.; Vhora, N.; Contractor, D.; Shard, A.; Kumar, D.; Kalia, K.; Jain, A. Computational drug repurposing study elucidating simultaneous inhibition of entry and replication of novel corona virus by Grazoprevir. Sci. Rep. 2021, 11, 7307. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scialò, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Baughn, L.B.; Sharma, N.; Elhaik, E.; Sekulic, A.; Bryce, A.H.; Fonseca, R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin. Proc. 2020, 95, 1989–1999. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-f.; Xu, W.; Liu, S.-w. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Tahir Ul Qamar, M.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.L. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Chen, C.Z.; Gorshkov, K.; Xu, M.; Lo, D.C.; Zheng, W. RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery. SLAS DISCOVERY Adv. Sci. Drug Discov. 2020, 25, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; McCullagh, M. Role of ATP in the RNA Translocation Mechanism of SARS-CoV-2 NSP13 Helicase. J. Phys. Chem. B 2021, 125, 8787–8796. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, S.; Nabavi, S.F.; Banach, M.; Berindan-Neagoe, I.; Sarkar, K.; Sil, P.C.; Nabavi, S.M. Should We Try SARS-CoV-2 Helicase Inhibitors for COVID-19 Therapy? Arch. Med. Res. 2020, 51, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Spratt, A.; Gallazzi, F.; Quinn, T.; Lorson, C.; Sönnerborg, A.; Singh, K. Coronavirus helicases: Attractive and unique targets of antiviral drug-development and therapeutic patents. Expert Opin. Ther. Pat. 2021, 31, 339–350. [Google Scholar] [CrossRef]
- Ekblad, B.; Kristiansen, P.E. NMR structures and mutational analysis of the two peptides constituting the bacteriocin plantaricin S. Sci. Rep. 2019, 9, 2333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekblad, B.; Kyriakou, P.K.; Oppegård, C.; Nissen-Meyer, J.; Kaznessis, Y.N.; Kristiansen, P.E. Structure–Function Analysis of the Two-Peptide Bacteriocin Plantaricin EF. Biochemistry 2016, 55, 5106–5116. [Google Scholar] [CrossRef]
- Fimland, N.; Rogne, P.; Fimland, G.; Nissen-Meyer, J.; Kristiansen, P.E. Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin plantaricin EF. Biochim. Biophys. Acta 2008, 1784, 1711–1719. [Google Scholar] [CrossRef]
- Selegård, R.; Musa, A.; Nyström, P.; Aili, D.; Bengtsson, T.; Khalaf, H. Plantaricins markedly enhance the effects of traditional antibiotics against Staphylococcus epidermidis. Future Microbiol. 2019, 14, 195–205. [Google Scholar] [CrossRef] [Green Version]
- AlphaFold Colab. 2021. Available online: https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb (accessed on 28 November 2021).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a platform for computational drug design. WIREs Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| No. | Identifier | Title | Treatment | Probiotic Strain | Number Enrolled | Status |
|---|---|---|---|---|---|---|
| 1 | NCT04517422 | Efficacy and safety of Lactobacillus plantarum and Pediococcus acidilactici as co-adjuvant therapy for reducing the risk of severe disease in adults with SARS-CoV-2 and its modulation of the fecal microbiota: A randomized clinical trial | Once per day, administered for 30 days | Combination of 4 probiotic strains, including 3 Lactobacillus plantarum strains CECT30292, CECT7484, CECT7485, and Pediococcus acidilactici strain CECT7483 | 300 | Completed |
| 2 | NCT04458519 | Clinical study of efficacy of intranasal probiotic treatment to reduce severity of symptoms in COVID-19 infection | Twice per day, administered for 14 days | Lactococcus lactis W136 | 23 | Completed |
| 3 | NCT04854941 | Efficacy of probiotics (Lactobacillus rhamnosus, Bifidobacterium bifidum, Bifidobacterium longum subsp. infantis and Bifidobacterium longum) in the treatment of hospitalized patients with novel coronavirus infection | 3 times per day, administered for 14 days | Combination of 4 probiotic strains, including Lactobacillus rhamnosus PDV 1705, Bifidobacterium bifidum PDV 0903, Bifidobacterium longum subsp. infantis PDV 1911 and Bifidobacterium longum PDV 2301 | 200 | Completed |
| 4 | NCT04399252 | A randomized trial of the effect of Lactobacillus on the microbiome of household contacts exposed to COVID-19 | 2 capsules per day, administered for 28 days | Lactobaciltus rhamnosus GG | 182 | Completed |
| 5 | NCT04734886 | Exploratory study for the probiotic supplementation effects on SARS-CoV-2 antibody response in healthy adults | 2 capsules per day, administered for 6 months | Lactobacillus reuteri DSM 17938 | 161 | Completed |
| 6 | NCT05043376 | A randomized, open-label, and controlled clinical trial to study the adjuvant treatment benefits of probiotic Streptococcus salivarius K12 to prevent/reduce lung inflammation in mild-to-moderate hospitalized patients with COVID-19 | 2 tablets per day, administered for up to 14 days | Streptococcus salivarius K12 | 50 | Completed |
| 7 | NCT04366180 | Multicentric study to assess the effect of consumption of Lactobacillus coryniformis K8 on healthcare personnel exposed to COVID-19 | Once per day, administered for 8 weeks | Lactobacillus coryniformis K8 | 314 | Ongoing |
| Method | Description | Tools | URL | Reference |
|---|---|---|---|---|
| Metagenomics | Metagenomics is the culture-independent analysis of a collection of genomes from microbial communities contained in natural living environments. | MetaPhlAn2 | https://huttenhower.sph.harvard.edu/metaphlan2/ | [110] |
| MG-RAST | https://www.mg-rast.org/ | [113] | ||
| MEGAHIT | https://github.com/voutcn/megahit | [114] | ||
| HUMAnN2 | https://huttenhower.sph.harvard.edu/humann2/ | [115] | ||
| QIIME | http://qiime.org/ | [116] | ||
| mothur | https://mothur.org/ | [117] | ||
| SPades | https://cab.spbu.ru/software/spades/ | [118] | ||
| Metatranscriptomics | Metatranscriptomics, also a culture-independent method, allows studying of the expressed RNA transcripts in the microbiome. | SOAPdenovo | http://soapdenovo2.sourceforge.net/ | [119] |
| SAMSA2 | https://github.com/transcript/samsa2 | [120] | ||
| mOTUs2 | https://motu-tool.org/ https://github.com/motu-tool/mOTUs | [121] |
| Superimpose | RMSD (Å) | |
|---|---|---|
| PlnE | AlphaFold Colab/SWISS-MODEL | 3.07 |
| AlphaFold Colab/Template | 2.91 | |
| SWISS-MODEL/Template | 0.62 | |
| PlnF | AlphaFold Colab/SWISS-MODEL | 1.48 |
| AlphaFold Colab/Template | 1.75 | |
| SWISS-MODEL/Template | 0.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, Q.V.; Chong, L.C.; Hor, Y.-Y.; Lew, L.-C.; Rather, I.A.; Choi, S.-B. Role of Probiotics in the Management of COVID-19: A Computational Perspective. Nutrients 2022, 14, 274. https://doi.org/10.3390/nu14020274
Nguyen QV, Chong LC, Hor Y-Y, Lew L-C, Rather IA, Choi S-B. Role of Probiotics in the Management of COVID-19: A Computational Perspective. Nutrients. 2022; 14(2):274. https://doi.org/10.3390/nu14020274
Chicago/Turabian StyleNguyen, Quang Vo, Li Chuin Chong, Yan-Yan Hor, Lee-Ching Lew, Irfan A. Rather, and Sy-Bing Choi. 2022. "Role of Probiotics in the Management of COVID-19: A Computational Perspective" Nutrients 14, no. 2: 274. https://doi.org/10.3390/nu14020274
APA StyleNguyen, Q. V., Chong, L. C., Hor, Y.-Y., Lew, L.-C., Rather, I. A., & Choi, S.-B. (2022). Role of Probiotics in the Management of COVID-19: A Computational Perspective. Nutrients, 14(2), 274. https://doi.org/10.3390/nu14020274







