Automated Classification of 6-n-Propylthiouracil Taster Status with Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Experimental Procedure
2.3. Subjects
2.4. Sensory Assessments
2.4.1. PROP and NaCl Intensity Ratings
2.4.2. Scores for the Five Basic Qualities and Taste Sensitivity Status Determination
2.5. Papilla Density
2.6. Molecular Analysis
2.7. Machine Learning
2.7.1. Data Processing Operations
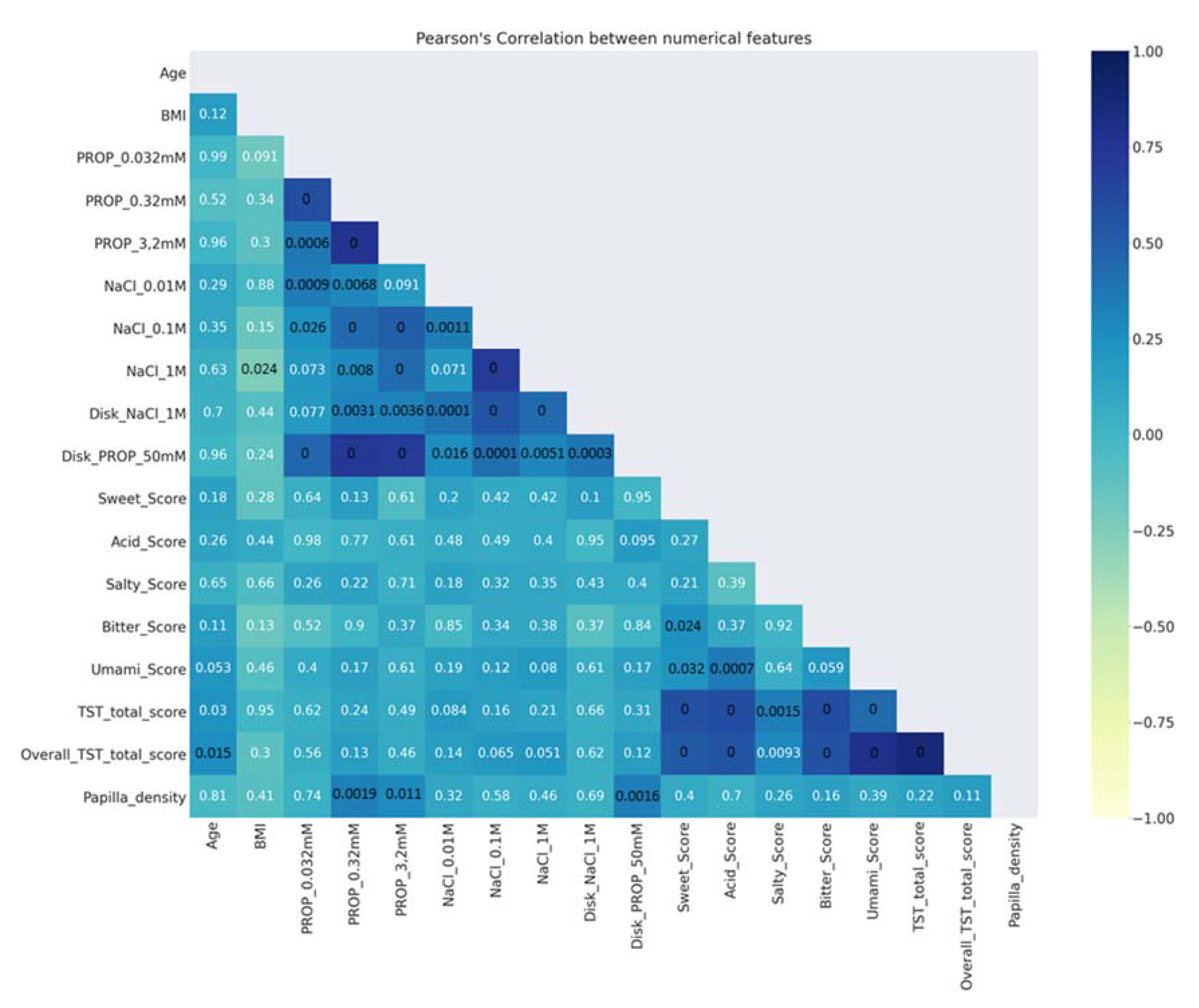
- Description of data analysis: A Pearson’s (r) coefficient analysis [97] was made to verify the correlations between numerical features. Figure 2 shows that the ratings determined with the solutions in the three-solution tests, as well as those determined with impregnated paper disks, were strongly correlated with one another (r > 0.43; p < 0.001). Moreover, a strong correlation was found between papilla density and the ratings of PROP paper disks (r = 0.34; p = 0.0016). In addition, the scores of the TST and the overall TST were strongly correlated with the scores of sweet, acid, bitter, and umami (r > 0.43; p < 0.001). The p-values of the significant correlations are shown in black (Figure 2);
- Preprocessing the data—This operation includes the handling of missing values: (1) removal of the columns/rows with more than 60% missing values; (2) estimation of the missing values (when less than 60%) with the mean or median value of the column; and (3) elimination of the duplicated values from the dataset. After analysis of the dataset, we had to eliminate four variables (sucrose threshold, and intensity ratings of three suprathreshold solutions of sucrose). In addition, in six rows in which the BMI value was lacking, it was estimated by the mean values of the column;
- Processing the features requires transforming the content of the dataset into a language readable to be processed by an algorithm. This operation includes one-hot encoding (encoding categorical data into numerical data) and normalization of the numerical data (through transforming an actual range of numerical values into a standard range of values between 0 and 1). Finally, the synthetic minority oversampling technique (SMOTE) [98] permits the balancing of the numbers of PROP taster status categories.
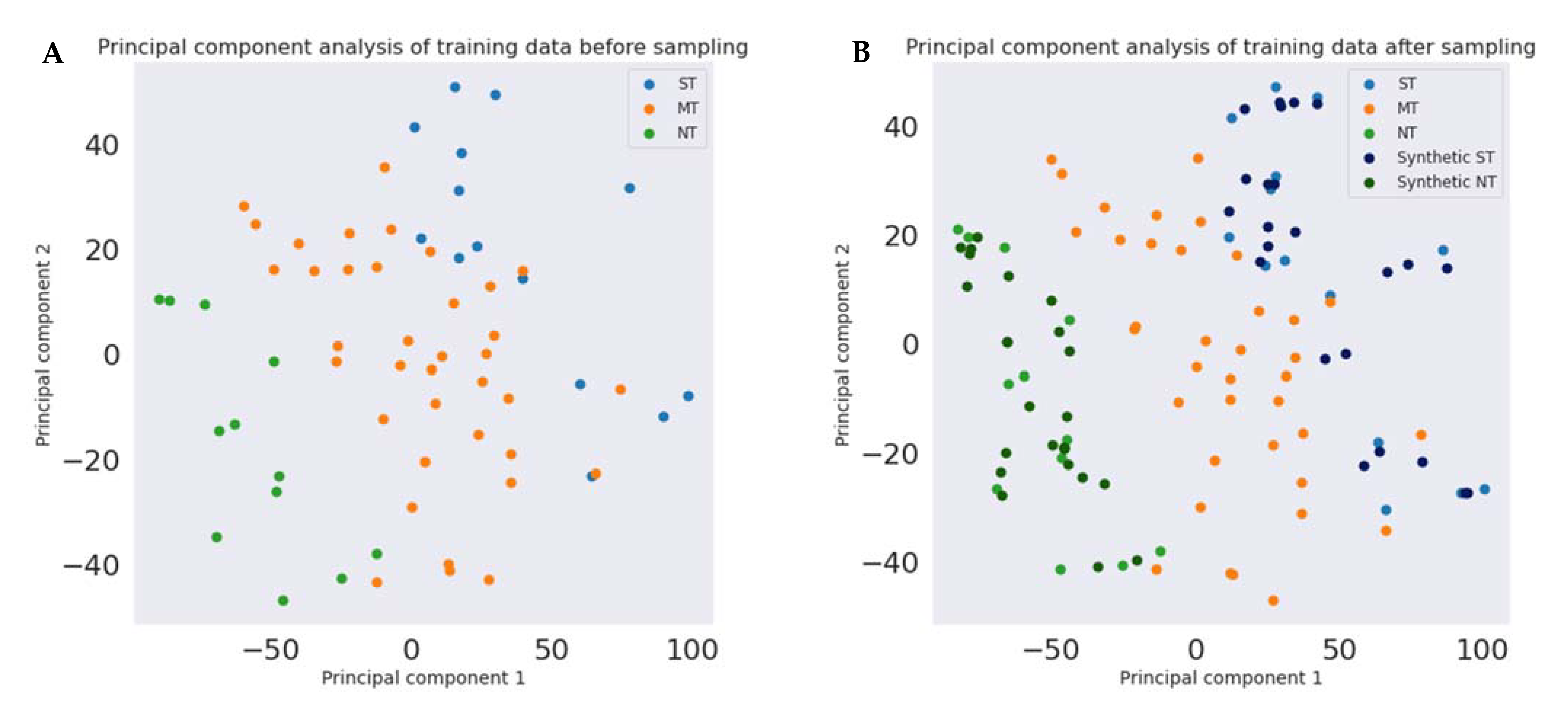
2.7.2. Principal Component Analysis
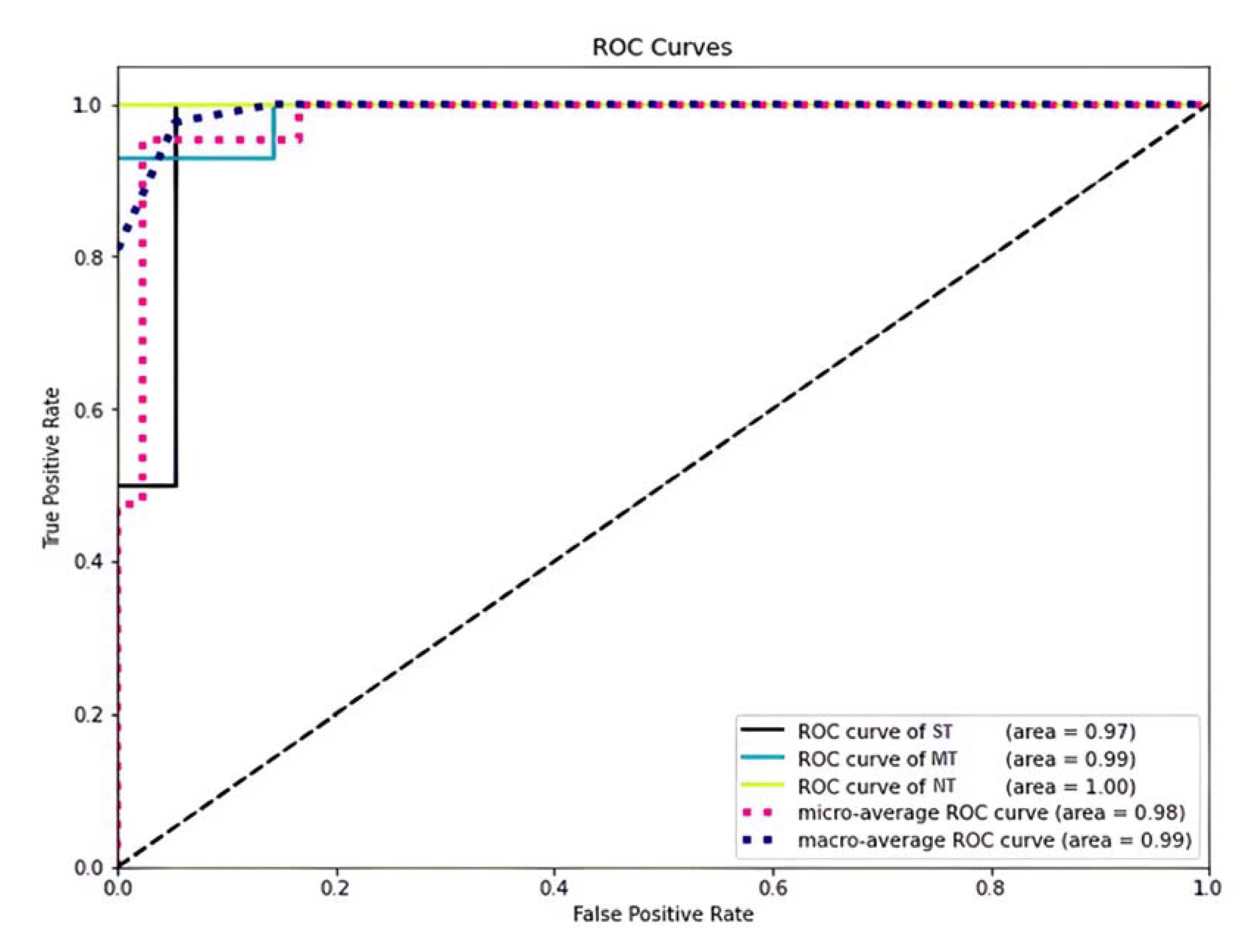
2.7.3. Model Training
2.7.4. Hyperparameter Tuning
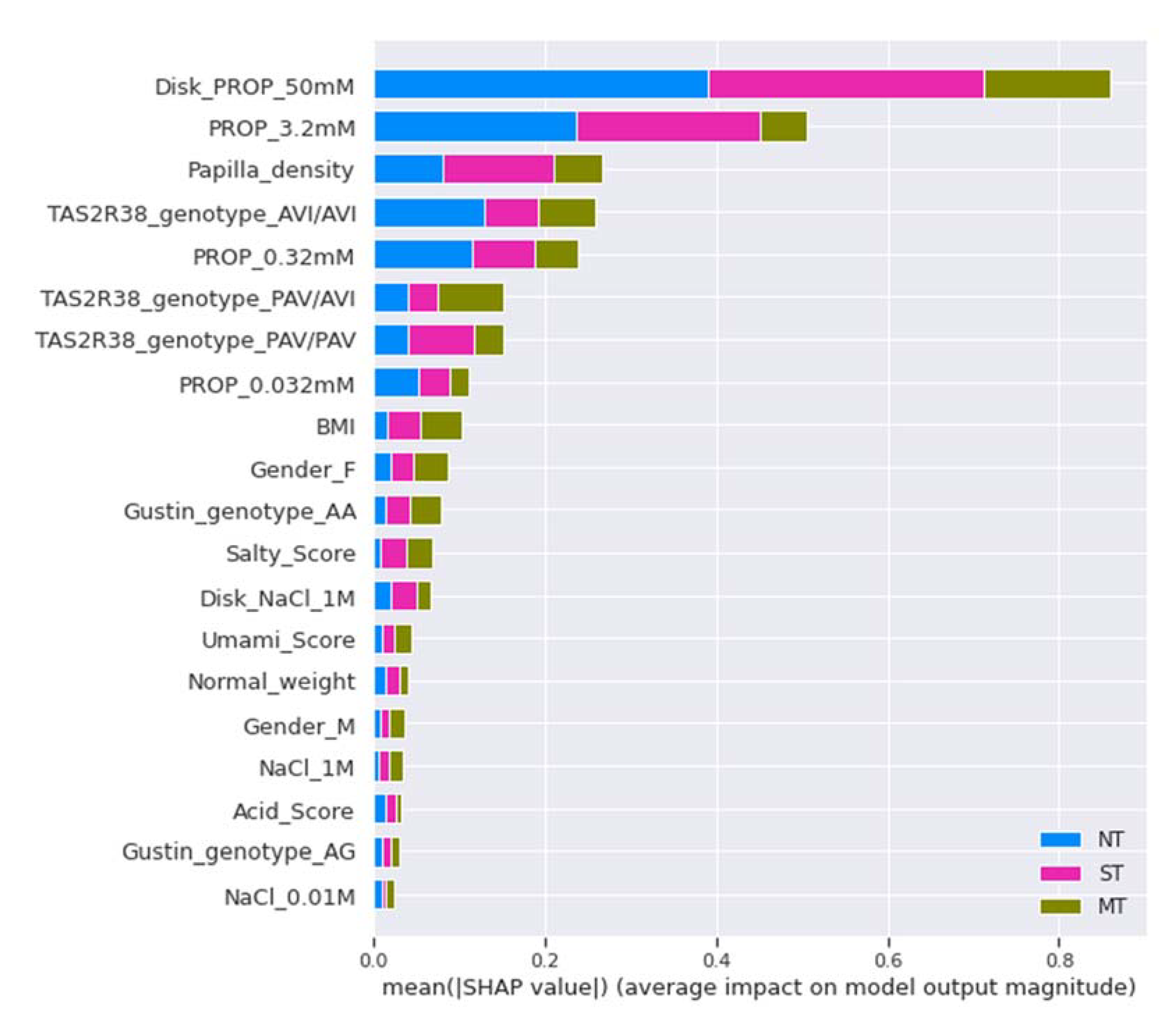
2.7.5. Model Explainability and Feature Importance
2.8. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scott, K. Taste recognition: Food for thought. Neuron 2005, 48, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.; Roper, S.D. The cell biology of taste. J. Cell Biol. 2010, 190, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Tepper, B.J.; Banni, S.; Melis, M.; Crnjar, R.; Tomassini Barbarossa, I. Genetic sensitivity to the bitter taste of 6-n-propylthiouracil (PROP) and its association with physiological mechanisms controlling body mass index (BMI). Nutrients 2014, 6, 3363–3381. [Google Scholar] [CrossRef] [PubMed]
- Tepper, B.J. Nutritional implications of genetic taste variation: The role of PROP sensitivity and other taste phenotypes. Annu. Rev. Nutr. 2008, 28, 367–388. [Google Scholar] [CrossRef]
- Tepper, B.J.; Christensen, C.M.; Cao, J. Development of brief methods to classify individuals by PROP taster status. Physiol. Behav. 2001, 73, 571–577. [Google Scholar] [CrossRef]
- Bartoshuk, L.M.; Duffy, V.B.; Miller, I.J. PTC/PROP tasting: Anatomy, psychophysics, and sex effects. Physiol. Behav. 1994, 56, 1165–1171. [Google Scholar] [CrossRef]
- Zhao, L.; Kirkmeyer, S.V.; Tepper, B.J. A paper screening test to assess genetic taste sensitivity to 6-n-propylthiouracil. Physiol. Behav. 2003, 78, 625–633. [Google Scholar] [CrossRef]
- Hayes, J.E.; Bartoshuk, L.M.; Kidd, J.R.; Duffy, V.B. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem. Senses 2008, 33, 255–265. [Google Scholar] [CrossRef]
- Tepper, B.J.; Nurse, R.J. PROP taster status is related to fat perception and preference. Ann. N. Y. Acad. Sci. 1998, 855, 802–804. [Google Scholar] [CrossRef]
- Bufe, B.; Breslin, P.A.; Kuhn, C.; Reed, D.R.; Tharp, C.D.; Slack, J.P.; Kim, U.K.; Drayna, D.; Meyerhof, W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005, 15, 322–327. [Google Scholar] [CrossRef]
- Kim, U.K.; Jorgenson, E.; Coon, H.; Leppert, M.; Risch, N.; Drayna, D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 2003, 299, 1221–1225. [Google Scholar] [CrossRef]
- Wooding, S.; Kim, U.K.; Bamshad, M.J.; Larsen, J.; Jorde, L.B.; Drayna, D. Natural Selection and Molecular Evolution in PTC, a Bitter-Taste Receptor Gene. Am. J. Hum. Genet. 2004, 74, 637–646. [Google Scholar] [CrossRef]
- Tepper, B.J.; Koelliker, Y.; Zhao, L.; Ullrich, N.V.; Lanzara, C.; d’Adamo, P.; Ferrara, A.; Ulivi, S.; Esposito, L.; Gasparini, P. Variation in the bitter-taste receptor gene TAS2R38, and adiposity in a genetically isolated population in Southern Italy. Obesity 2008, 16, 2289–2295. [Google Scholar] [CrossRef]
- Prodi, D.A.; Drayna, D.; Forabosco, P.; Palmas, M.A.; Maestrale, G.B.; Piras, D.; Pirastu, M.; Angius, A. Bitter taste study in a Sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem. Senses 2004, 29, 697–702. [Google Scholar] [CrossRef][Green Version]
- Melis, M.; Atzori, E.; Cabras, S.; Zonza, A.; Calò, C.; Muroni, P.; Nieddu, M.; Padiglia, A.; Sogos, V.; Tepper, B.J.; et al. The gustin (CA6) gene polymorphism, rs2274333 (A/G), as a mechanistic link between PROP tasting and fungiform taste papilla density and maintenance. PLoS ONE 2013, 8, e74151. [Google Scholar] [CrossRef]
- Tepper, B.J.; Nurse, R.J. Fat perception is related to PROP taster status. Physiol. Behav. 1997, 61, 949–954. [Google Scholar] [CrossRef]
- Yackinous, C.; Guinard, J.X. Relation between PROP taster status and fat perception, touch, and olfaction. Physiol. Behav. 2001, 72, 427–437. [Google Scholar]
- Essick, G.; Chopra, A.; Guest, S.; McGlone, F. Lingual tactile acuity, taste perception, and the density and diameter of fungiform papillae in female subjects. Physiol. Behav. 2003, 80, 289–302. [Google Scholar] [CrossRef]
- Bartoshuk, L.M.; Rifkin, B.; Marks, L.E.; Hooper, J.E. Bitterness of KCl and benzoate: Related to genetic status for sensitivity to PTC/PROP. Chem. Senses 1988, 13, 517–528. [Google Scholar] [CrossRef]
- Gent, J.; Bartoshuk, L. Sweetness of sucrose, neohesperidin dihydrochalcone, and saccharin is related to genetic ability to taste the bitter substance 6-n-propylthiouracil. Chem. Senses 1983, 7, 265–272. [Google Scholar] [CrossRef]
- Bartoshuk, L.M. Bitter taste of saccharin related to the genetic ability to taste the bitter substance 6-n-propylthiouracil. Science 1979, 205, 934–935. [Google Scholar] [CrossRef]
- Prescott, J.; Swain-Campbell, N. Responses to repeated oral irritation by capsaicin, cinnamaldehyde and ethanol in PROP tasters and non-tasters. Chem. Senses 2000, 25, 239–246. [Google Scholar] [CrossRef]
- Prescott, J.; Soo, J.; Campbell, H.; Roberts, C. Responses of PROP taster groups to variations in sensory qualities within foods and beverages. Physiol. Behav. 2004, 82, 459–469. [Google Scholar] [CrossRef]
- Duffy, V.B.; Peterson, J.M.; Bartoshuk, L.M. Associations between taste genetics, oral sensation and alcohol intake. Physiol. Behav. 2004, 82, 435–445. [Google Scholar] [CrossRef]
- Yeomans, M.R.; Tepper, B.J.; Rietzschel, J.; Prescott, J. Human hedonic responses to sweetness: Role of taste genetics and anatomy. Physiol. Behav. 2007, 91, 264–273. [Google Scholar] [CrossRef]
- Melis, M.; Tomassini Barbarossa, I. Taste Perception of Sweet, Sour, Salty, Bitter, and Umami and Changes Due to l-Arginine Supplementation, as a Function of Genetic Ability to Taste 6-n-Propylthiouracil. Nutrients 2017, 9, 541–558. [Google Scholar] [CrossRef]
- Melis, M.; Yousaf, N.Y.; Mattes, M.Z.; Cabras, T.; Messana, I.; Crnjar, R.; Tomassini Barbarossa, I.; Tepper, B.J. Sensory perception of and salivary protein response to astringency as a function of the 6-n-propylthioural (PROP) bitter-taste phenotype. Physiol. Behav. 2017, 173, 163–173. [Google Scholar] [CrossRef]
- Melis, M.; Sollai, G.; Muroni, P.; Crnjar, R.; Barbarossa, I.T. Associations between orosensory perception of oleic acid, the common single nucleotide polymorphisms (rs1761667 and rs1527483) in the CD36 gene, and 6-n-propylthiouracil (PROP) tasting. Nutrients 2015, 7, 2068–2084. [Google Scholar] [CrossRef]
- Kirkmeyer, S.V.; Tepper, B.J. Understanding creaminess perception of dairy products using free-choice profiling and genetic responsivity to 6-n-propylthiouracil. Chem. Senses 2003, 28, 527–536. [Google Scholar] [CrossRef]
- Keller, K.L.; Steinmann, L.; Nurse, R.J.; Tepper, B.J. Genetic taste sensitivity to 6-n-propylthiouracil influences food preference and reported intake in preschool children. Appetite 2002, 38, 3–12. [Google Scholar] [CrossRef]
- Bell, K.I.; Tepper, B.J. Short-term vegetable intake by young children classified by 6-n-propylthoiuracil bitter-taste phenotype. Am. J. Clin. Nutr. 2006, 84, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Dinehart, M.E.; Hayes, J.E.; Bartoshuk, L.M.; Lanier, S.L.; Duffy, V.B. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol. Behav. 2006, 87, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Calò, C.; Padiglia, A.; Zonza, A.; Corrias, L.; Contu, P.; Tepper, B.J.; Barbarossa, I.T. Polymorphisms in TAS2R38 and the taste bud trophic factor, gustin gene co-operate in modulating PROP taste phenotype. Physiol. Behav. 2011, 104, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Padiglia, A.; Zonza, A.; Atzori, E.; Chillotti, C.; Calò, C.; Tepper, B.J.; Barbarossa, I.T. Sensitivity to 6-n-propylthiouracil is associated with gustin (carbonic anhydrase VI) gene polymorphism, salivary zinc, and body mass index in humans. Am. J. Clin. Nutr. 2010, 92, 539–545. [Google Scholar] [CrossRef]
- Genick, U.K.; Kutalik, Z.; Ledda, M.; Destito, M.C.S.; Souza, M.M.; Cirillo, C.A.; Godinot, N.; Martin, N.; Morya, E.; Sameshima, K.; et al. Sensitivity of Genome-Wide-Association Signals to Phenotyping Strategy: The PROP-TAS2R38 Taste Association as a Benchmark. PLoS ONE 2011, 6, e27745. [Google Scholar] [CrossRef]
- Feeney, E.L.; Hayes, J.E. Exploring associations between taste perception, oral anatomy and polymorphisms in the carbonic anhydrase (gustin) gene CA6. Physiol. Behav. 2014, 128, 148–154. [Google Scholar] [CrossRef]
- Drayna, D.; Coon, H.; Kim, U.K.; Elsner, T.; Cromer, K.; Otterud, B.; Baird, L.; Peiffer, A.P.; Leppert, M. Genetic analysis of a complex trait in the Utah Genetic Reference Project: A major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum. Genet. 2003, 112, 567–572. [Google Scholar] [CrossRef]
- Reed, D.R.; Nanthakumar, E.; North, M.; Bell, C.; Bartoshuk, L.M.; Price, R.A. Localization of a gene for bitter-taste perception to human chromosome 5p15. Am. J. Hum. Genet. 1999, 64, 1478–1480. [Google Scholar] [CrossRef]
- Cabras, T.; Melis, M.; Castagnola, M.; Padiglia, A.; Tepper, B.J.; Messana, I.; Tomassini Barbarossa, I. Responsiveness to 6-n-propylthiouracil (PROP) is associated with salivary levels of two specific basic proline-rich proteins in humans. PLoS ONE 2012, 7, e30962. [Google Scholar] [CrossRef]
- Melis, M.; Aragoni, M.C.; Arca, M.; Cabras, T.; Caltagirone, C.; Castagnola, M.; Crnjar, R.; Messana, I.; Tepper, B.J.; Barbarossa, I.T. Marked increase in PROP taste responsiveness following oral supplementation with selected salivary proteins or their related free amino acids. PLoS ONE 2013, 8, e59810. [Google Scholar] [CrossRef]
- Melis, M.; Arca, M.; Aragoni, M.C.; Cabras, T.; Caltagirone, C.; Castagnola, M.; Crnjar, R.; Messana, I.; Tepper, B.J.; Tomassini Barbarossa, I. Dose-Dependent Effects of L-Arginine on PROP Bitterness Intensity and Latency and Characteristics of the Chemical Interaction between PROP and L-Arginine. PLoS ONE 2015, 10, e0131104. [Google Scholar] [CrossRef]
- Goldstein, G.L.; Daun, H.; Tepper, B.J. Influence of PROP taster status and maternal variables on energy intake and body weight of pre-adolescents. Physiol. Behav. 2007, 90, 809–817. [Google Scholar] [CrossRef]
- Whissell-Buechy, D.; Wills, C. Male and female correlations for taster (P.T.C.) phenotypes and rate of adolescent development. Ann. Hum. Biol. 1989, 16, 131–146. [Google Scholar] [CrossRef]
- Prutkin, J.; Fisher, E.M.; Etter, L.; Fast, K.; Gardner, E.; Lucchina, L.A.; Snyder, D.J.; Tie, K.; Weiffenbach, J.; Bartoshuk, L.M. Genetic variation and inferences about perceived taste intensity in mice and men. Physiol. Behav. 2000, 69, 161–173. [Google Scholar] [CrossRef]
- Zuniga, J.R.; Davis, S.H.; Englehardt, R.A.; Miller, I.J.; Schiffrman, S.S.; Phillips, C. Taste performance on the anterior human tongue varles with fungiform taste bud density. Chem. Senses 1993, 18, 449–460. [Google Scholar] [CrossRef]
- Correa, M.; Hutchinson, I.; Laing, D.G.; Jinks, A.L. Changes in Fungiform Papillae Density During Development in Humans. Chem. Senses 2013, 38, 519–527. [Google Scholar] [CrossRef]
- Tepper, B.J. 6-n-Propylthiouracil: A genetic marker for taste, with implications for food preference and dietary habits. Am. J. Hum. Genet. 1998, 63, 1271–1276. [Google Scholar] [CrossRef]
- Bartoshuk, L.M. The biological basis of food perception and acceptance. Food Qual. Prefer. 1993, 4, 21–32. [Google Scholar] [CrossRef]
- Bartoshuk, L.; Fast, K.; Karrer, T.; Marino, S.; Price, R.; Reed, D. PROP supertasters and the perception of sweetness and bitterness. Chem. Senses 1992, 17, 594. [Google Scholar]
- Bartoshuk, L.M.; Rifkin, B.; Marks, L.E.; Bars, P. Taste and aging. J. Gerontol. 1986, 41, 51–57. [Google Scholar] [CrossRef]
- Melis, M.; Errigo, A.; Crnjar, R.; Pes, G.M.; Tomassini Barbarossa, I. TAS2R38 bitter taste receptor and attainment of exceptional longevity. Sci. Rep. 2019, 9, 18047. [Google Scholar] [CrossRef]
- Whissell-Buechy, D. Effects of age and sex on taste sensitivity to phenylthiocarbamide (PTC) in the Berkeley Guidance sample. Chem. Senses 1990, 15, 39–57. [Google Scholar] [CrossRef]
- Mennella, J.; Pepino, M.Y.; Duke, F.; Reed, D. Age modifies the genotype-phenotype relationship for the bitter receptor TAS2R38. BMC Genet. 2010, 11, 60. [Google Scholar] [CrossRef]
- Tepper, B.J.; Williams, T.Z.; Burgess, J.R.; Antalis, C.J.; Mattes, R.D. Genetic variation in bitter taste and plasma markers of anti-oxidant status in college women. Int. J. Food Sci. Nutr. 2009, 60 (Suppl. 2), 35–45. [Google Scholar] [CrossRef]
- Tepper, B.J.; Neilland, M.; Ullrich, N.V.; Koelliker, Y.; Belzer, L.M. Greater energy intake from a buffet meal in lean, young women is associated with the 6-n-propylthiouracil (PROP) non-taster phenotype. Appetite 2011, 56, 104–110. [Google Scholar] [CrossRef]
- Tepper, B.J.; Ullrich, N.V. Influence of genetic taste sensitivity to 6-n-propylthiouracil (PROP), dietary restraint and disinhibition on body mass index in middle-aged women. Physiol. Behav. 2002, 75, 305–312. [Google Scholar] [CrossRef]
- Lumeng, J.C.; Cardinal, T.M.; Sitto, J.R.; Kannan, S. Ability to Taste 6-n-propylthiouracil and Body Mass Index in Low-Income Preschool-Aged Children. Obesity (Silver Spring) 2008, 16, 1522–1528. [Google Scholar] [CrossRef]
- Carta, G.; Melis, M.; Pintus, S.; Pintus, P.; Piras, C.A.; Muredda, L.; Demurtas, D.; Di Marzo, V.; Banni, S.; Barbarossa, I.T. Participants with Normal Weight or with Obesity Show Different Relationships of 6-n-Propylthiouracil (PROP) Taster Status with BMI and Plasma Endocannabinoids. Sci. Rep. 2017, 7, 1361. [Google Scholar] [CrossRef]
- Tomassini Barbarossa, I.; Carta, G.; Murru, E.; Melis, M.; Zonza, A.; Vacca, C.; Muroni, P.; Di Marzo, V.; Banni, S. Taste sensitivity to 6-n-propylthiouracil is associated with endocannabinoid plasma levels in normal-weight individuals. Nutrition 2013, 29, 531–536. [Google Scholar] [CrossRef]
- Enoch, M.A.; Harris, C.R.; Goldman, D. Does a reduced sensitivity to bitter taste increase the risk of becoming nicotine addicted? Addict. Behav. 2001, 26, 399–404. [Google Scholar] [CrossRef]
- Mangold, J.E.; Payne, T.J.; Ma, J.Z.; Chen, G.; Li, M.D. Bitter taste receptor gene polymorphisms are an important factor in the development of nicotine dependence in African Americans. J. Med. Genet. 2008, 45, 578–582. [Google Scholar] [CrossRef]
- Risso, D.S.; Kozlitina, J.; Sainz, E.; Gutierrez, J.; Wooding, S.; Getachew, B.; Luiselli, D.; Berg, C.J.; Drayna, D. Genetic Variation in the TAS2R38 Bitter Taste Receptor and Smoking Behaviors. PLoS ONE 2016, 11, e0164157. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Cohen, N.A. Role of the bitter taste receptor T2R38 in upper respiratory infection and chronic rhinosinusitis. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Xiong, G.; Kofonow, J.M.; Chen, B.; Lysenko, A.; Jiang, P.; Abraham, V.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Investig. 2012, 122, 4145–4159. [Google Scholar] [CrossRef] [PubMed]
- Melis, M.; Grzeschuchna, L.; Sollai, G.; Hummel, T.; Tomassini Barbarossa, I. Taste disorders are partly genetically determined: Role of the TAS2R38 gene, a pilot study. Laryngoscope 2019, 129, E307–E312. [Google Scholar] [CrossRef]
- Basson, M.D.; Bartoshuk, L.M.; Dichello, S.Z.; Panzini, L.; Weiffenbach, J.M.; Duffy, V.B. Association between 6-n-propylthiouracil (PROP) bitterness and colonic neoplasms. Dig. Dis. Sci. 2005, 50, 483–489. [Google Scholar] [CrossRef]
- Carrai, M.; Steinke, V.; Vodicka, P.; Pardini, B.; Rahner, N.; Holinski-Feder, E.; Morak, M.; Schackert, H.K.; Gorgens, H.; Stemmler, S.; et al. Association between TAS2R38 gene polymorphisms and colorectal cancer risk: A case-control study in two independent populations of Caucasian origin. PLoS ONE 2011, 6, e20464. [Google Scholar] [CrossRef]
- Cossu, G.; Melis, M.; Sarchioto, M.; Melis, M.; Melis, M.; Morelli, M.; Tomassini Barbarossa, I. 6-n-propylthiouracil taste disruption and TAS2R38 nontasting form in Parkinson’s disease. Mov. Disord. 2018, 33, 1331–1339. [Google Scholar] [CrossRef]
- Gorovic, N.; Afzal, S.; Tjonneland, A.; Overvad, K.; Vogel, U.; Albrechtsen, C.; Poulsen, H.E. Genetic variation in the hTAS2R38 taste receptor and brassica vegetable intake. Scand. J. Clin. Lab. Investig. 2011, 71, 274–279. [Google Scholar] [CrossRef]
- Feeney, E.; O’Brien, S.; Scannell, A.; Markey, A.; Gibney, E.R. Genetic variation in taste perception: Does it have a role in healthy eating? Proc. Nutr. Soc. 2011, 70, 135–143. [Google Scholar] [CrossRef]
- Baranowski, T.; Baranowski, J.C.; Watson, K.B.; Jago, R.; Islam, N.; Beltran, A.; Martin, S.J.; Nguyen, N.; Tepper, B.J. 6-n-propylthiouracil taster status not related to reported cruciferous vegetable intake among ethnically diverse children. Nutr. Res. 2011, 31, 594–600. [Google Scholar] [CrossRef]
- Drewnowski, A.; Henderson, S.A.; Cockroft, J.E. Genetic Sensitivity to 6-n-Propylthiouracil Has No Influence on Dietary Patterns, Body Mass Indexes, or Plasma Lipid Profiles of Women. J. Am. Diet. Assoc. 2007, 107, 1340–1348. [Google Scholar] [CrossRef]
- Duffy, V.B.; Bartoshuk, L.M. Food acceptance and genetic variation in taste. J. Am. Diet Assoc. 2000, 100, 647–655. [Google Scholar] [CrossRef]
- Mennella, J.A.; Pepino, M.Y.; Reed, D.R. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics 2005, 115, e216–e222. [Google Scholar] [CrossRef]
- O’Brien, S.A.; Feeney, E.L.; Scannell, A.G.; Markey, A.; Gibney, E.R. Bitter taste perception and dietary intake patterns in irish children. J. Nutrigenet. Nutr. 2013, 6, 43–58. [Google Scholar] [CrossRef]
- Kaminski, L.C.; Henderson, S.A.; Drewnowski, A. Young women’s food preferences and taste responsiveness to 6-n-propylthiouracil (PROP). Physiol. Behav. 2000, 68, 691–697. [Google Scholar] [CrossRef]
- Timpson, N.J.; Christensen, M.; Lawlor, D.A.; Gaunt, T.R.; Day, I.N.; Ebrahim, S.; Davey Smith, G. TAS2R38 (phenylthiocarbamide) haplotypes, coronary heart disease traits, and eating behavior in the British Women’s Heart and Health Study. Am. J. Clin. Nutr. 2005, 81, 1005–1011. [Google Scholar] [CrossRef]
- Yackinous, C.A.; Guinard, J.X. Relation between PROP (6-n-propylthiouracil) taster status, taste anatomy and dietary intake measures for young men and women. Appetite 2002, 38, 201–209. [Google Scholar] [CrossRef]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion. 2019, 50, 71–91. [Google Scholar] [CrossRef]
- Tepper, B.J.; Melis, M.; Koelliker, Y.; Gasparini, P.; Ahijevych, K.L.; Tomassini Barbarossa, I. Factors Influencing the Phenotypic Characterization of the Oral Marker, PROP. Nutrients 2017, 9, 1275. [Google Scholar] [CrossRef]
- Barbarossa, I.T.; Melis, M.; Mattes, M.Z.; Calò, C.; Muroni, P.; Crnjar, R.; Tepper, B.J. The gustin (CA6) gene polymorphism, rs2274333 (A/G), is associated with fungiform papilla density, whereas PROP bitterness is mostly due to TAS2R38 in an ethnically-mixed population. Physiol. Behav. 2015, 138, 6–12. [Google Scholar] [CrossRef]
- Glanville, E.V.; Kaplan, A.R. Taste Perception and the Menstrual Cycle. Nature 1965, 205, 930–931. [Google Scholar] [CrossRef]
- Landis, B.N.; Welge-Luessen, A.; Bramerson, A.; Bende, M.; Mueller, C.A.; Nordin, S.; Hummel, T. “Taste Strips”—A rapid, lateralized, gustatory bedside identification test based on impregnated filter papers. J. Neurol. 2009, 256, 242–248. [Google Scholar] [CrossRef]
- Mueller, C.; Kallert, S.; Renner, B.; Stiassny, K.; Temmel, A.F.; Hummel, T.; Kobal, G. Quantitative assessment of gustatory function in a clinical context using impregnated “taste strips”. Rhinology 2003, 41, 2–6. [Google Scholar]
- Mueller, C.A.; Pintscher, K.; Renner, B. Clinical Test of Gustatory Function Including Umami Taste. Ann. Otol. Rhinol. Laryngol. 2011, 120, 358–362. [Google Scholar] [CrossRef]
- Sollai, G.; Melis, M.; Mastinu, M.; Pani, D.; Cosseddu, P.; Bonfiglio, A.; Crnjar, R.; Tepper, B.J.; Tomassini Barbarossa, I. Human Tongue Electrophysiological Response to Oleic Acid and Its Associations with PROP Taster Status and the CD36 Polymorphism (rs1761667). Nutrients 2019, 11, 315. [Google Scholar] [CrossRef]
- Sollai, G.; Melis, M.; Pani, D.; Cosseddu, P.; Usai, I.; Crnjar, R.; Bonfiglio, A.; Tomassini Barbarossa, I. First objective evaluation of taste sensitivity to 6-n-propylthiouracil (PROP), a paradigm gustatory stimulus in humans. Sci. Rep. 2017, 7, 40353. [Google Scholar] [CrossRef]
- Melis, M.; Sollai, G.; Mastinu, M.; Pani, D.; Cosseddu, P.; Bonfiglio, A.; Crnjar, R.; Tepper, B.J.; Barbarossa, I.T. Electrophysiological Responses from the Human Tongue to the Six Taste Qualities and Their Relationships with PROP Taster Status. Nutrients 2020, 12, 2017. [Google Scholar] [CrossRef]
- Green, B.G.; Shaffer, G.S.; Gilmore, M.M. Derivation and evaluation of a semantic scale of oral sensation magnitude with apparent ratio properties. Chem. Senses 1993, 18, 683–702. [Google Scholar] [CrossRef]
- Bartoshuk, L.M.; Duffy, V.B.; Green, B.G.; Hoffman, H.J.; Ko, C.W.; Lucchina, L.A.; Marks, L.E.; Snyder, D.J.; Weiffenbach, J.M. Valid across-group comparisons with labeled scales: The gLMS versus magnitude matching. Physiol. Behav. 2004, 82, 109–114. [Google Scholar] [CrossRef]
- Lipchock, S.V.; Mennella, J.A.; Spielman, A.I.; Reed, D.R. Human bitter perception correlates with bitter receptor messenger RNA expression in taste cells. Am. J. Clin. Nutr. 2013, 98, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Reedy, F.E. Variations in human taste bud density and taste intensity perception. Physiol. Behav. 1990, 47, 1213–1219. [Google Scholar] [CrossRef]
- Boxer, E.E.; Garneau, N.L. Rare haplotypes of the gene TAS2R38 confer bitter taste sensitivity in humans. SpringerPlus 2015, 4, 505. [Google Scholar] [CrossRef] [PubMed]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 3 December 2018; pp. 6639–6649. [Google Scholar]
- Jiang, J.; Wang, R.; Wang, M.; Gao, K.; Nguyen, D.D.; Wei, G.-W. Boosting Tree-Assisted Multitask Deep Learning for Small Scientific Datasets. J. Chem. Inf. Modeling 2020, 60, 1235–1244. [Google Scholar] [CrossRef]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Barua, S.; Islam, M.M.; Murase, K. A Novel Synthetic Minority Oversampling Technique for Imbalanced Data Set Learning. In Neural Information Processing. ICONIP 2011. Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2011; Volume 7063. [Google Scholar]
- Browne, M.W. Cross-Validation Methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef]
- Wu, J.; Chen, X.-Y.; Zhang, H.; Xiong, L.-D.; Lei, H.; Deng, S.-H. Hyperparameter Optimization for Machine Learning Models Based on Bayesian Optimization. J. Electron. Sci. Technol. 2019, 17, 26–40. [Google Scholar]
- Zahedi, L.; Mohammadi, F.; Rezapour, S.; Ohland, M.W.; Amini, M.H. Search Algorithms for Automated Hyper-Parameter Tuning. arXiv Prepr. 2021, arXiv:2104.14677. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4 December 2017; pp. 4768–4777. [Google Scholar]
- Rousset, F. GENEPOP′007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Duffy, V.B.; Davidson, A.C.; Kidd, J.R.; Kidd, K.K.; Speed, W.C.; Pakstis, A.J.; Reed, D.R.; Snyder, D.J.; Bartoshuk, L.M. Bitter Receptor Gene (TAS2R38), 6-n-Propylthiouracil (PROP) Bitterness and Alcohol Intake. Alcohol. Clin. Exp. Res. 2004, 28, 1629–1637. [Google Scholar] [CrossRef]
- Althnian, A.; AlSaeed, D.; Al-Baity, H.; Samha, A.; Dris, A.B.; Alzakari, N.; Abou Elwafa, A.; Kurdi, H. Impact of Dataset Size on Classification Performance: An Empirical Evaluation in the Medical Domain. Appl. Sci. 2021, 11, 796. [Google Scholar] [CrossRef]

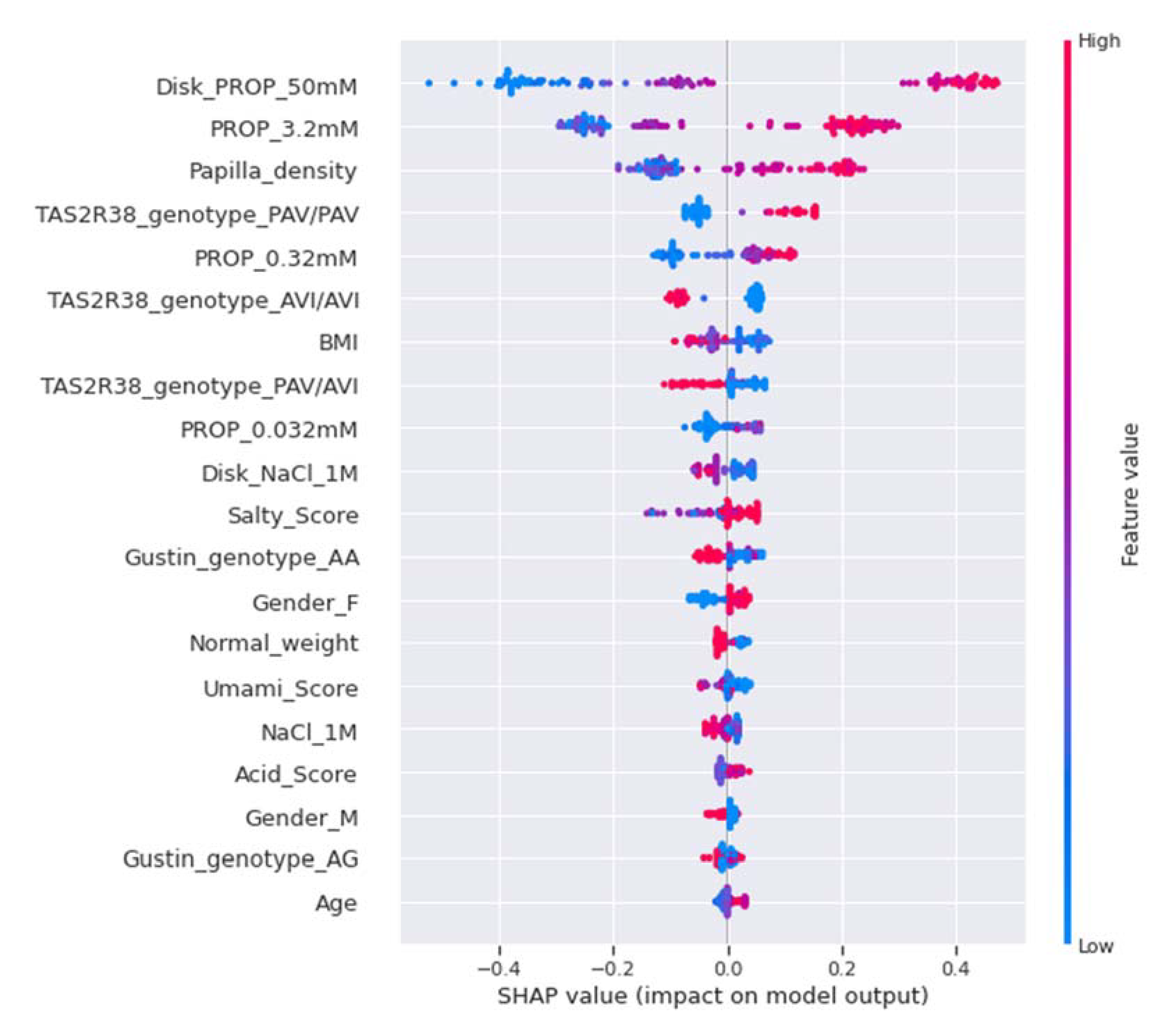
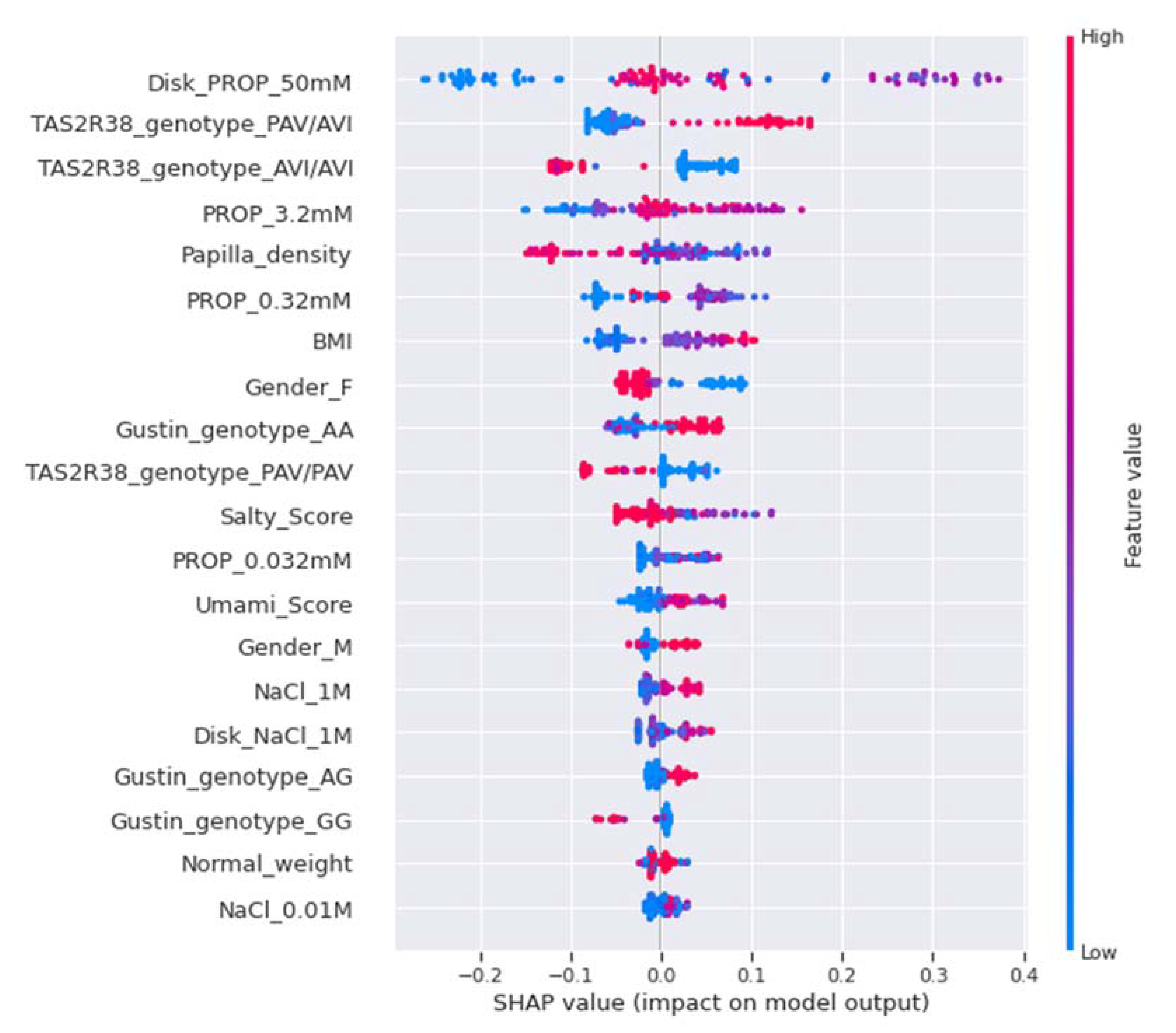

| Features | Overall | ST (n = 16) | MT (n = 51) | NT (n = 17) |
|---|---|---|---|---|
| Age (year) | 25.07 ± 0.46 | 24.94 ± 1.07 | 24.95 ± 0.60 | 25.59 ± 1.03 |
| BMI (kg/m2) | 21.82 ± 0.36 | 20.83 ± 0.82 | 22.27 ± 0.46 | 21.41 ± 0.79 |
| Papilla density/cm2 | 30.63 ± 1.58 | 39.04 ± 3.39 a | 30.98 ± 1.90 b | 21.68 ± 3.29 c |
| Male/female (n) | 35/49 | 5/11 | 24/27 | 6/11 |
| Smokers/non-smokers (n) | 19/65 | 3/13 | 13/38 | 3/14 |
| Genotypes | ||||
| TAS2R38 | ||||
| PP/PA/AA (n) | 20/43/21 | 10/6/0 x | 10/35/6 y | 0/2/15 z |
| Gustin gene | ||||
| AA/AG/GG (n) | 49/29/6 | 9/4/3 | 27/22/2 | 13/3/1 |
| Taste scores | ||||
| Sweet | 3.43 ± 0.07 | 3.37 ± 0.17 | 3.43 ± 0.09 | 3.47 ± 0.16 |
| Salty | 3.57 ± 0.08 | 3.81 ± 0.17 | 3.51 ± 0.10 | 3.53 ± 0.17 |
| Sour | 2.38 ± 0.10 | 2.44 ± 0.24 | 2.41 ± 0.13 | 2.23 ± 0.23 |
| Bitter | 3.21 ± 0.11 | 3.50 ± 0.26 | 3.15 ± 0.15 | 3.11 ± 0.25 |
| Umami | 1.32 ± 0.16 | 1.12 ± 0.38 | 1.43 ± 0.21 | 1.17 ± 0.37 |
| TST | 12.79 ± 0.22 | 13.19 ± 0.50 | 12.80 ± 0.28 | 12.41 ± 0.49 |
| Overall TST | 13.92 ± 0.31 | 14.25 ± 0.72 | 13.94 ± 0.40 | 13.53 ± 0.70 |
| Classifiers | Precision | Recall | F1-Score |
|---|---|---|---|
| Logistic regression | 83% | 81% | 81% |
| Gradient boosting | 90% | 86% | 87% |
| Decision trees | 92% | 90% | 91% |
| Random forests | 96% | 95% | 95% |
| CatBoost | 97% | 95% | 96% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naciri, L.C.; Mastinu, M.; Crnjar, R.; Tomassini Barbarossa, I.; Melis, M. Automated Classification of 6-n-Propylthiouracil Taster Status with Machine Learning. Nutrients 2022, 14, 252. https://doi.org/10.3390/nu14020252
Naciri LC, Mastinu M, Crnjar R, Tomassini Barbarossa I, Melis M. Automated Classification of 6-n-Propylthiouracil Taster Status with Machine Learning. Nutrients. 2022; 14(2):252. https://doi.org/10.3390/nu14020252
Chicago/Turabian StyleNaciri, Lala Chaimae, Mariano Mastinu, Roberto Crnjar, Iole Tomassini Barbarossa, and Melania Melis. 2022. "Automated Classification of 6-n-Propylthiouracil Taster Status with Machine Learning" Nutrients 14, no. 2: 252. https://doi.org/10.3390/nu14020252
APA StyleNaciri, L. C., Mastinu, M., Crnjar, R., Tomassini Barbarossa, I., & Melis, M. (2022). Automated Classification of 6-n-Propylthiouracil Taster Status with Machine Learning. Nutrients, 14(2), 252. https://doi.org/10.3390/nu14020252










