Abstract
The combination of multiple omics approaches has emerged as an innovative holistic scope to provide a more comprehensive view of the molecular and physiological events underlying human diseases (including obesity, dyslipidemias, fatty liver, insulin resistance, and inflammation), as well as for elucidating unique and specific metabolic phenotypes. These omics technologies include genomics (polymorphisms and other structural genetic variants), epigenomics (DNA methylation, histone modifications, long non-coding RNA, telomere length), metagenomics (gut microbiota composition, enterotypes), transcriptomics (RNA expression patterns), proteomics (protein quantities), and metabolomics (metabolite profiles), as well as interactions with dietary/nutritional factors. Although more evidence is still necessary, it is expected that the incorporation of integrative omics could be useful not only for risk prediction and early diagnosis but also for guiding tailored dietary treatments and prognosis schemes. Some challenges include ethical and regulatory issues, the lack of robust and reproducible results due to methodological aspects, the high cost of omics methodologies, and high-dimensional data analyses and interpretation. In this review, we provide examples of system biology studies using multi-omics methodologies to unravel novel insights into the mechanisms and pathways connecting the genotype to clinically relevant traits and therapy outcomes for precision nutrition applications in health and disease.
1. Introduction
Precision nutrition integrates information at scale by taking into account endogenous individuals’ backgrounds, but also exogenous factors including lifestyle aspects, cultural, socioeconomic and psychosocial characteristics, and food environments [1]. Thus, precision nutrition adopts a whole and dynamic scope to develop comprehensive tailored dietary recommendations for individuals and population subgroups centered on potentiating human health and nutritional wellbeing, as well as the prevention and management of chronic diseases [2,3].
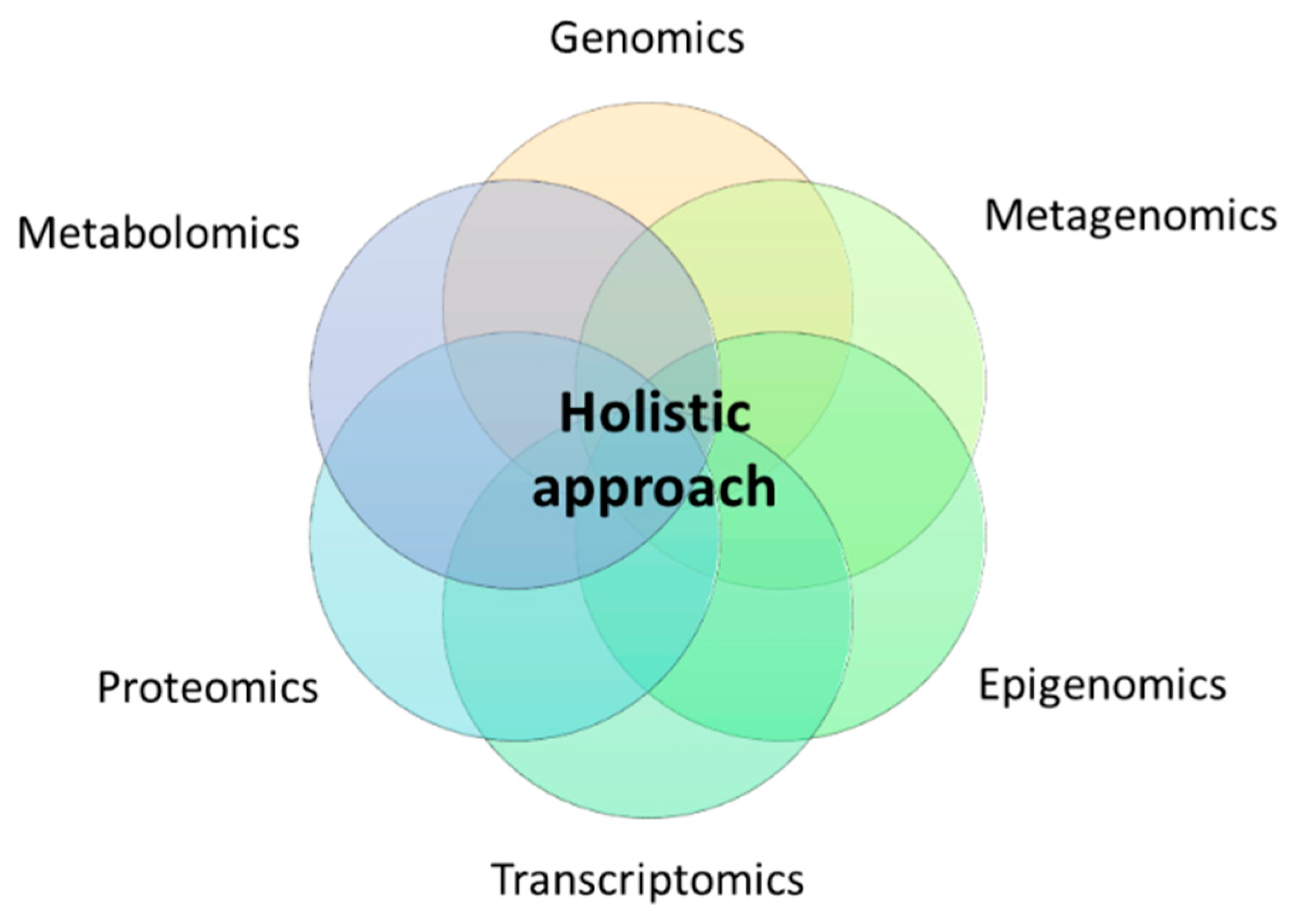
Thus, the notion of precision nutrition should contemplate in-depth metabolic phenotyping using high-throughput omics technologies such as genomics (polymorphisms and other structural genetic variants), epigenomics (DNA methylation, histone modifications, long non-coding RNA, telomere length), metagenomics (gut microbiota composition, enterotypes), transcriptomics (RNA expression patterns), proteomics (protein signatures), and metabolomics (metabolite profiles) under a holistic approach (Figure 1).
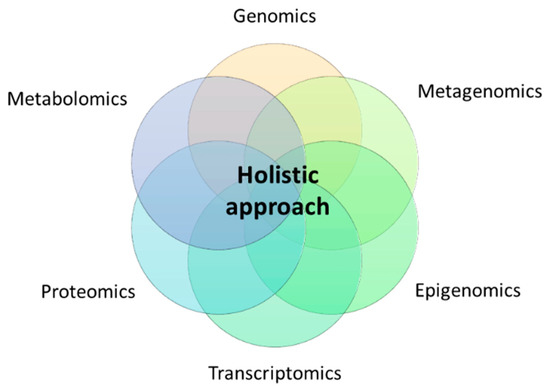
Figure 1.
Holistic approach for precision nutrition using multi-omics technologies.
However, because each method cannot individually explain entirely metabolic fingerprints, the bioinformatic integration of multiple omics skills has emerged as an innovative scope to provide a more comprehensive view of the molecular and physiological events leading to human disease [4]. Moreover, the concurrent application of these tools is helping to elucidate unique and specific phenotypes, enabling the design and implementation of personalized medicine schemes for precision health [5].
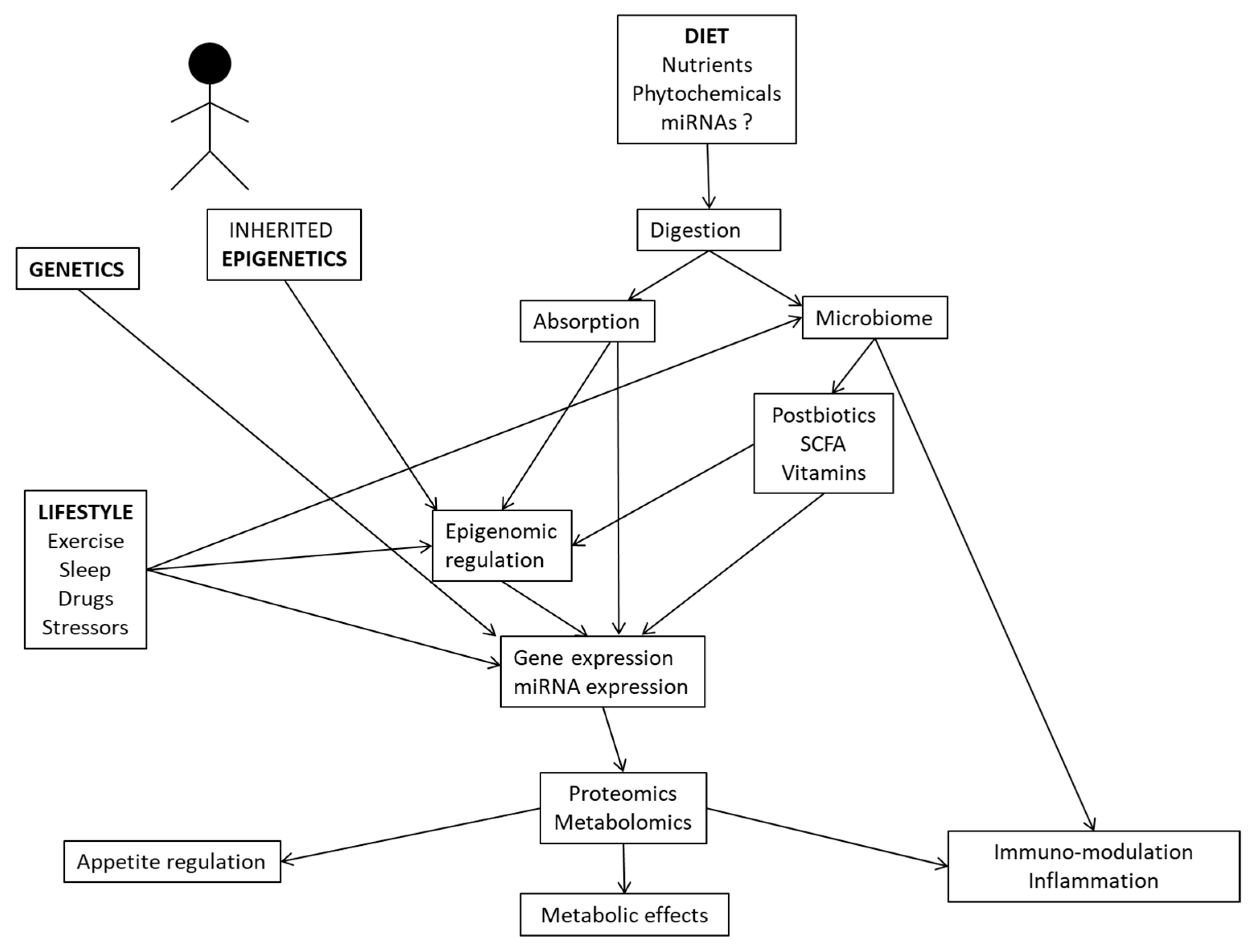
In this review, we provide examples of system biology studies using multi-omics methodologies (comprising genomics, metagenomics, epigenomics, transcriptomics, proteomics, and metabolomics) and interactions with dietary/nutritional factors to unravel novel insights into the mechanisms and pathways connecting the genotype to clinically relevant traits and therapy outcomes for precision nutrition applications in health and disease. In this regard, complex relationships between the genetic background, inherited epigenetics, nutrient utilization, microbiome-derived metabolites, and lifestyle factors may modulate gene and protein expressions, with implications for physiological processes, inflammation and metabolic phenotypes (Figure 2).
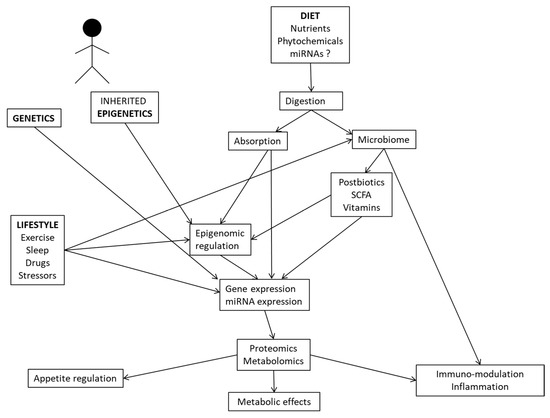
Figure 2.
Interrelationships between multi-omics tools and environmental factors influencing human metabolism and physiological functions.
2. Genomics in Combination Epigenomics, Metagenomics, Transcriptomics, Proteomics or Metabolomics Tools
The integration of genomics with other functional omics analyses may significantly improve the accuracy of host genetic data for explaining health outcomes [6]. For example, using multiple machine learning algorithms, best predictors of obesity status were identified, and included single-nucleotide polymorphisms (mapped to genes such as STXBP6, BBX, PLXDC2, PCDH15, TPH2, PCDH15, CALN1, FGF14, LRRN1, ACTBP2, RBMXP1, and ZNF32) together with differentially methylated sites (in proximity to CPT1A, ABCG1, SLC7A11, RNF145, and SREBF1 genes) and interactions with dietary factors encompassing specific foods, micronutrients, and bioactive compounds [7]. Additionally, the consortium of single-nucleotide polymorphisms in genes related to obesity and cardiometabolic diseases, low adherence to the Mediterranean diet, and harboring specific urolithin metabotypes (as biomarkers of the gut microbiota), was able to predict obesity in childhood and adolescence [8].
Intriguingly, higher milk intake in genetically lactase non-persistent individuals was reported to be associated with increased quantities of gut Bifidobacterium and serum concentrations of indolepropionate, a microbial-derived tryptophan metabolite inversely associated with type 2 diabetes risk [9]. Comparably, PNPLA3 gene variants, gut bacterial features (low abundances of Faecalibacterium or Prevotella, and high abundances of Gemmiger taxa), and specific dietary factors (low in fiber and vitamins as well as enriched in amino acids, uric acid and purine) were all associated with different histology features in non-alcoholic fatty liver disease [10]. Moreover, relevant interrelationships between gut Prevotellaceae and an obesity-related genetic risk score determined interindividual BMI differences in women [11]. Meanwhile, it was demonstrated that the microbiomes of subjects with low copy numbers of the AMY1 locus had enhanced capacity to break down dietary complex carbohydrates [12].
Notably, the methylation status of the APOA2 gene was associated with the intake of saturated fat and the APOA2 -265T>C genotype, promoting a differential APOA2 mRNA expression between APOA2 genotypes and modulating tryptophan and branched-chain amino acid (BCAAs) metabolic pathways [13]. Meanwhile, significant combined and interactive effects between two dietary factors related to gut microbiota (allium vegetables and overnight meal) and polymorphisms in the miRNA binding site of IL13 gene were detected in relation to the susceptibility to colorectal cancer risk [14]. Additionally, the LPL variant rs13702 induced an allele-specific regulation of the LPL gene, affecting blood lipid traits through the disruption of miR-410 binding sites, where interactions with dietary polyunsaturated fatty acid (PUFAs) played an important role [15].
Although caution should be exercised, an exploratory analysis suggested that the maternal FADS2 rs174575 genotype, combined with DNA methylation status in this gene, could be related to plasma fatty acid concentrations in toddlers [16]. Similarly, a meta-analysis revealed that higher ABCA1 promoter cg14019050 methylation correlated with lower ABCA1 expression and was concomitantly associated with the C allele of the ABCA1 rs2246293 variant and lower circulating eicosapentaenoic acid [17]. Genomic, epigenomic, and lipidomic analyses also showed that polymorphisms and methylation sites within the FADS1/2 region influenced the plasma levels of arachidonic acid in response to a high-fat meal in humans [18]. Of note, genomic assays of post-prandial lipidomic features after dietary fat intake identified potential biomarkers of cardiovascular risk including two polymorphisms in the SORBS1 gene (rs12247017 and rs12240292) affecting b-Sitosterol plasma concentrations [19].
The AA genotype of the Cdx-2 VDR polymorphism was associated with higher methylation of the VDR gene promoter and lower serum levels of 25-hydroxyvitamin D in infertile men [20]. Consistently, correlations between vitamin D intake and the expression of miRNA let-7a/b varied with VDR BsmI genotype in an elderly cohort [21]. Moreover, the blood levels of homocysteine were influenced by the dietary intakes of methyl group donors (methionine and 5-methyltetrahydrofolate), whose interactions with methylation-related gene polymorphisms (such as 2R3R-TS, C1420T-SHMT, A2756G-MS, and 844ins68-CBS) modified the risk of adenomatous polyps, a precursor of colorectal cancer [22]. In this regard, it was demonstrated that an miRNA binding site polymorphism (rs1062980) in the iron regulatory pathway, together with dietary iron intake, may modify the risk of lung cancer tumorigenesis [23]. Likewise, the MTHFR C677T polymorphism influenced genomic DNA methylation in peripheral blood mononuclear cells depending on folate status, where only subjects carrying the T/T genotype with low serum levels of folate accounted for reduced DNA methylation [24]. Accordingly, DNA methylation was directly related to folate concentrations in red blood cells in subjects carrying the T/T genotype of the MTHFR C677T polymorphism, but not in those with wild-type [25]. Choline intake also interacted with the MTHFR C677T genotype to influence changes in genomic DNA methylation and DNA damage in folate-compromised Mexican American men [26].
Remarkably, prospective analyses in different population cohorts showed that the habitual intake of food source B-vitamins may modify the effects of DNA methylation-related variants at SREBF1 and HIF3A genes on long-term adiposity changes [27,28]. In fact, interactions of fat intake with genetic (rs11150675), transcriptional (ILMN_1725441), and methylation (cg26663590) variations at the NFATC2IP locus mediated weight loss rates in response to dietary intervention [29]. However, an integrative model using microbiota and genetic information was proposed to prescribe two hypocaloric diets with different micronutrient distribution for a successful weight loss in individuals with excess of body weight [30]. Moreover, diet induced weight loss led to polymorphism-dependent modulation of miRNAs from the miR 25/93/106 gene cluster in humans [31].
Additionally, protein quantitative trait locus analyses provided evidence for distinct genetic mechanisms regulating BMI-associated proteins during diet-induced weight loss, including those associated with leptin protein expression changes [32]. Consistently, multivariate proteomic analysis using data from two clinical cohorts with obesity identified genetically driven proteins associated with low-grade inflammation, insulin resistance and dyslipidemia, which could act as endophenotypes for metabolic diseases [33]. Furthermore, BMI was associated with widespread changes in the human plasma proteome under substantial genetic control, impacting clinically relevant pathways of adiposity such as lipid metabolism and inflammation [34].
Lastly, a high dietary intake of antioxidants (α, β-carotene and α-tocopherol) protected buccal cells from telomere length (TL) shortening, depending on the genetic background of antioxidant vitamin-related genes (BCMO1 and ISX) in healthy Japanese adults [35]. In addition, a higher adherence to the Mediterranean dietary pattern prevented leukocyte TL shortening among Ala allele carriers of the PPARγ2 (rs1801282) polymorphism in subjects with high cardiovascular risk [36]. Additionally, inverse associations between TL and plasma zinc were found, especially in children carrying the homozygous mutant genotype of the RFC G80A (rs1051266) gene polymorphism [37]. Furthermore, telomerase RNA component genetic variants interacted with plasma monounsaturated fatty acids (MUFAs) levels, improving inflammation status and telomere attrition related with coronary heath disease [38].
3. Metagenomics Integrating Epigenomics, Transcriptomics, Proteomics or Metabolomics Methodologies
Metagenomic sequencing techniques have contributed to identify a number of microbial communities in the gut under different physiological and disease conditions [39]. Additional multi-omic tools analyzing gut microbial mRNA (metatranscriptomics), proteins (metaproteomics) and metabolites (metabolomics) are complementing information about the gut microbial ecology, the biological roles of uncultured microbes, and complex interactions between host, gut microbes, and environment affecting health status [40].
For instance, a clinical trial demonstrated that the daily consumption of 12 g of a prebiotic fiber supplement for 4 weeks significantly increased the abundance of several beneficial Bifidobacterium species and the production of health-promoting bacteria-derived metabolites in healthy individuals, with abundance of genes associated with prebiotic utilization, acetate production, and choline to betaine oxidation [41]. Additionally, normal diets of participants supplemented with either pea- or orange-fiber-containing snacks for 10 weeks correlated with abundances of genes encoding carbohydrate-active enzymes in the fecal microbiome, whose changes in turn correlated with levels of plasma proteins involved in vascular function, fibrotic responses, immune cell signaling, and obesity-associated hormonal regulators [42]. However, a very low-calorie diet intervention (800 kcal/day) in obese postmenopausal women induced changes in individual microbial taxa correlating with variations in the plasma metabolome, fecal bile acid composition, and altered gene expression pathways in adipose tissue [43]. Improvements in metabolic alterations were linked to specific microbiota genera (relative abundances of Lachnospiraceae NK4A136 and uncultured genera of Ruminococcaceae) and fecal metabolites (cholate and cadaverine) after following a Mediterranean dietary pattern for 2 months instead of only nut consumption [44]. Consistently, the consumption of a vegetarian diet for 4 weeks significantly improved cardiometabolic risk factors and altered the relative abundance of gut microbes (dominated by several genera of Ruminococcaceae) and plasma metabolites (including l-carnitine, acylcarnitine metabolites, and phospholipids) in patients with ischemic heart disease [45]. Additionally, differential gut microbial protein expression was detected in stool samples of individuals consuming diets varying in fiber content and glycemic index for 28 days, including those implicated in production and degradation of fatty acids [46]. Interestingly, replacing beef with a chicken-based diet for two weeks largely affected the abundance of Bacteroides genus, and thus probably induced downregulation of immunoglobulins in feces, especially in high- and middle-BMI Chinese volunteers [47].
At the cross-sectional level, the intakes of plant-derived nutrients or artificial sweeteners in healthy individuals were associated with relevant differences in circulating metabolites (particularly bile acids) depending on gut enterotypes [48]. Similarly, gut microbiome composition influenced the relationships between soy isoflavone intake and plasma and stool metabolites, including 2-hydroxybutyrate, glycine, and liquiritigenin, with relevance in hypertension and diabetes pathogenesis [49]. Remarkably, yoghurt consumption was associated with reduced visceral adiposity and changes in gut microbiome (transient increases of S. thermophilus and B. lactis species) and fecal metabolome (elevated concentrations of 3-hydroxyoctanoic acid) in female twins [50]. Of note, chicken eaters had more diverse gut microbiota and higher abundances of Prevotella 2 and 9 than pork eaters, which positively correlated with fecal levels of skatole and indole [51]. Moreover, the levels of circulatory or gut metabolites were concurrently influenced by gut microbiome alterations shaped by the quality of diet consumed [52]. In addition, dietary differences among vegans and omnivores correlated with large variations in the metabolome, including co-metabolites produced by the gut microbiota [53]. In general, higher occurrence of potentially beneficial host microbiome metabolites (i.e., short- and medium-chain fatty acids and their derivatives) have been found in vegans compared to omnivores [54]. Likewise, vegan and vegetarian diets were associated with increased abundance of microbial genes/proteins involved in cell motility, nutrient breakdown and transport, and the synthesis of essential amino acids and vitamins [55]. Specifically, vegetarians showed low levels of circulating BCAAs and upregulation of the gut microbial pathway implicated in the degradation and utilization of BCAAs [56]. Accordingly, multi-omics analyses revealed that Indian subjects presented unique gut microbiome and serum metabolome profiles compared to other populations, which were associated with specific dietary patterns [57]. Meanwhile, high-level adherence to a Mediterranean diet (based on plant foodstuffs) was associated with increased levels of fecal short-chain fatty acids and favorable microbiome-related metabolomic profiles [58].
Furthermore, it has been demonstrated that gut microbiota may mediate the effects of diet on the host health via mechanisms targeting the epigenome [59]. Thus, microbial metabolites of diet (i.e., phenolic acids, isothiocyanates, and short chain fatty acids) may influence epigenome status may altering the expression of epigenetically active enzymes including DNA methyltransferases, histone acetyltransferases, deacetylases and demethylases [60]. In this context, the impact of common dietary patterns on the gut microbiota composition and the host epigenome status has been recently reviewed [61]. However, because most evidence comes from animal studies and in vitro assays, further clinical trials are still required to analyze the nutrition-microbiota-epigenetic axis and applications in precision nutrition approaches.
Additionally, it was evidenced that imbalances in intestinal microbiota due to diet may contribute to the development of different pathologies by impairing the expression of miRNAs [62]. Accordingly, a poor diet quality was associated with a higher risk of mild cognitive impairment, which could have been mediated by microbiota composition (abundance of Proteobacteria and Gammaproteobacteria) and miRNA expression (hsa-let-7g-5p, hsa-miR-107, and hsa-miR-186-3p) in middle-aged and elderly Chinese population [63]. Of note, interactions between the abundance of bacterial species (i.e., Bacteroides eggerthi) and circulating miRNAs (miR-130b-3p, miR-185-5p and miR-21-5p) were found in humans in relation to obesity [64]. Moreover, an integrated analysis using 25 miRNAs, 25 taxa and 7 dietary nutrients was able to clearly discriminate vegan, vegetarian, and omnivore dietary patterns in healthy individuals [65]. Lastly, dietary plant-derived miRNAs (xenomiRs) appear to modulate gut microbiota composition, influencing gut epithelial barrier permeability and related gastrointestinal health [66].
4. Nutritional Relationships between the Epigenome, Transcriptome, and the Metabolome
Epigenome landscapes play an important role in determining cell phenotypes via regulation of gene expression [67]. In turn, certain nutrients may induce epigenetic modifications such as DNA methylation, probably modifying the expression of key genes associated with physiologic and pathologic processes [68]. Thus, some observational and intervention studies have analyzed interactions between nutrition, epigenome, and transcriptomic signatures on health outcomes. For example, associations between dietary folate deficiency, CAMKK2 methylation and expression levels, and insulin resistance status were reported in subjects with obesity [69]. Additionally, gestational fish intake was related to changes in the methylation and expression levels of the FADS1/2 and ELOVL5 genes, with impact on allergy development in early childhood [70].
Consistently, fatty acid supplementation with 4 g/day of either n-3 PUFAs or olive oil (OO) for 8 weeks altered the methylation and the transcript levels of the FADS2 and ELOVL5 genes in peripheral blood mononuclear cells (PBMCs) of adults suffering renal disease [71]. Likewise, it was demonstrated that dietary supplementation of 5.7 g/day of n-3 PUFAs or 6 g/day of extra virgin olive oil (EVOO) for 4 weeks induced DNA methylation changes in leukocytes in trained male cyclists, potentially via the modulation of DNA methyltransferases (DNMTs) mRNA expression [72]. By contrast, daily supplementation with 200 mg of monomeric and oligomeric flavanols from grape seeds for 8 weeks modulated the expression of genes associated with cardiovascular disease pathways, without parallel changes in DNA methylation states [73].
Interestingly, an integrated transcriptomic and epigenomic analysis identified differential DNA methylation and expression levels of the CD44 gene on PBMCs depending on the success to the RESMENA (moderately high-protein content) weight-loss program [74]. In addition, the beneficial effects of consuming a very low-calorie ketogenic diet (600–800 kcal/day) for six months on obesity measurements involved methylome and transcriptome changes in ZNF331 and FGFRL1 genes in blood leukocytes [75].
Furthermore, epigenomics act as a mechanistic link between energy metabolism and control of gene expression (known as metaboloepigenetics), where a number of dietary metabolites (including SAM, acetyl-CoA, NAD+, and ATP) modulate the activity of epigenetic enzymes regulating transcriptional rates as necessary to maintain cell homeostasis [76]. Therefore, epigenetic marks (such as DNA methylation, posttranslational histone modifications, and nucleosome position) have the capacity to integrate the expression state of chromatin with the metabolic state of the cell [77]. In this regard, maternal nutrient availability of diet-derived methyl donors (folate, coline, betaine, and methionine) and cofactors (vitamins B2, B6, and B12) is critical for DNA methylation reactions through 1-carbon metabolism, which impacts gene expression and health outcomes in offspring [78]. Moreover, the risk for non-communicable diseases (NCDs) in adulthood can be programmed by early nutrition through alterations in methylation/expression patterns of key genes implicated in various metabolic pathways during development, and persisting into adulthood [79]. Hence, the importance of applying personalized nutrition approaches targeting the epigenome, transcriptome, and metabolome to prevent and treat NCDs since early stages of fetal life has been highlighted [80].
5. Metabolomics, Proteomics, and Transcriptomics Interplays
The analysis of proteins and metabolites associated with nutritional features may provide insights into the molecular mechanisms mediating diet-related disease, with preventive and management applications [81]. For instance, three dietary patterns, the Mediterranean-style diet, the Dietary Approaches to Stop Hypertension diet, and the Alternative Healthy Eating Index were unique metabolome and proteome signatures involved in important physiological pathways such as cellular metabolism and immune response within the Framingham cohort [81]. Additionally, plasma metabolomics and proteomics analyses revealed subtle multiple processes related to metabolism, oxidation and inflammation after a postprandial dietary challenge as demonstrated by changes in the concentrations of metabolites, proteins and clinical chemistry parameters in overweight subjects [82]. Interestingly, it was demonstrated that a nutrigenomic intervention with a nutritional supplement containing selected bioactive compounds (including polyphenols, alpha-tocopherol, vitamin C, and n-3 PUFAs) for five weeks affected inflammatory processes, oxidative stress, and metabolism in healthy overweight men, as evidenced by an integrated analysis of plasma metabolites/proteins, and gene expression profiles [83].
6. Conclusions
The combination of diverse types of biological data from genomics, epigenomics, metagenomics, transcriptomics, proteomics and metabolomics is expanding our current understanding of the complexity and diversity of human metabolism, as well as yielding profound insights into disease pathogenesis. Hence, this knowledge is allowing for the identification and characterization of potential molecular targets and active biomarkers involved in many nutritional disorders, including obesity, dyslipidemias, fatty liver, insulin resistance, and inflammation. Additionally, it is expected that the application of integrative omics approaches could be useful not only for stratifying patients for risk prediction and early diagnosis purposes but also for guiding precision disease treatments and prognosis under a holistic scope. For instance, the prediction of obesity risk and weight loss has considerably improved when using genomics, epigenomics, metagenomics and transcriptomics signatures instead of single omics approaches. Additionally, integrative genomic, epigenomic, and metabolomic analyses have better characterized post-prandial lipidomic features as potential biomarkers of nutrient intakes (including polyunsaturated fatty acids and B-complex vitamins) and subsequent cardiovascular and cancer risks. Moreover, the combined application of metagenomics, epigenomics, metabolomics, and proteomics methodologies has allowed us to discriminate different dietary patterns including vegan, vegetarian, and omnivore regimes and their implications for health status. In addition, the early programmation of chronic diseases in adulthood can be tackled by precision nutrition strategies targeting the epigenome, transcriptome, and the metabolome in preconception and pregnancy stages. However, despite these scientific advances, more evidence in these research areas is still necessary before precision nutrition can be implemented in clinical practice and public health settings around the world. Furthermore, some challenges include ethical and regulatory issues, the lack of robust and reproducible results due to methodological aspects (type of samples analyzed, standardization of procedures, population characteristics), the high cost of omics methodologies, and high-dimensional data analyses and interpretation.
Author Contributions
Conceptualization, O.R.-L., F.I.M. and J.A.M.; writing—original draft preparation, O.R.-L.; writing—review and editing, O.R.-L., F.I.M. and J.A.M.; supervision, F.I.M. and J.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CIBER (CB12/03/30002) and Gobierno de Navarra: Obekit (PT024), Microbiota (PI035), and Nutribiota (0011-1411-2018-000040) projects. The PRODEP-Mexico program (UABC-PTC-796) is also gratefully acknowledged. Support from Synergic R&D Projects in New and Emerging Scientific Areas on the Frontier of Science and Interdisciplinary Nature of The Community of Madrid (METAINFLAMATION-Y2020/BIO-6600) is also credited.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Toro-Martín, J.; Arsenault, B.J.; Després, J.P.; Vohl, M.C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for Current Nutrigenetic, Nutrigenomic, and Nutriepigenetic Approaches for Precision Nutrition Involving the Prevention and Management of Chronic Diseases Associated with Obesity. J. Nutrigenet. Nutr. 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, K.M.; Ramos-Lopez, O.; Pérusse, L.; Kato, H.; Ordovas, J.M.; Martínez, J.A. Precision nutrition: A review of current approaches and future endeavors. Trends Food Sci. Technol. 2022, 128, 253–264. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milton-Laskibar, I.; Martínez, J.A. Precision nutrition based on phenotypical traits and the (epi)genotype: Nutrigenetic and nutrigenomic approaches for obesity care. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 315–325. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Lee, Y.C.; Christensen, J.J.; Parnell, L.D.; Smith, C.E.; Shao, J.; McKeown, N.M.; Ordovás, J.M.; Lai, C.Q. Using Machine Learning to Predict Obesity Based on Genome-Wide and Epigenome-Wide Gene-Gene and Gene-Diet Interactions. Front Genet. 2022, 12, 783845. [Google Scholar] [CrossRef]
- Cortés-Martín, A.; Colmenarejo, G.; Selma, M.V.; Espín, J.C. Genetic Polymorphisms, Mediterranean Diet and Microbiota-Associated Urolithin Metabotypes can Predict Obesity in Childhood-Adolescence. Sci. Rep. 2020, 10, 7850. [Google Scholar] [CrossRef]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.Y.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut 2022, 71, 1095–1105. [Google Scholar] [CrossRef]
- Lang, S.; Martin, A.; Zhang, X.; Farowski, F.; Wisplinghoff, H.; Vehreschild, M.J.G.T.; Krawczyk, M.; Nowag, A.; Kretzschmar, A.; Scholz, C.; et al. Combined analysis of gut microbiota, diet and PNPLA3 polymorphism in biopsy-proven non-alcoholic fatty liver disease. Liver Int. 2021, 41, 1576–1591. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Riezu-Boj, J.I.; Guruceaga, E.; Milagro, F.I.; Martínez, J.A. Sex-Specific Associations between Gut Prevotellaceae and Host Genetics on Adiposity. Microorganisms 2020, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.C.; Goodrich, J.K.; Youngblut, N.D.; Luque, G.G.; Ruaud, A.; Sutter, J.L.; Waters, J.L.; Shi, Q.; El-Hadidi, M.; Johnson, L.M.; et al. Human Salivary Amylase Gene Copy Number Impacts Oral and Gut Microbiomes. Cell Host Microbe 2019, 25, 553–564.e7. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Q.; Smith, C.E.; Parnell, L.D.; Lee, Y.C.; Corella, D.; Hopkins, P.; Hidalgo, B.A.; Aslibekyan, S.; Province, M.A.; Absher, D.; et al. Epigenomics and metabolomics reveal the mechanism of the APOA2-saturated fat intake interaction affecting obesity. Am. J. Clin. Nutr. 2018, 108, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhou, J.; Gong, C.; Long, Z.; Tian, J.; Zhu, L.; Li, J.; Yu, H.; Wang, F.; Zhao, Y. Dietary factors and microRNA-binding site polymorphisms in the IL13 gene: Risk and prognosis analysis of colorectal cancer. Oncotarget 2017, 8, 47379–47388. [Google Scholar] [CrossRef] [PubMed]
- Richardson, K.; Nettleton, J.A.; Rotllan, N.; Tanaka, T.; Smith, C.E.; Lai, C.Q.; Parnell, L.D.; Lee, Y.C.; Lahti, J.; Lemaitre, R.N.; et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am. J. Hum. Genet. 2013, 92, 5–14. [Google Scholar] [CrossRef]
- Lupu, D.S.; Cheatham, C.L.; Corbin, K.D.; Niculescu, M.D. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part I: Maternal FADS2 genotype and DNA methylation correlate with polyunsaturated fatty acid status in toddlers: An exploratory analysis. Nutr. Res. 2015, 35, 939–947. [Google Scholar] [CrossRef]
- Ma, Y.; Follis, J.L.; Smith, C.E.; Tanaka, T.; Manichaikul, A.W.; Chu, A.Y.; Samieri, C.; Zhou, X.; Guan, W.; Wang, L.; et al. Interaction of methylation-related genetic variants with circulating fatty acids on plasma lipids: A meta-analysis of 7 studies and methylation analysis of 3 studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Am. J. Clin. Nutr. 2016, 103, 567–578. [Google Scholar] [CrossRef]
- Irvin, M.R.; Montasser, M.E.; Kind, T.; Fan, S.; Barupal, D.K.; Patki, A.; Tanner, R.M.; Armstrong, N.D.; Ryan, K.A.; Claas, S.A.; et al. Genomics of Postprandial Lipidomics in the Genetics of Lipid-Lowering Drugs and Diet Network Study. Nutrients 2021, 13, 4000. [Google Scholar] [CrossRef]
- Irvin, M.R.; Zhi, D.; Aslibekyan, S.; Claas, S.A.; Absher, D.M.; Ordovas, J.M.; Tiwari, H.K.; Watkins, S.; Arnett, D.K. Genomics of post-prandial lipidomic phenotypes in the Genetics of Lipid lowering Drugs and Diet Network (GOLDN) study. PLoS ONE 2014, 9, e99509. [Google Scholar] [CrossRef]
- Vladoiu, S.; Botezatu, A.; Anton, G.; Manda, D.; Paun, D.L.; Oros, S.; Rosca, R.; Dinu Draganescu, D. The involvement of vdr promoter methylation, CDX-2 VDR polymorphism and vitamin d levels in male infertility. Acta Endocrinol. 2017, 13, 294–301. [Google Scholar] [CrossRef]
- Beckett, E.L.; Martin, C.; Duesing, K.; Jones, P.; Furst, J.; Yates, Z.; Veysey, M.; Lucock, M. Vitamin D Receptor Genotype Modulates the Correlation between Vitamin D and Circulating Levels of let-7a/b and Vitamin D Intake in an Elderly Cohort. J. Nutrigenet. Nutr. 2014, 7, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Lucock, M.; Yates, Z.; Martin, C.; Choi, J.H.; Beckett, E.; Boyd, L.; LeGras, K.; Ng, X.; Skinner, V.; Wai, R.; et al. Methylation diet and methyl group genetics in risk for adenomatous polyp occurrence. BBA Clin. 2015, 3, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ye, Y.; Tu, H.; Hildebrandt, M.A.; Zhao, L.; Heymach, J.V.; Roth, J.A.; Wu, X. MicroRNA-related genetic variants in iron regulatory genes, dietary iron intake, microRNAs and lung cancer risk. Ann. Oncol. 2017, 28, 1124–1129. [Google Scholar] [CrossRef] [PubMed]
- Friso, S.; Choi, S.W.; Girelli, D.; Mason, J.B.; Dolnikowski, G.G.; Bagley, P.J.; Olivieri, O.; Jacques, P.F.; Rosenberg, I.H.; Corrocher, R.; et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 2002, 99, 5606–5611. [Google Scholar] [CrossRef]
- Stern, L.L.; Mason, J.B.; Selhub, J.; Choi, S.W. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol. Biomark. Prev. 2000, 9, 849–853. [Google Scholar]
- Shin, W.; Yan, J.; Abratte, C.M.; Vermeylen, F.; Caudill, M.A. Choline intake exceeding current dietary recommendations preserves markers of cellular methylation in a genetic subgroup of folate-compromised men. J. Nutr. 2010, 140, 975–980. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Zhao, M.; Huang, T.; Sun, D.; Han, L.; Nisa, H.; Shang, X.; Heianza, Y.; Qi, L. DNA methylation variant, B-vitamins intake and longitudinal change in body mass index. Int. J. Obes. 2019, 43, 468–474. [Google Scholar] [CrossRef]
- Huang, T.; Zheng, Y.; Qi, Q.; Xu, M.; Ley, S.H.; Li, Y.; Kang, J.H.; Wiggs, J.; Pasquale, L.R.; Chan, A.T.; et al. DNA Methylation Variants at HIF3A Locus, B-Vitamin Intake, and Long-term Weight Change: Gene-Diet Interactions in Two U.S. Cohorts. Diabetes 2015, 64, 3146–3154. [Google Scholar] [CrossRef]
- Sun, D.; Heianza, Y.; Li, X.; Shang, X.; Smith, S.R.; Bray, G.A.; Sacks, F.M.; Qi, L. Genetic, epigenetic and transcriptional variations at NFATC2IP locus with weight loss in response to diet interventions: The POUNDS Lost Trial. Diabetes Obes. Metab. 2018, 20, 2298–2303. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Milagro, F.I.; Guruceaga, E.; Cuervo, M.; Goni, L.; García-Granero, M.; Martinez, J.A.; Riezu-Boj, J.I. A weight-loss model based on baseline microbiota and genetic scores for selection of dietary treatments in overweight and obese population. Clin. Nutr. 2022, 41, 1712–1723. [Google Scholar] [CrossRef]
- Müller, S.; Wallner, S.; Schmitz, G.; Loew, T.; Stempfl, T.; Möhle, C.; Strack, C.; Sag, S.; Baessler, A.; Fischer, M. SNP dependent modulation of circulating miRNAs from the miR25/93/106 cluster in patients undergoing weight loss. Gene 2020, 753, 144787. [Google Scholar] [PubMed]
- Carayol, J.; Chabert, C.; Di Cara, A.; Armenise, C.; Lefebvre, G.; Langin, D.; Viguerie, N.; Metairon, S.; Saris, W.H.M.; Astrup, A.; et al. Protein quantitative trait locus study in obesity during weight-loss identifies a leptin regulator. Nat. Commun. 2017, 8, 2084. [Google Scholar] [CrossRef] [PubMed]
- Ruffieux, H.; Carayol, J.; Popescu, R.; Harper, M.E.; Dent, R.; Saris, W.H.M.; Astrup, A.; Hager, J.; Davison, A.C.; Valsesia, A. A fully joint Bayesian quantitative trait locus mapping of human protein abundance in plasma. PLoS Comput. Biol. 2020, 16, e1007882. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.B.; Sharma, S.; Molnar, M.; Matías-García, P.R.; Elhadad, M.A.; Waldenberger, M.; Peters, A.; Rathmann, W.; Graumann, J.; Gieger, C.; et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat. Commun. 2021, 12, 1279. [Google Scholar] [CrossRef]
- Yabuta, S.; Masaki, M.; Shidoji, Y. Associations of Buccal Cell Telomere Length with Daily Intake of β-Carotene or α-Tocopherol Are Dependent on Carotenoid Metabolism-related Gene Polymorphisms in Healthy Japanese Adults. J. Nutr. Health Aging 2016, 20, 267–274. [Google Scholar] [CrossRef]
- García-Calzón, S.; Martínez-González, M.A.; Razquin, C.; Corella, D.; Salas-Salvadó, J.; Martínez, J.A.; Zalba, G.; Marti, A. Pro12Ala polymorphism of the PPARγ2 gene interacts with a mediterranean diet to prevent telomere shortening in the PREDIMED-NAVARRA randomized trial. Circ. Cardiovasc. Genet. 2015, 8, 91–99. [Google Scholar] [CrossRef]
- Milne, E.; O’Callaghan, N.; Ramankutty, P.; de Klerk, N.H.; Greenop, K.R.; Armstrong, B.K.; Miller, M.; Fenech, M. Plasma micronutrient levels and telomere length in children. Nutrition 2015, 31, 331–336. [Google Scholar] [CrossRef]
- Gomez-Delgado, F.; Delgado-Lista, J.; Lopez-Moreno, J.; Rangel-Zuñiga, O.A.; Alcala-Diaz, J.F.; Leon-Acuña, A.; Corina, A.; Yubero-Serrano, E.; Torres-Peña, J.D.; Camargo, A.; et al. Telomerase RNA Component Genetic Variants Interact With the Mediterranean Diet Modifying the Inflammatory Status and its Relationship With Aging: CORDIOPREV Study. J. Gerontol. Biol. Sci. Med. Sci. 2018, 73, 327–332. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Daliri, E.B.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.H. Challenges and Perspective in Integrated Multi-Omics in Gut Microbiota Studies. Biomolecules 2021, 11, 300. [Google Scholar] [CrossRef]
- Kang, J.W.; Tang, X.; Walton, C.J.; Brown, M.J.; Brewer, R.A.; Maddela, R.L.; Zheng, J.J.; Agus, J.K.; Zivkovic, A.M. Multi-Omic Analyses Reveal Bifidogenic Effect and Metabolomic Shifts in Healthy Human Cohort Supplemented With a Prebiotic Dietary Fiber Blend. Front. Nutr. 2022, 9, 908534. [Google Scholar] [CrossRef] [PubMed]
- Delannoy-Bruno, O.; Desai, C.; Castillo, J.J.; Couture, G.; Barve, R.A.; Lombard, V.; Henrissat, B.; Cheng, J.; Han, N.; Hayashi, D.K.; et al. An approach for evaluating the effects of dietary fiber polysaccharides on the human gut microbiome and plasma proteome. Proc. Natl. Acad. Sci. USA 2022, 119, e2123411119. [Google Scholar] [CrossRef] [PubMed]
- Alemán, J.O.; Bokulich, N.A.; Swann, J.R.; Walker, J.M.; De Rosa, J.C.; Battaglia, T.; Costabile, A.; Pechlivanis, A.; Liang, Y.; Breslow, J.L.; et al. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J. Transl. Med. 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Galié, S.; García-Gavilán, J.; Camacho-Barcía, L.; Atzeni, A.; Muralidharan, J.; Papandreou, C.; Arcelin, P.; Palau-Galindo, A.; Garcia, D.; Basora, J.; et al. Effects of the Mediterranean Diet or Nut Consumption on Gut Microbiota Composition and Fecal Metabolites and their Relationship with Cardiometabolic Risk Factors. Mol. Nutr. Food Res. 2021, 65, e2000982. [Google Scholar] [CrossRef] [PubMed]
- Djekic, D.; Shi, L.; Brolin, H.; Carlsson, F.; Särnqvist, C.; Savolainen, O.; Cao, Y.; Bäckhed, F.; Tremaroli, V.; Landberg, R.; et al. Effects of a Vegetarian Diet on Cardiometabolic Risk Factors, Gut Microbiota, and Plasma Metabolome in Subjects With Ischemic Heart Disease: A Randomized, Crossover Study. J. Am. Heart Assoc. 2020, 9, e016518. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Hullar, M.A.J.; Lai, L.A.; Peng, H.; May, D.H.; Noble, W.S.; Raftery, D.; Navarro, S.L.; Neuhouser, M.L.; Lampe, P.D.; et al. Gut Microbial Protein Expression in Response to Dietary Patterns in a Controlled Feeding Study: A Metaproteomic Approach. Microorganisms 2020, 8, 379. [Google Scholar] [CrossRef]
- Zhao, D.; Shan, K.; Xie, Y.; Zhang, G.; An, Q.; Yu, X.; Zhou, G.; Li, C. Body weight index indicates the responses of the fecal microbiota, metabolome and proteome to beef/chicken-based diet alterations in Chinese volunteers. NPJ Biofilms Microbiomes 2022, 8, 56. [Google Scholar] [CrossRef]
- Tang, Z.Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Shah, R.D.; Tang, Z.Z.; Chen, G.; Huang, S.; Ferguson, J.F. Soy food intake associates with changes in the metabolome and reduced blood pressure in a gut microbiota dependent manner. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 1500–1511. [Google Scholar] [CrossRef]
- Le Roy, C.I.; Kurilshikov, A.; Leeming, E.R.; Visconti, A.; Bowyer, R.C.E.; Menni, C.; Falchi, M.; Koutnikova, H.; Veiga, P.; Zhernakova, A.; et al. Yoghurt consumption is associated with changes in the composition of the human gut microbiome and metabolome. BMC Microbiol. 2022, 22, 39. [Google Scholar]
- Shi, J.; Zhao, D.; Zhao, F.; Wang, C.; Zamaratskaia, G.; Li, C. Chicken-eaters and pork-eaters have different gut microbiota and tryptophan metabolites. Sci. Rep. 2021, 11, 11934. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; Shah, R.D.; Mosley, J.D.; Ferguson, J.F. A metabolome and microbiome wide association study of healthy eating index points to the mechanisms linking dietary pattern and metabolic status. Eur. J. Nutr. 2021, 60, 4413–4427. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, M.; Budinska, E.; Kuzma, M.; Pelantova, H.; Hradecky, J.; Heczkova, M.; Daskova, N.; Bratova, M.; Modos, I.; Videnska, P.; et al. Vegan Diet Is Associated With Favorable Effects on the Metabolic Performance of Intestinal Microbiota: A Cross-Sectional Multi-Omics Study. Front. Nutr. 2022, 8, 783302. [Google Scholar] [CrossRef]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wan, Y.; Yin, K.; Wei, Y.; Wang, B.; Yu, X.; Ni, Y.; Zheng, J.; Huang, T.; Song, M.; et al. Lower Circulating Branched-Chain Amino Acid Concentrations Among Vegetarians are Associated with Changes in Gut Microbial Composition and Function. Mol. Nutr. Food Res. 2019, 63, e1900612. [Google Scholar] [CrossRef]
- Dhakan, D.B.; Maji, A.; Sharma, A.K.; Saxena, R.; Pulikkan, J.; Grace, T.; Gomez, A.; Scaria, J.; Amato, K.R.; Sharma, V.K. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience 2019, 8, giz004. [Google Scholar] [CrossRef]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Hullar, M.A.; Fu, B.C. Diet, the gut microbiome, and epigenetics. Cancer J. 2014, 20, 170–175. [Google Scholar] [CrossRef]
- Gerhauser, C. Impact of dietary gut microbial metabolites on the epigenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170359. [Google Scholar] [CrossRef]
- D’Aquila, P.; Carelli, L.L.; De Rango, F.; Passarino, G.; Bellizzi, D. Gut Microbiota as Important Mediator Between Diet and DNA Methylation and Histone Modifications in the Host. Nutrients 2020, 12, 597. [Google Scholar] [CrossRef] [PubMed]
- Guz, M.; Jeleniewicz, W.; Malm, A.; Korona-Glowniak, I. A Crosstalk between Diet, Microbiome and microRNA in Epigenetic Regulation of Colorectal Cancer. Nutrients 2021, 13, 2428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Liu, W.; Wang, T.; Wang, L.; Hao, L.; Ju, M.; Xiao, R. Diet quality, gut microbiota, and microRNAs associated with mild cognitive impairment in middle-aged and elderly Chinese population. Am. J. Clin. Nutr. 2021, 114, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Cuevas-Sierra, A.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Comprehensive Analysis Reveals Novel Interactions between Circulating MicroRNAs and Gut Microbiota Composition in Human Obesity. Int. J. Mol. Sci. 2020, 21, 9509. [Google Scholar] [CrossRef]
- Tarallo, S.; Ferrero, G.; De Filippis, F.; Francavilla, A.; Pasolli, E.; Panero, V.; Cordero, F.; Segata, N.; Grioni, S.; Pensa, R.G.; et al. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut 2022, 71, 1302–1314. [Google Scholar] [CrossRef]
- Díez-Sainz, E.; Lorente-Cebrián, S.; Aranaz, P.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I. Potential Mechanisms Linking Food-Derived MicroRNAs, Gut Microbiota and Intestinal Barrier Functions in the Context of Nutrition and Human Health. Front. Nutr. 2021, 8, 586564. [Google Scholar] [CrossRef]
- Choi, S.W.; Friso, S. Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef]
- Milagro, F.I.; Mansego, M.L.; De Miguel, C.; Martínez, J.A. Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. Mol. Asp. Med. 2013, 34, 782–812. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Samblas, M.; Milagro, F.I.; Zulet, M.A.; Mansego, M.L.; Riezu-Boj, J.I.; Martinez, J.A. Association of low dietary folate intake with lower CAMKK2 gene methylation, adiposity, and insulin resistance in obese subjects. Nutr. Res. 2018, 50, 53–62. [Google Scholar] [CrossRef]
- Losol, P.; Rezwan, F.I.; Patil, V.K.; Venter, C.; Ewart, S.; Zhang, H.; Arshad, S.H.; Karmaus, W.; Holloway, J.W. Effect of gestational oily fish intake on the risk of allergy in children may be influenced by FADS1/2, ELOVL5 expression and DNA methylation. Genes Nutr. 2019, 14, 20. [Google Scholar] [CrossRef]
- Hoile, S.P.; Clarke-Harris, R.; Huang, R.C.; Calder, P.C.; Mori, T.A.; Beilin, L.J.; Lillycrop, K.A.; Burdge, G.C. Supplementation with N-3 long-chain polyunsaturated fatty acids or olive oil in men and women with renal disease induces differential changes in the DNA methylation of FADS2 and ELOVL5 in peripheral blood mononuclear cells. PLoS ONE 2014, 9, e109896. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; James, L.; Hussey, B.; Wadley, A.J.; Lindley, M.R.; Mastana, S.S. Impact of aerobic exercise and fatty acid supplementation on global and gene-specific DNA methylation. Epigenetics 2019, 14, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Vanden Berghe, W.; Boby, C.; Leroux, C.; Declerck, K.; Szarc vel Szic, K.; Heyninck, K.; Laukens, K.; Bizet, M.; Defrance, M.; et al. Dietary flavanols modulate the transcription of genes associated with cardiovascular pathology without changes in their DNA methylation state. PLoS ONE 2014, 9, e95527. [Google Scholar] [CrossRef]
- Samblas, M.; Mansego, M.L.; Zulet, M.A.; Milagro, F.I.; Martinez, J.A. An integrated transcriptomic and epigenomic analysis identifies CD44 gene as a potential biomarker for weight loss within an energy-restricted program. Eur. J. Nutr. 2019, 58, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Crujeiras, A.B.; Izquierdo, A.G.; Primo, D.; Milagro, F.I.; Sajoux, I.; Jácome, A.; Fernandez-Quintela, A.; Portillo, M.P.; Martínez, J.A.; Martinez-Olmos, M.A.; et al. Epigenetic landscape in blood leukocytes following ketosis and weight loss induced by a very low calorie ketogenic diet (VLCKD) in patients with obesity. Clin. Nutr. 2021, 40, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Bultman, S.J. Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell Physiol. 2012, 227, 3169–3177. [Google Scholar] [CrossRef]
- Janke, R.; Dodson, A.E.; Rine, J. Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 2015, 31, 473–496. [Google Scholar] [CrossRef]
- Dominguez-Salas, P.; Cox, S.E.; Prentice, A.M.; Hennig, B.J.; Moore, S.E. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: A review of current evidence in human subjects. Proc. Nutr. Soc. 2012, 71, 154–165. [Google Scholar] [CrossRef]
- Randunu, R.S.; Bertolo, R.F. The Effects of Maternal and Postnatal Dietary Methyl Nutrients on Epigenetic Changes that Lead to Non-Communicable Diseases in Adulthood. Int. J. Mol. Sci. 2020, 21, 3290. [Google Scholar] [CrossRef]
- Alabduljabbar, S.; Zaidan, S.A.; Lakshmanan, A.P.; Terranegra, A. Personalized Nutrition Approach in Pregnancy and Early Life to Tackle Childhood and Adult Non-Communicable Diseases. Life 2021, 11, 467. [Google Scholar] [CrossRef]
- Walker, M.E.; Song, R.J.; Xu, X.; Gerszten, R.E.; Ngo, D.; Clish, C.B.; Corlin, L.; Ma, J.; Xanthakis, V.; Jacques, P.F.; et al. Proteomic and Metabolomic Correlates of Healthy Dietary Patterns: The Framingham Heart Study. Nutrients 2020, 12, 1476. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.; Hendriks, H.F.; Cnubben, N.H.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M.; et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Bakker, G.C.; van Erk, M.J.; Pellis, L.; Wopereis, S.; Rubingh, C.M.; Cnubben, N.H.; Kooistra, T.; van Ommen, B.; Hendriks, H.F. An antiinflammatory dietary mix modulates inflammation and oxidative and metabolic stress in overweight men: A nutrigenomics approach. Am. J. Clin. Nutr. 2010, 91, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).