Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease
Abstract
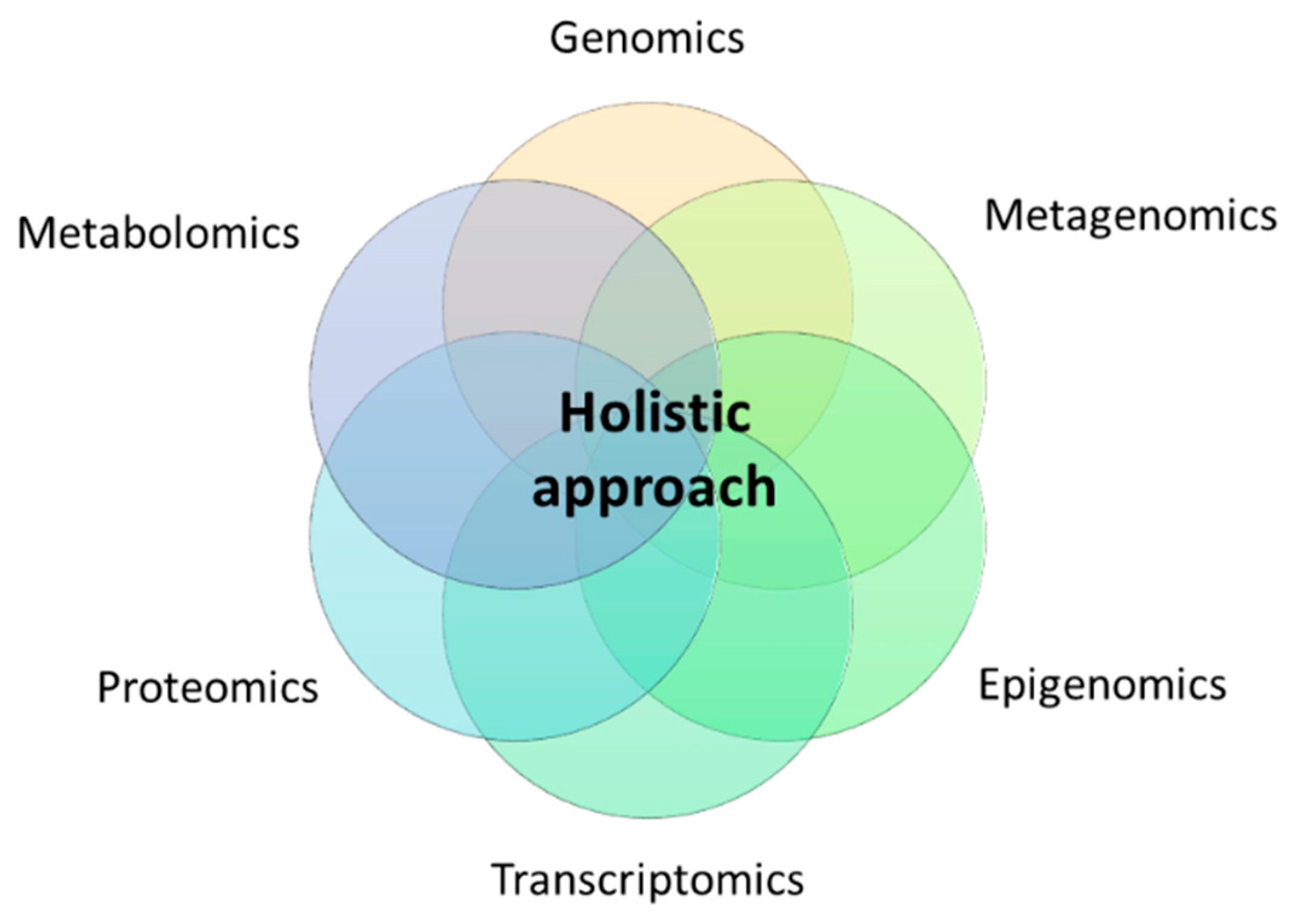
1. Introduction
2. Genomics in Combination Epigenomics, Metagenomics, Transcriptomics, Proteomics or Metabolomics Tools
3. Metagenomics Integrating Epigenomics, Transcriptomics, Proteomics or Metabolomics Methodologies
4. Nutritional Relationships between the Epigenome, Transcriptome, and the Metabolome
5. Metabolomics, Proteomics, and Transcriptomics Interplays
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Toro-Martín, J.; Arsenault, B.J.; Després, J.P.; Vohl, M.C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for Current Nutrigenetic, Nutrigenomic, and Nutriepigenetic Approaches for Precision Nutrition Involving the Prevention and Management of Chronic Diseases Associated with Obesity. J. Nutrigenet. Nutr. 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, K.M.; Ramos-Lopez, O.; Pérusse, L.; Kato, H.; Ordovas, J.M.; Martínez, J.A. Precision nutrition: A review of current approaches and future endeavors. Trends Food Sci. Technol. 2022, 128, 253–264. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milton-Laskibar, I.; Martínez, J.A. Precision nutrition based on phenotypical traits and the (epi)genotype: Nutrigenetic and nutrigenomic approaches for obesity care. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 315–325. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Lee, Y.C.; Christensen, J.J.; Parnell, L.D.; Smith, C.E.; Shao, J.; McKeown, N.M.; Ordovás, J.M.; Lai, C.Q. Using Machine Learning to Predict Obesity Based on Genome-Wide and Epigenome-Wide Gene-Gene and Gene-Diet Interactions. Front Genet. 2022, 12, 783845. [Google Scholar] [CrossRef]
- Cortés-Martín, A.; Colmenarejo, G.; Selma, M.V.; Espín, J.C. Genetic Polymorphisms, Mediterranean Diet and Microbiota-Associated Urolithin Metabotypes can Predict Obesity in Childhood-Adolescence. Sci. Rep. 2020, 10, 7850. [Google Scholar] [CrossRef]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.Y.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut 2022, 71, 1095–1105. [Google Scholar] [CrossRef]
- Lang, S.; Martin, A.; Zhang, X.; Farowski, F.; Wisplinghoff, H.; Vehreschild, M.J.G.T.; Krawczyk, M.; Nowag, A.; Kretzschmar, A.; Scholz, C.; et al. Combined analysis of gut microbiota, diet and PNPLA3 polymorphism in biopsy-proven non-alcoholic fatty liver disease. Liver Int. 2021, 41, 1576–1591. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Riezu-Boj, J.I.; Guruceaga, E.; Milagro, F.I.; Martínez, J.A. Sex-Specific Associations between Gut Prevotellaceae and Host Genetics on Adiposity. Microorganisms 2020, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.C.; Goodrich, J.K.; Youngblut, N.D.; Luque, G.G.; Ruaud, A.; Sutter, J.L.; Waters, J.L.; Shi, Q.; El-Hadidi, M.; Johnson, L.M.; et al. Human Salivary Amylase Gene Copy Number Impacts Oral and Gut Microbiomes. Cell Host Microbe 2019, 25, 553–564.e7. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Q.; Smith, C.E.; Parnell, L.D.; Lee, Y.C.; Corella, D.; Hopkins, P.; Hidalgo, B.A.; Aslibekyan, S.; Province, M.A.; Absher, D.; et al. Epigenomics and metabolomics reveal the mechanism of the APOA2-saturated fat intake interaction affecting obesity. Am. J. Clin. Nutr. 2018, 108, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhou, J.; Gong, C.; Long, Z.; Tian, J.; Zhu, L.; Li, J.; Yu, H.; Wang, F.; Zhao, Y. Dietary factors and microRNA-binding site polymorphisms in the IL13 gene: Risk and prognosis analysis of colorectal cancer. Oncotarget 2017, 8, 47379–47388. [Google Scholar] [CrossRef] [PubMed]
- Richardson, K.; Nettleton, J.A.; Rotllan, N.; Tanaka, T.; Smith, C.E.; Lai, C.Q.; Parnell, L.D.; Lee, Y.C.; Lahti, J.; Lemaitre, R.N.; et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am. J. Hum. Genet. 2013, 92, 5–14. [Google Scholar] [CrossRef]
- Lupu, D.S.; Cheatham, C.L.; Corbin, K.D.; Niculescu, M.D. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part I: Maternal FADS2 genotype and DNA methylation correlate with polyunsaturated fatty acid status in toddlers: An exploratory analysis. Nutr. Res. 2015, 35, 939–947. [Google Scholar] [CrossRef]
- Ma, Y.; Follis, J.L.; Smith, C.E.; Tanaka, T.; Manichaikul, A.W.; Chu, A.Y.; Samieri, C.; Zhou, X.; Guan, W.; Wang, L.; et al. Interaction of methylation-related genetic variants with circulating fatty acids on plasma lipids: A meta-analysis of 7 studies and methylation analysis of 3 studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium. Am. J. Clin. Nutr. 2016, 103, 567–578. [Google Scholar] [CrossRef]
- Irvin, M.R.; Montasser, M.E.; Kind, T.; Fan, S.; Barupal, D.K.; Patki, A.; Tanner, R.M.; Armstrong, N.D.; Ryan, K.A.; Claas, S.A.; et al. Genomics of Postprandial Lipidomics in the Genetics of Lipid-Lowering Drugs and Diet Network Study. Nutrients 2021, 13, 4000. [Google Scholar] [CrossRef]
- Irvin, M.R.; Zhi, D.; Aslibekyan, S.; Claas, S.A.; Absher, D.M.; Ordovas, J.M.; Tiwari, H.K.; Watkins, S.; Arnett, D.K. Genomics of post-prandial lipidomic phenotypes in the Genetics of Lipid lowering Drugs and Diet Network (GOLDN) study. PLoS ONE 2014, 9, e99509. [Google Scholar] [CrossRef]
- Vladoiu, S.; Botezatu, A.; Anton, G.; Manda, D.; Paun, D.L.; Oros, S.; Rosca, R.; Dinu Draganescu, D. The involvement of vdr promoter methylation, CDX-2 VDR polymorphism and vitamin d levels in male infertility. Acta Endocrinol. 2017, 13, 294–301. [Google Scholar] [CrossRef]
- Beckett, E.L.; Martin, C.; Duesing, K.; Jones, P.; Furst, J.; Yates, Z.; Veysey, M.; Lucock, M. Vitamin D Receptor Genotype Modulates the Correlation between Vitamin D and Circulating Levels of let-7a/b and Vitamin D Intake in an Elderly Cohort. J. Nutrigenet. Nutr. 2014, 7, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Lucock, M.; Yates, Z.; Martin, C.; Choi, J.H.; Beckett, E.; Boyd, L.; LeGras, K.; Ng, X.; Skinner, V.; Wai, R.; et al. Methylation diet and methyl group genetics in risk for adenomatous polyp occurrence. BBA Clin. 2015, 3, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ye, Y.; Tu, H.; Hildebrandt, M.A.; Zhao, L.; Heymach, J.V.; Roth, J.A.; Wu, X. MicroRNA-related genetic variants in iron regulatory genes, dietary iron intake, microRNAs and lung cancer risk. Ann. Oncol. 2017, 28, 1124–1129. [Google Scholar] [CrossRef] [PubMed]
- Friso, S.; Choi, S.W.; Girelli, D.; Mason, J.B.; Dolnikowski, G.G.; Bagley, P.J.; Olivieri, O.; Jacques, P.F.; Rosenberg, I.H.; Corrocher, R.; et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 2002, 99, 5606–5611. [Google Scholar] [CrossRef]
- Stern, L.L.; Mason, J.B.; Selhub, J.; Choi, S.W. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol. Biomark. Prev. 2000, 9, 849–853. [Google Scholar]
- Shin, W.; Yan, J.; Abratte, C.M.; Vermeylen, F.; Caudill, M.A. Choline intake exceeding current dietary recommendations preserves markers of cellular methylation in a genetic subgroup of folate-compromised men. J. Nutr. 2010, 140, 975–980. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Zhao, M.; Huang, T.; Sun, D.; Han, L.; Nisa, H.; Shang, X.; Heianza, Y.; Qi, L. DNA methylation variant, B-vitamins intake and longitudinal change in body mass index. Int. J. Obes. 2019, 43, 468–474. [Google Scholar] [CrossRef]
- Huang, T.; Zheng, Y.; Qi, Q.; Xu, M.; Ley, S.H.; Li, Y.; Kang, J.H.; Wiggs, J.; Pasquale, L.R.; Chan, A.T.; et al. DNA Methylation Variants at HIF3A Locus, B-Vitamin Intake, and Long-term Weight Change: Gene-Diet Interactions in Two U.S. Cohorts. Diabetes 2015, 64, 3146–3154. [Google Scholar] [CrossRef]
- Sun, D.; Heianza, Y.; Li, X.; Shang, X.; Smith, S.R.; Bray, G.A.; Sacks, F.M.; Qi, L. Genetic, epigenetic and transcriptional variations at NFATC2IP locus with weight loss in response to diet interventions: The POUNDS Lost Trial. Diabetes Obes. Metab. 2018, 20, 2298–2303. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Milagro, F.I.; Guruceaga, E.; Cuervo, M.; Goni, L.; García-Granero, M.; Martinez, J.A.; Riezu-Boj, J.I. A weight-loss model based on baseline microbiota and genetic scores for selection of dietary treatments in overweight and obese population. Clin. Nutr. 2022, 41, 1712–1723. [Google Scholar] [CrossRef]
- Müller, S.; Wallner, S.; Schmitz, G.; Loew, T.; Stempfl, T.; Möhle, C.; Strack, C.; Sag, S.; Baessler, A.; Fischer, M. SNP dependent modulation of circulating miRNAs from the miR25/93/106 cluster in patients undergoing weight loss. Gene 2020, 753, 144787. [Google Scholar] [PubMed]
- Carayol, J.; Chabert, C.; Di Cara, A.; Armenise, C.; Lefebvre, G.; Langin, D.; Viguerie, N.; Metairon, S.; Saris, W.H.M.; Astrup, A.; et al. Protein quantitative trait locus study in obesity during weight-loss identifies a leptin regulator. Nat. Commun. 2017, 8, 2084. [Google Scholar] [CrossRef] [PubMed]
- Ruffieux, H.; Carayol, J.; Popescu, R.; Harper, M.E.; Dent, R.; Saris, W.H.M.; Astrup, A.; Hager, J.; Davison, A.C.; Valsesia, A. A fully joint Bayesian quantitative trait locus mapping of human protein abundance in plasma. PLoS Comput. Biol. 2020, 16, e1007882. [Google Scholar] [CrossRef] [PubMed]
- Zaghlool, S.B.; Sharma, S.; Molnar, M.; Matías-García, P.R.; Elhadad, M.A.; Waldenberger, M.; Peters, A.; Rathmann, W.; Graumann, J.; Gieger, C.; et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat. Commun. 2021, 12, 1279. [Google Scholar] [CrossRef]
- Yabuta, S.; Masaki, M.; Shidoji, Y. Associations of Buccal Cell Telomere Length with Daily Intake of β-Carotene or α-Tocopherol Are Dependent on Carotenoid Metabolism-related Gene Polymorphisms in Healthy Japanese Adults. J. Nutr. Health Aging 2016, 20, 267–274. [Google Scholar] [CrossRef]
- García-Calzón, S.; Martínez-González, M.A.; Razquin, C.; Corella, D.; Salas-Salvadó, J.; Martínez, J.A.; Zalba, G.; Marti, A. Pro12Ala polymorphism of the PPARγ2 gene interacts with a mediterranean diet to prevent telomere shortening in the PREDIMED-NAVARRA randomized trial. Circ. Cardiovasc. Genet. 2015, 8, 91–99. [Google Scholar] [CrossRef]
- Milne, E.; O’Callaghan, N.; Ramankutty, P.; de Klerk, N.H.; Greenop, K.R.; Armstrong, B.K.; Miller, M.; Fenech, M. Plasma micronutrient levels and telomere length in children. Nutrition 2015, 31, 331–336. [Google Scholar] [CrossRef]
- Gomez-Delgado, F.; Delgado-Lista, J.; Lopez-Moreno, J.; Rangel-Zuñiga, O.A.; Alcala-Diaz, J.F.; Leon-Acuña, A.; Corina, A.; Yubero-Serrano, E.; Torres-Peña, J.D.; Camargo, A.; et al. Telomerase RNA Component Genetic Variants Interact With the Mediterranean Diet Modifying the Inflammatory Status and its Relationship With Aging: CORDIOPREV Study. J. Gerontol. Biol. Sci. Med. Sci. 2018, 73, 327–332. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Daliri, E.B.; Ofosu, F.K.; Chelliah, R.; Lee, B.H.; Oh, D.H. Challenges and Perspective in Integrated Multi-Omics in Gut Microbiota Studies. Biomolecules 2021, 11, 300. [Google Scholar] [CrossRef]
- Kang, J.W.; Tang, X.; Walton, C.J.; Brown, M.J.; Brewer, R.A.; Maddela, R.L.; Zheng, J.J.; Agus, J.K.; Zivkovic, A.M. Multi-Omic Analyses Reveal Bifidogenic Effect and Metabolomic Shifts in Healthy Human Cohort Supplemented With a Prebiotic Dietary Fiber Blend. Front. Nutr. 2022, 9, 908534. [Google Scholar] [CrossRef] [PubMed]
- Delannoy-Bruno, O.; Desai, C.; Castillo, J.J.; Couture, G.; Barve, R.A.; Lombard, V.; Henrissat, B.; Cheng, J.; Han, N.; Hayashi, D.K.; et al. An approach for evaluating the effects of dietary fiber polysaccharides on the human gut microbiome and plasma proteome. Proc. Natl. Acad. Sci. USA 2022, 119, e2123411119. [Google Scholar] [CrossRef] [PubMed]
- Alemán, J.O.; Bokulich, N.A.; Swann, J.R.; Walker, J.M.; De Rosa, J.C.; Battaglia, T.; Costabile, A.; Pechlivanis, A.; Liang, Y.; Breslow, J.L.; et al. Fecal microbiota and bile acid interactions with systemic and adipose tissue metabolism in diet-induced weight loss of obese postmenopausal women. J. Transl. Med. 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Galié, S.; García-Gavilán, J.; Camacho-Barcía, L.; Atzeni, A.; Muralidharan, J.; Papandreou, C.; Arcelin, P.; Palau-Galindo, A.; Garcia, D.; Basora, J.; et al. Effects of the Mediterranean Diet or Nut Consumption on Gut Microbiota Composition and Fecal Metabolites and their Relationship with Cardiometabolic Risk Factors. Mol. Nutr. Food Res. 2021, 65, e2000982. [Google Scholar] [CrossRef] [PubMed]
- Djekic, D.; Shi, L.; Brolin, H.; Carlsson, F.; Särnqvist, C.; Savolainen, O.; Cao, Y.; Bäckhed, F.; Tremaroli, V.; Landberg, R.; et al. Effects of a Vegetarian Diet on Cardiometabolic Risk Factors, Gut Microbiota, and Plasma Metabolome in Subjects With Ischemic Heart Disease: A Randomized, Crossover Study. J. Am. Heart Assoc. 2020, 9, e016518. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Hullar, M.A.J.; Lai, L.A.; Peng, H.; May, D.H.; Noble, W.S.; Raftery, D.; Navarro, S.L.; Neuhouser, M.L.; Lampe, P.D.; et al. Gut Microbial Protein Expression in Response to Dietary Patterns in a Controlled Feeding Study: A Metaproteomic Approach. Microorganisms 2020, 8, 379. [Google Scholar] [CrossRef]
- Zhao, D.; Shan, K.; Xie, Y.; Zhang, G.; An, Q.; Yu, X.; Zhou, G.; Li, C. Body weight index indicates the responses of the fecal microbiota, metabolome and proteome to beef/chicken-based diet alterations in Chinese volunteers. NPJ Biofilms Microbiomes 2022, 8, 56. [Google Scholar] [CrossRef]
- Tang, Z.Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Shah, R.D.; Tang, Z.Z.; Chen, G.; Huang, S.; Ferguson, J.F. Soy food intake associates with changes in the metabolome and reduced blood pressure in a gut microbiota dependent manner. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 1500–1511. [Google Scholar] [CrossRef]
- Le Roy, C.I.; Kurilshikov, A.; Leeming, E.R.; Visconti, A.; Bowyer, R.C.E.; Menni, C.; Falchi, M.; Koutnikova, H.; Veiga, P.; Zhernakova, A.; et al. Yoghurt consumption is associated with changes in the composition of the human gut microbiome and metabolome. BMC Microbiol. 2022, 22, 39. [Google Scholar]
- Shi, J.; Zhao, D.; Zhao, F.; Wang, C.; Zamaratskaia, G.; Li, C. Chicken-eaters and pork-eaters have different gut microbiota and tryptophan metabolites. Sci. Rep. 2021, 11, 11934. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, M.; Shah, R.D.; Mosley, J.D.; Ferguson, J.F. A metabolome and microbiome wide association study of healthy eating index points to the mechanisms linking dietary pattern and metabolic status. Eur. J. Nutr. 2021, 60, 4413–4427. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Prochazkova, M.; Budinska, E.; Kuzma, M.; Pelantova, H.; Hradecky, J.; Heczkova, M.; Daskova, N.; Bratova, M.; Modos, I.; Videnska, P.; et al. Vegan Diet Is Associated With Favorable Effects on the Metabolic Performance of Intestinal Microbiota: A Cross-Sectional Multi-Omics Study. Front. Nutr. 2022, 8, 783302. [Google Scholar] [CrossRef]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wan, Y.; Yin, K.; Wei, Y.; Wang, B.; Yu, X.; Ni, Y.; Zheng, J.; Huang, T.; Song, M.; et al. Lower Circulating Branched-Chain Amino Acid Concentrations Among Vegetarians are Associated with Changes in Gut Microbial Composition and Function. Mol. Nutr. Food Res. 2019, 63, e1900612. [Google Scholar] [CrossRef]
- Dhakan, D.B.; Maji, A.; Sharma, A.K.; Saxena, R.; Pulikkan, J.; Grace, T.; Gomez, A.; Scaria, J.; Amato, K.R.; Sharma, V.K. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. Gigascience 2019, 8, giz004. [Google Scholar] [CrossRef]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Hullar, M.A.; Fu, B.C. Diet, the gut microbiome, and epigenetics. Cancer J. 2014, 20, 170–175. [Google Scholar] [CrossRef]
- Gerhauser, C. Impact of dietary gut microbial metabolites on the epigenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170359. [Google Scholar] [CrossRef]
- D’Aquila, P.; Carelli, L.L.; De Rango, F.; Passarino, G.; Bellizzi, D. Gut Microbiota as Important Mediator Between Diet and DNA Methylation and Histone Modifications in the Host. Nutrients 2020, 12, 597. [Google Scholar] [CrossRef] [PubMed]
- Guz, M.; Jeleniewicz, W.; Malm, A.; Korona-Glowniak, I. A Crosstalk between Diet, Microbiome and microRNA in Epigenetic Regulation of Colorectal Cancer. Nutrients 2021, 13, 2428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Liu, W.; Wang, T.; Wang, L.; Hao, L.; Ju, M.; Xiao, R. Diet quality, gut microbiota, and microRNAs associated with mild cognitive impairment in middle-aged and elderly Chinese population. Am. J. Clin. Nutr. 2021, 114, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Cuevas-Sierra, A.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Comprehensive Analysis Reveals Novel Interactions between Circulating MicroRNAs and Gut Microbiota Composition in Human Obesity. Int. J. Mol. Sci. 2020, 21, 9509. [Google Scholar] [CrossRef]
- Tarallo, S.; Ferrero, G.; De Filippis, F.; Francavilla, A.; Pasolli, E.; Panero, V.; Cordero, F.; Segata, N.; Grioni, S.; Pensa, R.G.; et al. Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut 2022, 71, 1302–1314. [Google Scholar] [CrossRef]
- Díez-Sainz, E.; Lorente-Cebrián, S.; Aranaz, P.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I. Potential Mechanisms Linking Food-Derived MicroRNAs, Gut Microbiota and Intestinal Barrier Functions in the Context of Nutrition and Human Health. Front. Nutr. 2021, 8, 586564. [Google Scholar] [CrossRef]
- Choi, S.W.; Friso, S. Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef]
- Milagro, F.I.; Mansego, M.L.; De Miguel, C.; Martínez, J.A. Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. Mol. Asp. Med. 2013, 34, 782–812. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Samblas, M.; Milagro, F.I.; Zulet, M.A.; Mansego, M.L.; Riezu-Boj, J.I.; Martinez, J.A. Association of low dietary folate intake with lower CAMKK2 gene methylation, adiposity, and insulin resistance in obese subjects. Nutr. Res. 2018, 50, 53–62. [Google Scholar] [CrossRef]
- Losol, P.; Rezwan, F.I.; Patil, V.K.; Venter, C.; Ewart, S.; Zhang, H.; Arshad, S.H.; Karmaus, W.; Holloway, J.W. Effect of gestational oily fish intake on the risk of allergy in children may be influenced by FADS1/2, ELOVL5 expression and DNA methylation. Genes Nutr. 2019, 14, 20. [Google Scholar] [CrossRef]
- Hoile, S.P.; Clarke-Harris, R.; Huang, R.C.; Calder, P.C.; Mori, T.A.; Beilin, L.J.; Lillycrop, K.A.; Burdge, G.C. Supplementation with N-3 long-chain polyunsaturated fatty acids or olive oil in men and women with renal disease induces differential changes in the DNA methylation of FADS2 and ELOVL5 in peripheral blood mononuclear cells. PLoS ONE 2014, 9, e109896. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; James, L.; Hussey, B.; Wadley, A.J.; Lindley, M.R.; Mastana, S.S. Impact of aerobic exercise and fatty acid supplementation on global and gene-specific DNA methylation. Epigenetics 2019, 14, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Vanden Berghe, W.; Boby, C.; Leroux, C.; Declerck, K.; Szarc vel Szic, K.; Heyninck, K.; Laukens, K.; Bizet, M.; Defrance, M.; et al. Dietary flavanols modulate the transcription of genes associated with cardiovascular pathology without changes in their DNA methylation state. PLoS ONE 2014, 9, e95527. [Google Scholar] [CrossRef]
- Samblas, M.; Mansego, M.L.; Zulet, M.A.; Milagro, F.I.; Martinez, J.A. An integrated transcriptomic and epigenomic analysis identifies CD44 gene as a potential biomarker for weight loss within an energy-restricted program. Eur. J. Nutr. 2019, 58, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Crujeiras, A.B.; Izquierdo, A.G.; Primo, D.; Milagro, F.I.; Sajoux, I.; Jácome, A.; Fernandez-Quintela, A.; Portillo, M.P.; Martínez, J.A.; Martinez-Olmos, M.A.; et al. Epigenetic landscape in blood leukocytes following ketosis and weight loss induced by a very low calorie ketogenic diet (VLCKD) in patients with obesity. Clin. Nutr. 2021, 40, 3959–3972. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Bultman, S.J. Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell Physiol. 2012, 227, 3169–3177. [Google Scholar] [CrossRef]
- Janke, R.; Dodson, A.E.; Rine, J. Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol. 2015, 31, 473–496. [Google Scholar] [CrossRef]
- Dominguez-Salas, P.; Cox, S.E.; Prentice, A.M.; Hennig, B.J.; Moore, S.E. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: A review of current evidence in human subjects. Proc. Nutr. Soc. 2012, 71, 154–165. [Google Scholar] [CrossRef]
- Randunu, R.S.; Bertolo, R.F. The Effects of Maternal and Postnatal Dietary Methyl Nutrients on Epigenetic Changes that Lead to Non-Communicable Diseases in Adulthood. Int. J. Mol. Sci. 2020, 21, 3290. [Google Scholar] [CrossRef]
- Alabduljabbar, S.; Zaidan, S.A.; Lakshmanan, A.P.; Terranegra, A. Personalized Nutrition Approach in Pregnancy and Early Life to Tackle Childhood and Adult Non-Communicable Diseases. Life 2021, 11, 467. [Google Scholar] [CrossRef]
- Walker, M.E.; Song, R.J.; Xu, X.; Gerszten, R.E.; Ngo, D.; Clish, C.B.; Corlin, L.; Ma, J.; Xanthakis, V.; Jacques, P.F.; et al. Proteomic and Metabolomic Correlates of Healthy Dietary Patterns: The Framingham Heart Study. Nutrients 2020, 12, 1476. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.; Hendriks, H.F.; Cnubben, N.H.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M.; et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Bakker, G.C.; van Erk, M.J.; Pellis, L.; Wopereis, S.; Rubingh, C.M.; Cnubben, N.H.; Kooistra, T.; van Ommen, B.; Hendriks, H.F. An antiinflammatory dietary mix modulates inflammation and oxidative and metabolic stress in overweight men: A nutrigenomics approach. Am. J. Clin. Nutr. 2010, 91, 1044–1059. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Lopez, O.; Martinez, J.A.; Milagro, F.I. Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease. Nutrients 2022, 14, 4074. https://doi.org/10.3390/nu14194074
Ramos-Lopez O, Martinez JA, Milagro FI. Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease. Nutrients. 2022; 14(19):4074. https://doi.org/10.3390/nu14194074
Chicago/Turabian StyleRamos-Lopez, Omar, J. Alfredo Martinez, and Fermin I. Milagro. 2022. "Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease" Nutrients 14, no. 19: 4074. https://doi.org/10.3390/nu14194074
APA StyleRamos-Lopez, O., Martinez, J. A., & Milagro, F. I. (2022). Holistic Integration of Omics Tools for Precision Nutrition in Health and Disease. Nutrients, 14(19), 4074. https://doi.org/10.3390/nu14194074






