Wogonin, a Compound in Scutellaria baicalensis, Activates ATF4–FGF21 Signaling in Mouse Hepatocyte AML12 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Plasmids and Small Interfering RNA (siRNA)
2.4. Screening Analysis to Increase Fgf21 Expression Using the WAKANYAKU Library
2.5. Luciferase Analysis
2.6. Quantitative Polymerase Chain Reaction (qPCR)
2.7. Western Blotting
2.8. Statistical Analyses
3. Results
3.1. Cell-Based Screening Using a Natural Medicine Library Identified That SBE Induced Fgf21 Expression
3.2. Wogonin, a Flavonoid in SBE, Induces Fgf21 Expression in AML12 Cells
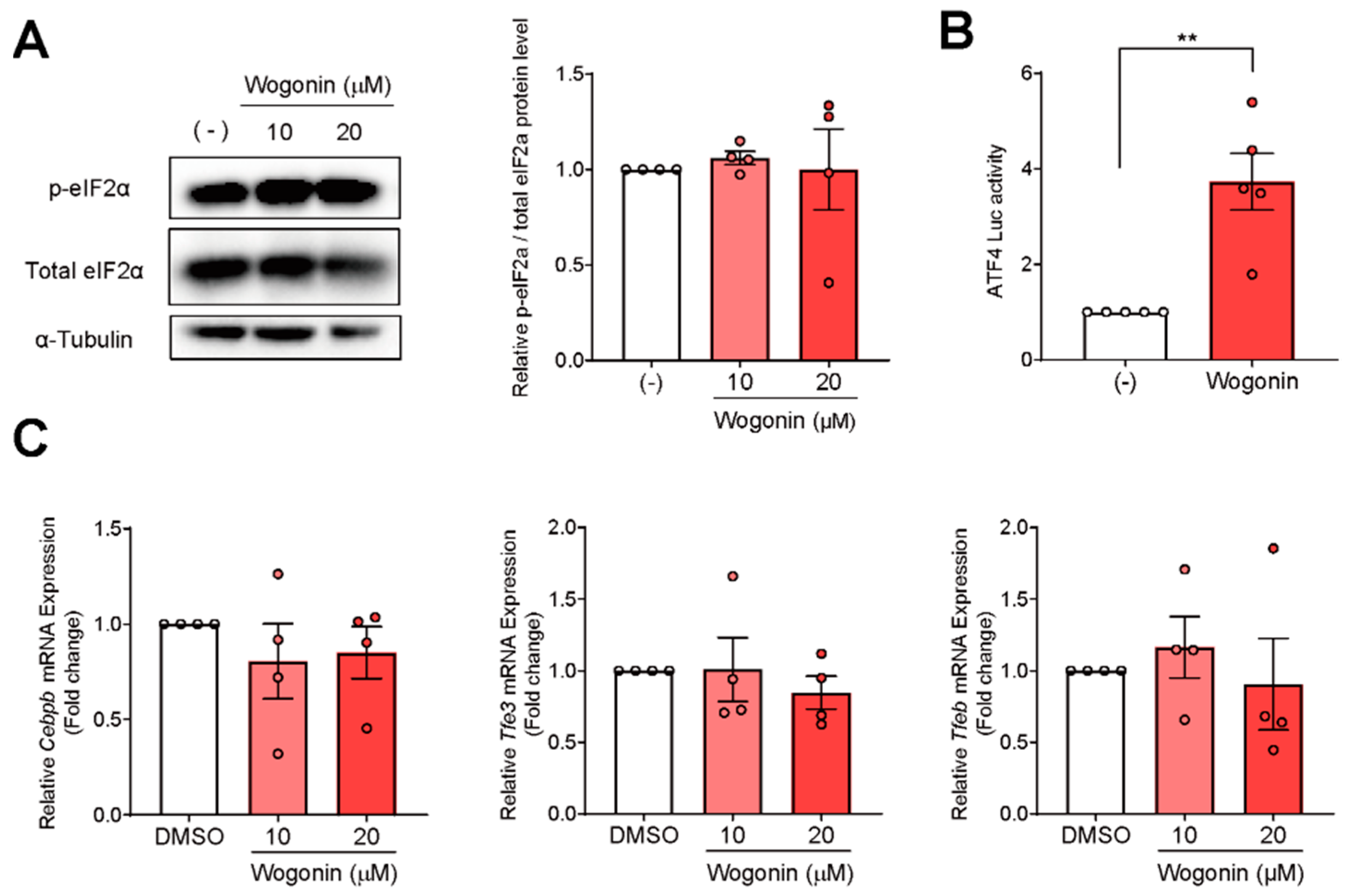
3.3. ATF4 Increased Fgf21 Expression in Response to Wogonin
3.4. Wogonin Controls ATF4 at the Transcription Level
3.5. Knockdown of Atf4 Suppresses Wogonin-Induced Fgf21 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fon Tacer, K.; Bookout, A.L.; Ding, X.; Kurosu, H.; John, G.B.; Wang, L.; Goetz, R.; Mohammadi, M.; Kuro-o, M.; Mangelsdorf, D.J.; et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 2010, 24, 2050–2064. [Google Scholar] [CrossRef] [PubMed]
- Foltz, I.N.; Hu, S.; King, C.; Wu, X.; Yang, C.; Wang, W.; Weiszmann, J.; Stevens, J.; Chen, J.S.; Nuanmanee, N.; et al. Treating diabetes and obesity with an FGF21-mimetic antibody activating the betaKlotho/FGFR1c receptor complex. Sci. Transl. Med. 2012, 4, 162ra153. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.C.; Yang, C.; Coskun, T.; Cheng, C.C.; Gimeno, R.E.; Luo, Y.; Kharitonenkov, A. The breadth of FGF21’s metabolic actions are governed by FGFR1 in adipose tissue. Mol. Metab. 2012, 2, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Markan, K.R.; Naber, M.C.; Ameka, M.K.; Anderegg, M.D.; Mangelsdorf, D.J.; Kliewer, S.A.; Mohammadi, M.; Potthoff, M.J. Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes 2014, 63, 4057–4063. [Google Scholar] [CrossRef] [PubMed]
- Kharitonenkov, A.; Shiyanova, T.L.; Koester, A.; Ford, A.M.; Micanovic, R.; Galbreath, E.J.; Sandusky, G.E.; Hammond, L.J.; Moyers, J.S.; Owens, R.A.; et al. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 2005, 115, 1627–1635. [Google Scholar] [CrossRef]
- Fisher, F.M.; Kleiner, S.; Douris, N.; Fox, E.C.; Mepani, R.J.; Verdeguer, F.; Wu, J.; Kharitonenkov, A.; Flier, J.S.; Maratos-Flier, E.; et al. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012, 26, 271–281. [Google Scholar] [CrossRef]
- Xu, J.; Lloyd, D.J.; Hale, C.; Stanislaus, S.; Chen, M.; Sivits, G.; Vonderfecht, S.; Hecht, R.; Li, Y.S.; Lindberg, R.A.; et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes 2009, 58, 250–259. [Google Scholar] [CrossRef]
- Inagaki, T.; Dutchak, P.; Zhao, G.; Ding, X.; Gautron, L.; Parameswara, V.; Li, Y.; Goetz, R.; Mohammadi, M.; Esser, V.; et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007, 5, 415–425. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Satoh, A.; Yabe, S.; Furusawa, M.; Tokushige, N.; Tezuka, H.; Mikami, M.; Iwata, W.; Shingyouchi, A.; Matsuzaka, T.; et al. Hepatic CREB3L3 controls whole-body energy homeostasis and improves obesity and diabetes. Endocrinology 2014, 155, 4706–4719. [Google Scholar] [CrossRef]
- De Sousa-Coelho, A.L.; Marrero, P.F.; Haro, D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. Biochem. J. 2012, 443, 165–171. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, F.; Gong, Q.; Cui, A.; Zhuo, S.; Hu, Z.; Han, Y.; Gao, J.; Sun, Y.; Liu, Z.; et al. Hepatic ATF6 Increases Fatty Acid Oxidation to Attenuate Hepatic Steatosis in Mice Through Peroxisome Proliferator-Activated Receptor alpha. Diabetes 2016, 65, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Takeda, J.; Horikawa, Y. Glucose induces FGF21 mRNA expression through ChREBP activation in rat hepatocytes. FEBS Lett. 2009, 583, 2882–2886. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; He, J.; Li, S.; Song, L.; Guo, X.; Yao, W.; Zou, D.; Gao, X.; Liu, Y.; Bai, F.; et al. Fibroblast growth factor 21 (FGF21) inhibits macrophage-mediated inflammation by activating Nrf2 and suppressing the NF-kappaB signaling pathway. Int. Immunopharmacol. 2016, 38, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yan, C.; Fang, Q.C.; Shao, M.L.; Zhang, Y.L.; Liu, Y.; Deng, Y.P.; Shan, B.; Liu, J.Q.; Li, H.T.; et al. Fibroblast growth factor 21 is regulated by the IRE1alpha-XBP1 branch of the unfolded protein response and counteracts endoplasmic reticulum stress-induced hepatic steatosis. J. Biol. Chem. 2014, 289, 29751–29765. [Google Scholar] [CrossRef] [PubMed]
- B’Chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef]
- Kilberg, M.S.; Shan, J.; Su, N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009, 20, 436–443. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Ron, D.; Harding, H.P. Protein-folding homeostasis in the endoplasmic reticulum and nutritional regulation. Cold Spring Harb. Perspect. Biol. 2012, 4, a013177. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef]
- Afonyushkin, T.; Oskolkova, O.V.; Philippova, M.; Resink, T.J.; Erne, P.; Binder, B.R.; Bochkov, V.N. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: Novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Diab, H.I.; Brady, O.A.; Puertollano, R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016, 35, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Savant, S.; Teske, B.F.; Hatzoglou, M.; Calkhoven, C.F.; Wek, R.C. Transcriptional repression of ATF4 gene by CCAAT/enhancer-binding protein beta (C/EBPbeta) differentially regulates integrated stress response. J. Biol. Chem. 2012, 287, 21936–21949. [Google Scholar] [CrossRef]
- Kim, K.H.; Jeong, Y.T.; Oh, H.; Kim, S.H.; Cho, J.M.; Kim, Y.N.; Kim, S.S.; Kim, D.H.; Hur, K.Y.; Kim, H.K.; et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013, 19, 83–92. [Google Scholar] [CrossRef]
- Laeger, T.; Albarado, D.C.; Burke, S.J.; Trosclair, L.; Hedgepeth, J.W.; Berthoud, H.R.; Gettys, T.W.; Collier, J.J.; Munzberg, H.; Morrison, C.D. Metabolic Responses to Dietary Protein Restriction Require an Increase in FGF21 that Is Delayed by the Absence of GCN2. Cell Rep. 2016, 16, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Barroso, E.; Leiva, R.; Barniol-Xicota, M.; Pujol, E.; Escolano, C.; Vazquez, S.; Palomer, X.; Pardo, V.; Gonzalez-Rodriguez, A.; et al. Heme-Regulated eIF2alpha Kinase Modulates Hepatic FGF21 and Is Activated by PPARbeta/delta Deficiency. Diabetes 2016, 65, 3185–3199. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Lee, A.R.; Yang, C.H. Antioxidative and anti-inflammatory activities of polyhydroxyflavonoids of Scutellaria baicalensis GEORGI. Biosci. Biotechnol. Biochem. 2006, 70, 2371–2380. [Google Scholar] [CrossRef]
- Ye, F.; Xui, L.; Yi, J.; Zhang, W.; Zhang, D.Y. Anticancer activity of Scutellaria baicalensis and its potential mechanism. J. Altern. Complement. Med. 2002, 8, 567–572. [Google Scholar] [CrossRef]
- Kim, E.H.; Shim, B.; Kang, S.; Jeong, G.; Lee, J.S.; Yu, Y.B.; Chun, M. Anti-inflammatory effects of Scutellaria baicalensis extract via suppression of immune modulators and MAP kinase signaling molecules. J. Ethnopharmacol. 2009, 126, 320–331. [Google Scholar] [CrossRef]
- Nagai, T.; Moriguchi, R.; Suzuki, Y.; Tomimori, T.; Yamada, H. Mode of action of the anti-influenza virus activity of plant flavonoid, 5,7,4′-trihydroxy-8-methoxyflavone, from the roots of Scutellaria baicalensis. Antivir. Res. 1995, 26, 11–25. [Google Scholar] [CrossRef]
- Heo, H.J.; Kim, D.O.; Choi, S.J.; Shin, D.H.; Lee, C.Y. Potent Inhibitory effect of flavonoids in Scutellaria baicalensis on amyloid beta protein-induced neurotoxicity. J. Agric. Food Chem. 2004, 52, 4128–4132. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, M.; Yu, H.; Li, J.; Wang, S.; Zhang, Y.; Qiu, F.; Wang, T. Scutellaria baicalensis regulates FFA metabolism to ameliorate NAFLD through the AMPK-mediated SREBP signaling pathway. J. Nat. Med. 2018, 72, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Lee, S.H.; Kim, B.Y.; Park, A.Y.; Kim, J.Y. Extracts of Scutellaria baicalensis reduced body weight and blood triglyceride in db/db Mice. Phytother. Res. 2013, 27, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Waisundara, V.Y.; Hsu, A.; Huang, D.; Tan, B.K. Scutellaria baicalensis enhances the anti-diabetic activity of metformin in streptozotocin-induced diabetic Wistar rats. Am. J. Chin. Med. 2008, 36, 517–540. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.T.; Wang, C.Y.; Yang, R.C.; Chu, C.J.; Wu, H.T.; Pang, J.H. Wogonin, an active compound in Scutellaria baicalensis, induces apoptosis and reduces telomerase activity in the HL-60 leukemia cells. Phytomedicine 2010, 17, 47–54. [Google Scholar] [CrossRef]
- Bak, E.J.; Kim, J.; Choi, Y.H.; Kim, J.H.; Lee, D.E.; Woo, G.H.; Cha, J.H.; Yoo, Y.J. Wogonin ameliorates hyperglycemia and dyslipidemia via PPARalpha activation in db/db mice. Clin. Nutr. 2014, 33, 156–163. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Wang, Y.; Hu, X.; Zhou, F.; Hu, Y.; Yuan, Y.; Xu, Y. Wogonin mitigates nonalcoholic fatty liver disease via enhancing PPARalpha/AdipoR2, in vivo and in vitro. Biomed. Pharmacother. 2017, 91, 621–631. [Google Scholar] [CrossRef]
- Wang, Q.; Mora-Jensen, H.; Weniger, M.A.; Perez-Galan, P.; Wolford, C.; Hai, T.; Ron, D.; Chen, W.; Trenkle, W.; Wiestner, A.; et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2200–2205. [Google Scholar] [CrossRef]
- Makino, T.; Hishida, A.; Goda, Y.; Mizukami, H. Comparison of the major flavonoid content of S. baicalensis, S. lateriflora, and their commercial products. J. Nat. Med. 2008, 62, 294–299. [Google Scholar] [CrossRef]
- Baradaran Rahimi, V.; Askari, V.R.; Hosseinzadeh, H. Promising influences of Scutellaria baicalensis and its two active constituents, baicalin, and baicalein, against metabolic syndrome: A review. Phytother. Res. 2021, 35, 3558–3574. [Google Scholar] [CrossRef]
- Sun, W.; Liu, P.; Wang, T.; Wang, X.; Zheng, W.; Li, J. Baicalein reduces hepatic fat accumulation by activating AMPK in oleic acid-induced HepG2 cells and high-fat diet-induced non-insulin-resistant mice. Food Funct. 2020, 11, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Nomura, K.; Ikai, R.; Nakashima, K.I.; Inoue, M. Baicalein stimulates fibroblast growth factor 21 expression by up-regulating retinoic acid receptor-related orphan receptor alpha in C2C12 myotubes. Biomed. Pharmacother. 2019, 109, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Shimano, H. CREBH Regulates Systemic Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2018, 19, 1396. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Dang Do, A.N.; Kimball, S.R.; Cavener, D.R.; Jefferson, L.S. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol. Genom. 2009, 38, 328–341. [Google Scholar] [CrossRef]
- Matsuzawa, Y.; Funahashi, T.; Nakamura, T. Molecular mechanism of metabolic syndrome X: Contribution of adipocytokines adipocyte-derived bioactive substances. Ann. N. Y. Acad. Sci. 1999, 892, 146–154. [Google Scholar] [CrossRef]
- Yao, J.; Pan, D.; Zhao, Y.; Zhao, L.; Sun, J.; Wang, Y.; You, Q.D.; Xi, T.; Guo, Q.L.; Lu, N. Wogonin prevents lipopolysaccharide-induced acute lung injury and inflammation in mice via peroxisome proliferator-activated receptor gamma-mediated attenuation of the nuclear factor-kappaB pathway. Immunology 2014, 143, 241–257. [Google Scholar] [CrossRef]
- Dai, J.M.; Guo, W.N.; Tan, Y.Z.; Niu, K.W.; Zhang, J.J.; Liu, C.L.; Yang, X.M.; Tao, K.S.; Chen, Z.N.; Dai, J.Y. Wogonin alleviates liver injury in sepsis through Nrf2-mediated NF-kappaB signalling suppression. J. Cell. Mol. Med. 2021, 25, 5782–5798. [Google Scholar] [CrossRef]



| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| Asns | TTACCTGTCTCTGCCGCCAGAT | CACTGAAGGCTTCTTTGGGTCG |
| Atf3 | TTTGCTAACCTGACACCCTTTG | AGAGGACATCCGATGGCAGA |
| Atf4 | CCTGAACAGCGAAGTGTTGG | TGGAGAACCCATGAGGTTTCAA |
| Atf6 | GGACGAGGTGGTGTCAGAG | GACAGCTCTTCGCTTTGGAC |
| Cebpb | TACGAGCCCGACTGCCTG | TCGGAGAGGAAGTCGTGGTG |
| Chop | GGAGGTCCTGTCCTCAGATGAA | GCTCCTCTGTCAGCCAAGCTAG |
| Chrebp | AATGGGATGGTGTCTACCGC | GGCGAAGGGAATTCAGGACA |
| Fgf21 | GGCAAGATATACGGGCTGAT | TCCATTTCCTCCCTGAAGGT |
| CrebH | AGATCAGGGAGGATGGAACA | TCAAAGTGAGGCGATCCATA |
| Cyclophilin | TGGCTCACAGTTCTTCATAACCA | ATGACATCCTTCAGTGGCTTGTC |
| Nrf2 | CAAGACTTGGGCCACTTAAAAGAC | AGTAAGGCTTTCCATCCTCATCAC |
| Ppara | ACGCGAGTTCCTTAAGAACCTG | GTGTCATCTGGATGGTTGCTCT |
| Rora | GATGACCTCAGCACCTATATGGA | CGGGTTTGATCCCATTGATGTC |
| Tfe3 | AGGATCAAAGAGCTGGGCAC | CCGGCTCTCCAGGTCTTTG |
| Tfeb | CAGAAGCGAGAGCTAACAGAT | TGTGATTGTCTTTCTTCTGCCG |
| Xbp1s | CTGAGTCCGAATCAGGTGCAG | GTCCATGGGAAGATGTTCTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamada, Y.; Saito, H.; Araki, M.; Tsuchimoto, Y.; Muroi, S.-i.; Suzuki, K.; Toume, K.; Kim, J.-D.; Matsuzaka, T.; Sone, H.; et al. Wogonin, a Compound in Scutellaria baicalensis, Activates ATF4–FGF21 Signaling in Mouse Hepatocyte AML12 Cells. Nutrients 2022, 14, 3920. https://doi.org/10.3390/nu14193920
Yamada Y, Saito H, Araki M, Tsuchimoto Y, Muroi S-i, Suzuki K, Toume K, Kim J-D, Matsuzaka T, Sone H, et al. Wogonin, a Compound in Scutellaria baicalensis, Activates ATF4–FGF21 Signaling in Mouse Hepatocyte AML12 Cells. Nutrients. 2022; 14(19):3920. https://doi.org/10.3390/nu14193920
Chicago/Turabian StyleYamada, Yasunari, Hodaka Saito, Masaya Araki, Yuhei Tsuchimoto, Shin-ichi Muroi, Kyohei Suzuki, Kazufumi Toume, Jun-Dal Kim, Takashi Matsuzaka, Hirohito Sone, and et al. 2022. "Wogonin, a Compound in Scutellaria baicalensis, Activates ATF4–FGF21 Signaling in Mouse Hepatocyte AML12 Cells" Nutrients 14, no. 19: 3920. https://doi.org/10.3390/nu14193920
APA StyleYamada, Y., Saito, H., Araki, M., Tsuchimoto, Y., Muroi, S.-i., Suzuki, K., Toume, K., Kim, J.-D., Matsuzaka, T., Sone, H., Shimano, H., & Nakagawa, Y. (2022). Wogonin, a Compound in Scutellaria baicalensis, Activates ATF4–FGF21 Signaling in Mouse Hepatocyte AML12 Cells. Nutrients, 14(19), 3920. https://doi.org/10.3390/nu14193920






